The Over-Expression of Two R2R3-MYB Genes, PdMYB2R089 and PdMYB2R151, Increases the Drought-Resistant Capacity of Transgenic Arabidopsis
Abstract
:1. Introduction
2. Results
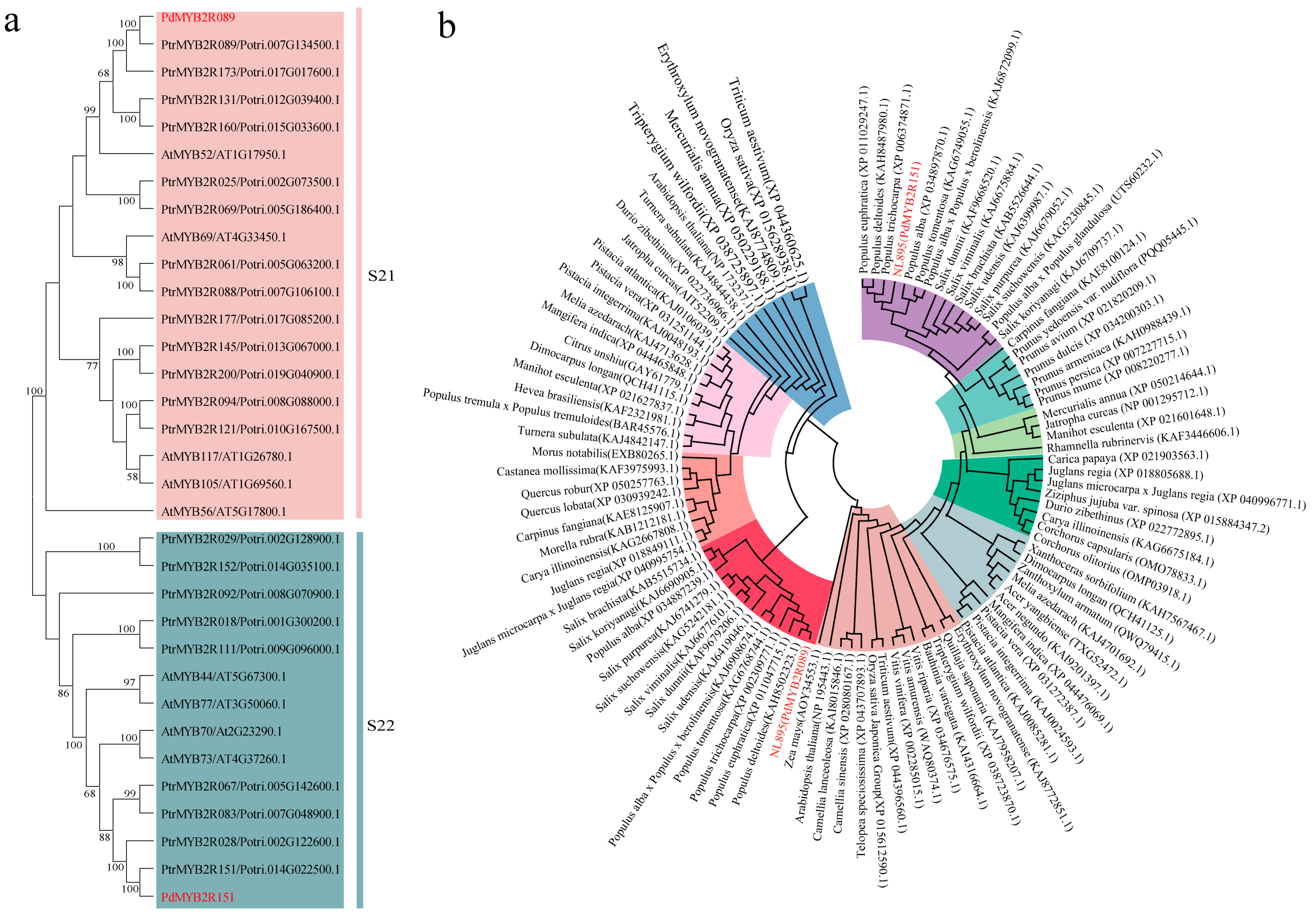
2.1. Phylogenetic Analysis and Physicochemical Properties of PdMYB2R089 and PdMYB2R151 Proteins
2.2. Analysis of Secondary Structure and Tertiary Structure of PdMYB2R089 and PdMYB2R151 Protein
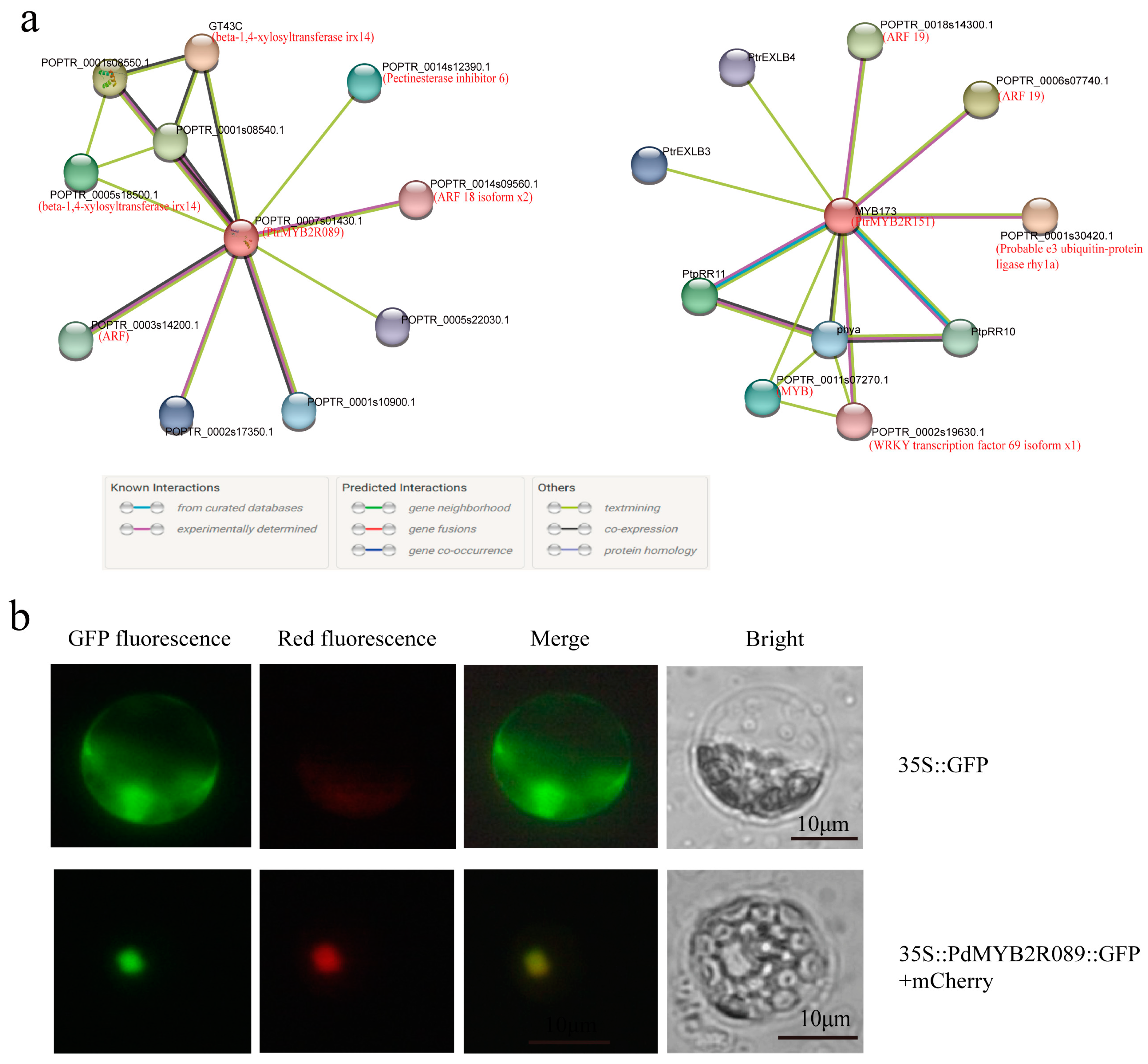
2.3. Function Domain Analysis and Protein–Protein Interaction Prediction
2.4. Function of PdMYB2R089 and PdMYB2R151 Gene under Drought Stress
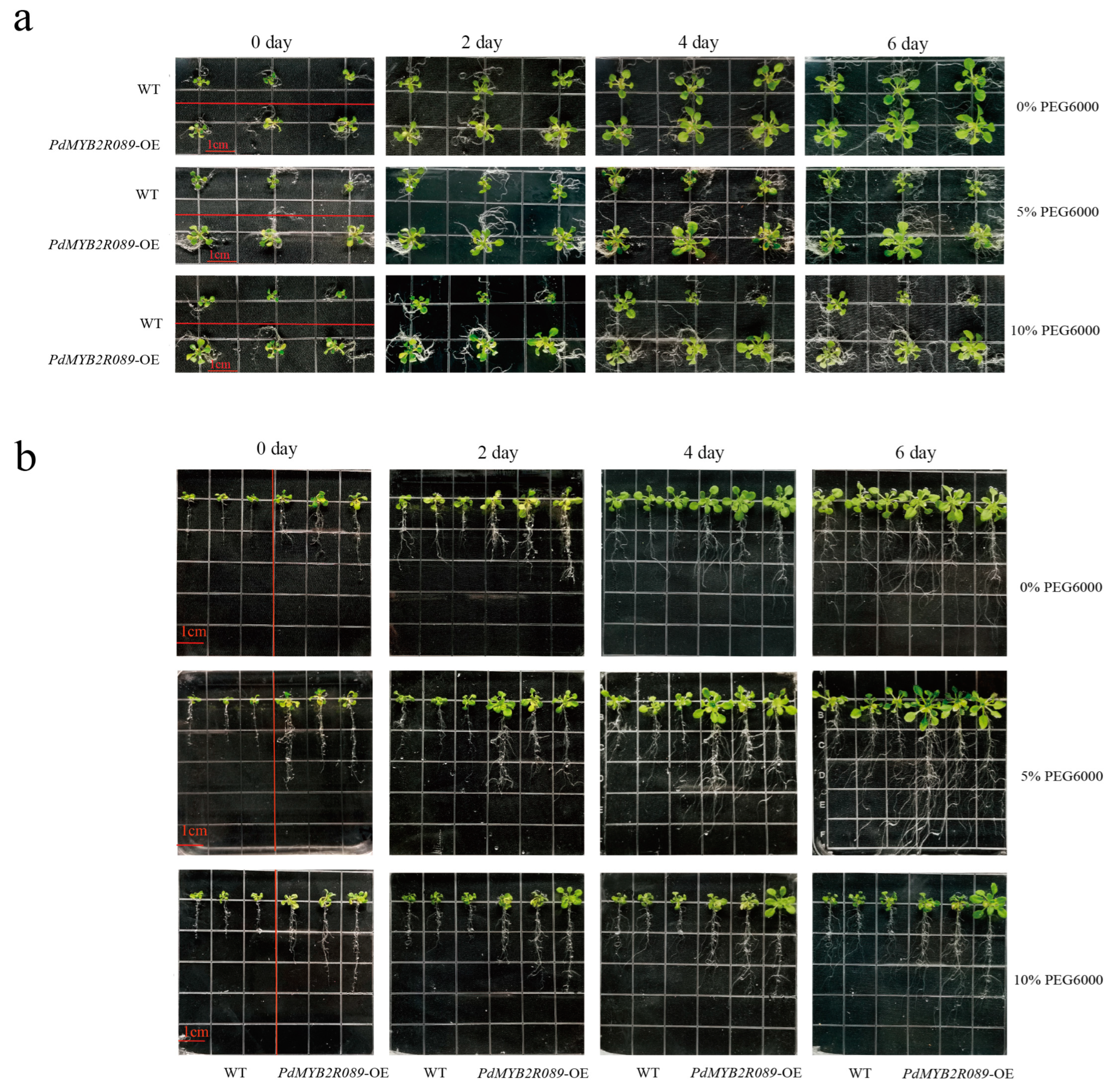
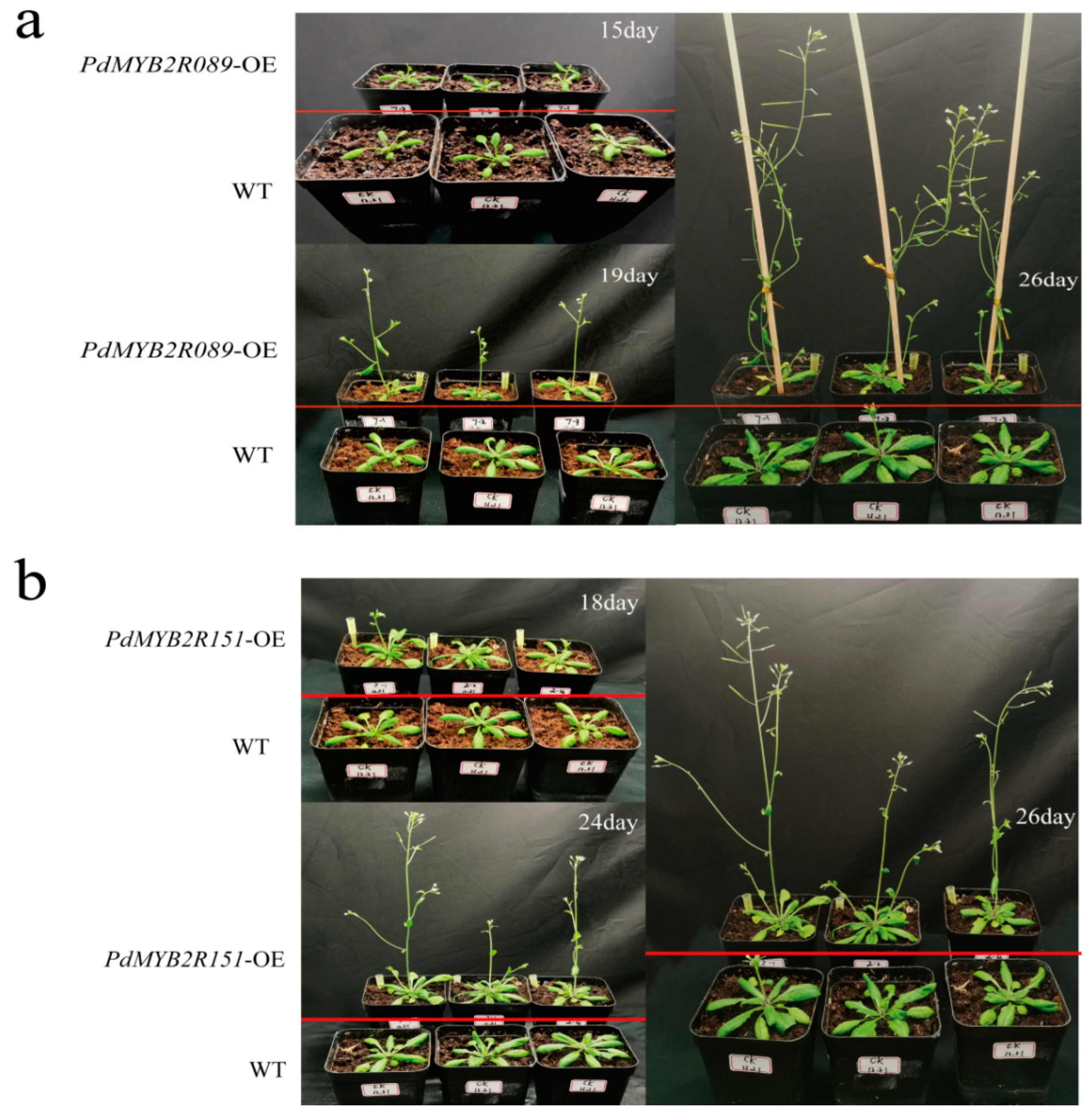
2.4.1. Overexpressed PdMYB2R089 and PdMYB2R151 Affected the Growth of Plants under Drought Stress
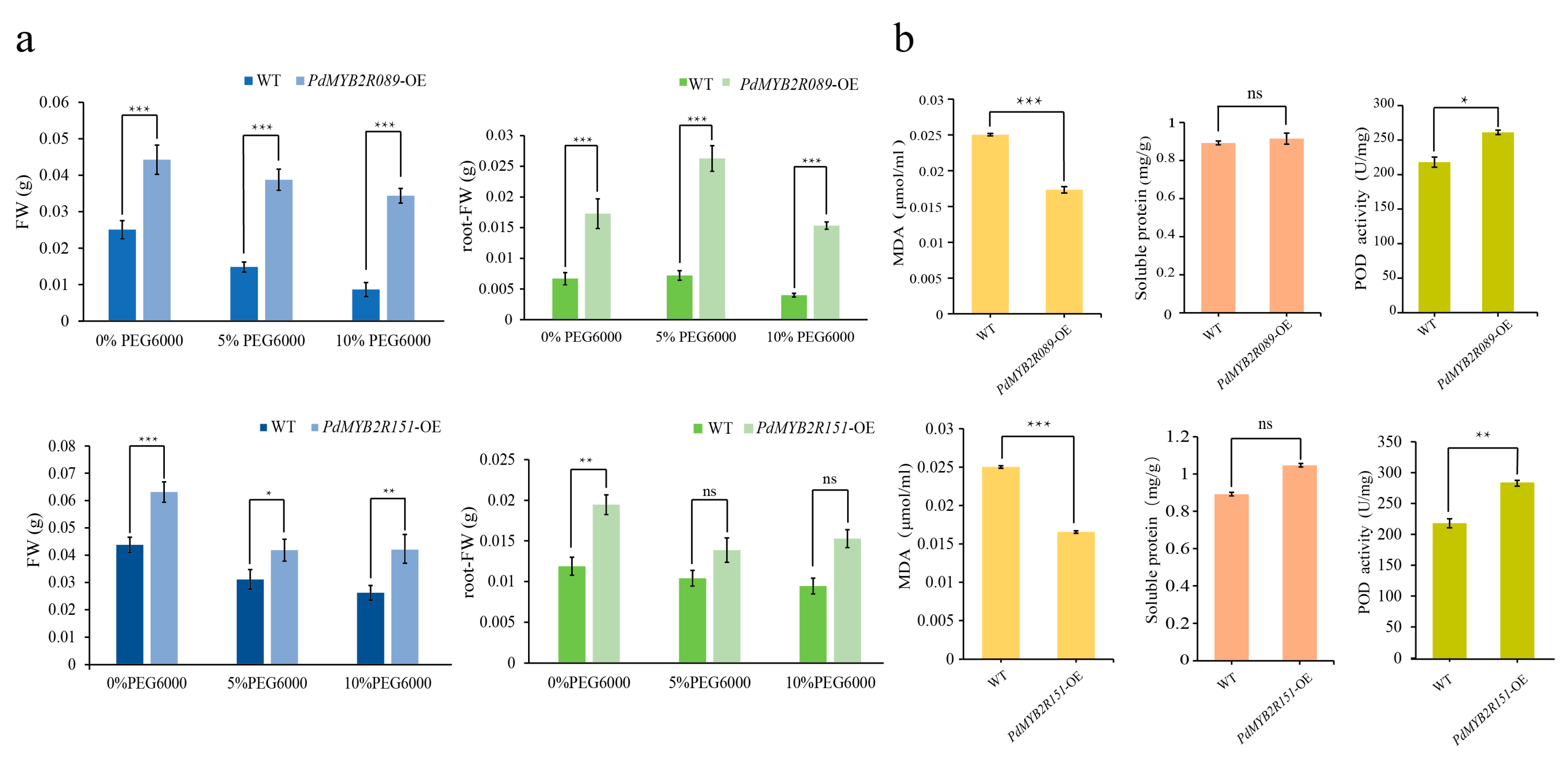
2.4.2. Overexpressed PdMYB2R089 and PdMYB2R151 Reduced Plant Drought Damage
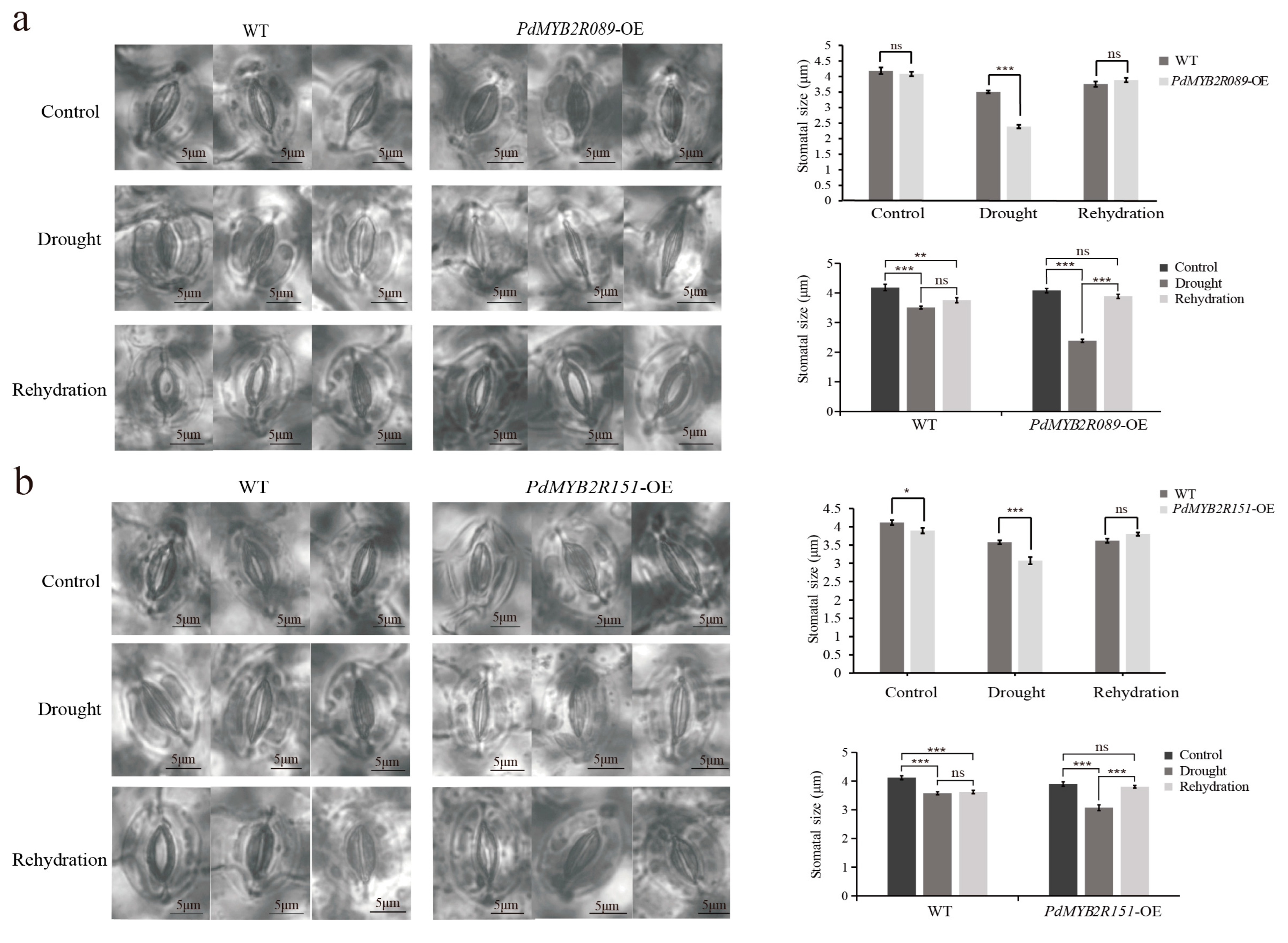
2.4.3. Overexpressed PdMYB2R089 and PdMYB2R151 Promoted Stomatal Movement under Drought Stress
2.4.4. Overexpressed PdMYB2R151 Promoted Seed Germination
2.5. Effect of PdMYB2R089 and PdMYB2R151 on the Flowering and Fruiting Time of Plants
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Phylogenetic Analysis
4.3. Structure Analysis and Protein Interaction Prediction
4.4. Subcellular Localization of the PdMYB2R089 in Poplar
4.5. Construction of Plant Overexpression Vectors
4.6. Genetic Transformation and Detection of Transgenic Positive Plants
4.7. Drought Stress Management and Seed Germination Phenotype Analysis
4.8. Plant Fresh Weight and Phenotype Analysis
4.9. Stomatal Variation and Life Cycle of Arabidopsis
4.10. Determination of Physiological Indicators Related to Drought
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sun, S.; He, C.; Qiu, L.; Li, C.; Zhang, J.; Meng, P. Stable isotope analysis reveals prolonged drought stress in poplar plantation mortality of the Three-North Shelter Forest in Northern China. Agric. For. Meteorol. 2018, 252, 39–48. [Google Scholar]
- Clifford, M.J.; Royer, P.D.; Cobb, N.S.; Breshears, D.D.; Ford, P.L. Precipitation thresholds and drought-induced tree die-off: Insights from patterns of Pinus edulis mortality along an environmental stress gradient. New Phytol. 2013, 200, 413–421. [Google Scholar]
- Bindu, Y.; Abhimanyu, J.; Samiur, R.M.; Prakash, N.O. Secondary metabolites in the drought stress tolerance of crop plants: A review. Gene Rep. 2021, 23, 101040. [Google Scholar]
- Lehrer, M.A.; Hawkins, J.S. Plant height shapes hydraulic architecture but does not predict metaxylem area under drought in Sorghum bicolor. Plant Direct. 2023, 7, e498. [Google Scholar] [PubMed]
- Abdel-Ghany, S.E.; Ullah, F.; Ben-Hur, A.; Reddy, A.S.N. Transcriptome analysis of drought-resistant and drought-sensitive Sorghum (Sorghum bicolor) genotypes in response to PEG-induced drought stress. Int. J. Mol. Sci. 2020, 21, 772. [Google Scholar]
- Ahmed, N.; Rahman, K.; Rahman, M.; Sathi, K.S.; Alam, M.M.; Nahar, K.; Islam, M.S.; Hasanuzzaman, M. Insight into the thiourea-induced drought tolerance in two chickpea varieties: Regulation of osmoprotection, reactive oxygen species metabolism and glyoxalase system. Plant Physiol. Biochem. 2021, 167, 449–458. [Google Scholar]
- Manna, M.; Thakur, T.; Chirom, O.; Mandlik, R.; Deshmukh, R.; Salvi, P. Transcription factors as key molecular target to strengthen the drought stress tolerance in plants. Physiol. Plant 2021, 172, 847–868. [Google Scholar]
- Gahlaut, V.; Jaiswal, V.; Kumar, A.; Gupta, P.K. Transcription factors involved in drought tolerance and their possible role in developing drought tolerant cultivars with emphasis on wheat (Triticum aestivum L.). Theor. Appl. Genet. 2016, 129, 2019–2042. [Google Scholar]
- Sekhwal, M.K.; Swami, A.K.; Sharma, V.; Sarin, R. Identification of drought-induced transcription factors in Sorghum bicolor using GO term semantic similarity. Cell Mol. Biol. Lett. 2015, 20, 1–23. [Google Scholar] [PubMed]
- Mun, B.G.; Lee, S.U.; Park, E.J.; Kim, H.H.; Hussain, A.; Imran, Q.M.; Lee, I.J.; Yun, B.W. Analysis of transcription factors among differentially expressed genes induced by drought stress in Populus davidiana. 3 Biotech. 2017, 7, 209. [Google Scholar]
- Saha, G.; Park, J.I.; Ahmed, N.U.; Kayum, M.A.; Kang, K.K.; Nou, I.S. Characterization and expression profiling of MYB transcription factors against stresses and during male organ development in Chinese cabbage (Brassica rapa ssp. pekinensis). Plant Physiol. Biochem. 2016, 104, 200–215. [Google Scholar] [PubMed]
- Cominelli, E.; Tonelli, C. A new role for plant R2R3-MYB transcription factors in cell cycle regulation. Cell Res. 2009, 19, 1231–1232. [Google Scholar] [PubMed]
- Zhou, F.; Chen, Y.; Wu, H.; Yin, T. Genome-Wide Comparative Analysis of R2R3 MYB Gene Family in Populus and Salix and Identification of Male Flower Bud Development-Related Genes. Front. Plant Sci. 2021, 12, 721558. [Google Scholar] [PubMed]
- Komatsuzaki, A.; Hoshino, A.; Otagaki, S.; Matsumoto, S.; Shiratake, K. Genome-wide analysis of R2R3-MYB transcription factors in Japanese morning glory. PLoS ONE 2022, 17, e0271012. [Google Scholar]
- Pratyusha, D.S.; Sarada, D.V.L. MYB transcription factors-master regulators of phenylpropanoid biosynthesis and diverse developmental and stress responses. Plant Cell Rep. 2022, 41, 2245–2260. [Google Scholar]
- Roy, S. Function of MYB domain transcription factors in abiotic stress and epigenetic control of stress response in plant genome. Plant Signal. Behav. 2016, 11, e1117723. [Google Scholar]
- Dubos, C.; Stracke, R.; Grotewold, E.; Weisshaar, B.; Martin, C.; Lepiniec, L. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010, 15, 573–581. [Google Scholar]
- Oh, J.E.; Kwon, Y.; Kim, J.H.; Noh, H.; Hong, S.W.; Lee, H. A dual role for MYB60 in stomatal regulation and root growth of Arabidopsis thaliana under drought stress. Plant Mol. Biol. 2011, 77, 91–103. [Google Scholar]
- Seo, P.J.; Xiang, F.; Qiao, M.; Park, J.Y.; Lee, Y.N.; Kim, S.G.; Lee, Y.H.; Park, W.J.; Park, C.M. The MYB96 transcription factor mediates abscisic acid signaling during drought stress response in Arabidopsis. Plant Physiol. 2009, 151, 275–289. [Google Scholar]
- Seo, P.J.; Lee, S.B.; Suh, M.C.; Park, M.J.; Go, Y.S.; Park, C.M. The MYB96 transcription factor regulates cuticular wax biosynthesis under drought conditions in Arabidopsis. Plant Cell 2011, 23, 1138–1152. [Google Scholar] [CrossRef]
- Lee, S.B.; Suh, M.C. Cuticular wax biosynthesis is up-regulated by the MYB94 transcription factor in Arabidopsis. Plant Cell Physiol. 2015, 56, 48–60. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Fu, X.; Liu, R.; Guo, L.; Ran, L.; Li, C.; Tian, Q.; Jiao, B.; Wang, B.; Luo, K. PtoMYB170 positively regulates lignin deposition during wood formation in poplar and confers drought tolerance in transgenic Arabidopsis. Tree Physiol. 2017, 37, 1713–1726. [Google Scholar] [CrossRef] [PubMed]
- Zhao, K.; Cheng, Z.; Guo, Q.; Yao, W.; Liu, H.; Zhou, B.; Jiang, T. Characterization of the Poplar R2R3-MYB Gene Family and Over-Expression of PsnMYB108 Confers Salt Tolerance in Transgenic Tobacco. Front. Plant Sci. 2020, 11, 571881. [Google Scholar] [CrossRef]
- Fang, Q.; Wang, X.; Wang, H.; Tang, X.; Liu, C.; Yin, H.; Ye, S.; Jiang, Y.; Duan, Y.; Luo, K. The poplar R2R3 MYB transcription factor PtrMYB94 coordinates with abscisic acid signaling to improve drought tolerance in plants. Tree Physiol. 2020, 40, 46–59. [Google Scholar] [CrossRef] [PubMed]
- Song, Q.; Kong, L.; Yang, X.; Jiao, B.; Hu, J.; Zhang, Z.; Xu, C.; Luo, K. PtoMYB142, a poplar R2R3-MYB transcription factor, contributes to drought tolerance by regulating wax biosynthesis. Tree Physiol. 2022, 42, 2133–2147. [Google Scholar] [CrossRef]
- Yanhui, C.; Xiaoyuan, Y.; Kun, H.; Meihua, L.; Jigang, L.; Zhaofeng, G.; Zhiqiang, L.; Yunfei, Z.; Xiaoxiao, W.; Xiaoming, Q.; et al. The MYB transcription factor superfamily of Arabidopsis: Expression analysis and phylogenetic comparison with the rice MYB family. Plant Mol. Biol. 2006, 60, 107–124. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, H.; Chen, Y.; Huang, M.; Zhu, S. Comprehensive Genome-Wide Analyses of Poplar R2R3-MYB Transcription Factors and Tissue-Specific Expression Patterns under Drought Stress. Int. J. Mol. Sci. 2023, 24, 5389. [Google Scholar] [CrossRef]
- Yu, Y.T.; Wu, Z.; Lu, K.; Bi, C.; Liang, S.; Wang, X.F.; Zhang, D.P. Overexpression of the MYB37 transcription factor enhances abscisic acid sensitivity, and improves both drought tolerance and seed productivity in Arabidopsis thaliana. Plant Mol. Biol. 2016, 90, 267–279. [Google Scholar] [CrossRef]
- Deng, M.; Wang, Y.; Kuzma, M.; Chalifoux, M.; Tremblay, L.; Yang, S.; Ying, J.; Sample, A.; Wang, H.M.; Griffiths, R.; et al. Activation tagging identifies Arabidopsis transcription factor AtMYB68 for heat and drought tolerance at yield determining reproductive stages. Plant J. 2020, 104, 1535–1550. [Google Scholar] [CrossRef]
- Li, X.; Zhong, M.; Qu, L.; Yang, J.; Liu, X.; Zhao, Q.; Liu, X.; Zhao, X. AtMYB32 regulates the ABA response by targeting ABI3, ABI4 and ABI5 and the drought response by targeting CBF4 in Arabidopsis. Plant Sci. 2021, 310, 110983. [Google Scholar] [CrossRef] [PubMed]
- Oshima, Y.; Shikata, M.; Koyama, T.; Ohtsubo, N.; Mitsuda, N.; Ohme-Takagi, M. MIXTA-like transcription factors and WAX INDUCER1/SHINE1 coordinately regulate cuticle development in Arabidopsis and Torenia fournieri. Plant Cell 2013, 25, 1609–1624. [Google Scholar] [CrossRef] [PubMed]
- Park, M.Y.; Kang, J.Y.; Kim, S.Y. Overexpression of AtMYB52 confers ABA hypersensitivity and drought tolerance. Mol. Cells. 2011, 31, 447–454. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Mao, Y.; Guo, Y.; Gao, J.; Liu, X.; Li, S.; Lin, Y.J.; Chen, H.; Wang, J.P.; Chiang, V.L.; et al. MYB Transcription Factor161 Mediates Feedback Regulation of Secondary wall-associated NAC-Domain1 Family Genes for Wood Formation. Plant Physiol. 2020, 184, 1389–1406. [Google Scholar] [CrossRef] [PubMed]
- Seo, J.S.; Sohn, H.B.; Noh, K.; Jung, C.; An, J.H.; Donovan, C.M.; Somers, D.A.; Kim, D.I.; Jeong, S.-C.; Kim, C.-G.; et al. Expression of the Arabidopsis AtMYB44 gene confers drought/salt-stress tolerance in transgenic soybean. Mol. Breed. 2012, 29, 601–608. [Google Scholar] [CrossRef]
- Che, Y.; Sun, Y.; Lu, S.; Hou, L.; Fan, X.; Liu, X. AtMYB77 Involves in lateral root development via regulating nitric oxide biosynthesis under drought stress in Arabidopsis thaliana. Chin. Bull. Bot. 2021, 56, 404–413. [Google Scholar]
- Fan, J. Preliminary Exploration for Mechanism of Arabidopsis thaliana AtMYB73 under Drought Stress. Master’s Thesis, Agricultural University of Hebei, Baoding, China, 2015. [Google Scholar]
- Wilkins, O.; Nahal, H.; Foong, J.; Provart, N.J.; Campbell, M.M. Expansion and diversification of the Populus R2R3-MYB family of transcription factors. Plant Physiol. 2009, 149, 981–993. [Google Scholar] [CrossRef]
- Millard, P.S.; Kragelund, B.B.; Burow, M. R2R3 MYB Transcription Factors—Functions outside the DNA-Binding Domain. Trends Plant Sci. 2019, 24, 934–946. [Google Scholar] [CrossRef]
- Muthusamy, M.; Kim, J.Y.; Yoon, E.K.; Kim, J.A.; Lee, S.I. BrEXLB1, a Brassica rapa Expansin-Like B1 Gene Is Associated with Root Development, Drought Stress Response, and Seed Germination. Genes 2020, 11, 404. [Google Scholar] [CrossRef]
- Nakamichi, N.; Takao, S.; Kudo, T.; Kiba, T.; Wang, Y.; Kinoshita, T.; Sakakibara, H. Improvement of Arabidopsis Biomass and Cold, Drought and Salinity Stress Tolerance by Modified Circadian Clock-Associated PSEUDO-RESPONSE REGULATORs. Plant Cell Physiol. 2016, 57, 1085–1097. [Google Scholar] [CrossRef]
- Khoso, M.A.; Hussain, A.; Ritonga, F.N.; Ali, Q.; Channa, M.M.; Alshegaihi, R.M.; Meng, Q.; Ali, M.; Zaman, W.; Brohi, R.D.; et al. WRKY transcription factors (TFs): Molecular switches to regulate drought, temperature, and salinity stresses in plants. Front. Plant Sci. 2022, 13, 1039329. [Google Scholar] [CrossRef]
- Verma, S.; Negi, N.P.; Pareek, S.; Mudgal, G.; Kumar, D. Auxin response factors in plant adaptation to drought and salinity stress. Physiol. Plant 2022, 174, e13714. [Google Scholar] [CrossRef]
- Shin, R.; Burch, A.Y.; Huppert, K.A.; Tiwari, S.B.; Murphy, A.S.; Guilfoyle, T.J.; Schachtman, D.P. The Arabidopsis transcription factor MYB77 modulates auxin signal transduction. Plant Cell 2007, 19, 2440–2453. [Google Scholar] [CrossRef]
- Blümel, M.; Dally, N.; Jung, C. Flowering time regulation in crops—What did we learn from Arabidopsis? Curr. Opin. Biotechnol. 2015, 32, 121–129. [Google Scholar] [CrossRef]
- Nakamichi, N.; Kudo, T.; Makita, N.; Kiba, T.; Kinoshita, T.; Sakakibara, H. Flowering time control in rice by introducing Arabidopsis clock-associated PSEUDO-RESPONSE REGULATOR 5. Biosci. Biotechnol. Biochem. 2020, 84, 970–979. [Google Scholar] [CrossRef] [PubMed]
- Prouse, M.B.; Campbell, M.M. The interaction between MYB proteins and their target DNA binding sites. Biochim. Biophys. Acta 2012, 1819, 67–77. [Google Scholar] [CrossRef] [PubMed]
- Bouzroud, S.; Gouiaa, S.; Hu, N.; Bernadac, A.; Mila, I.; Bendaou, N.; Smouni, A.; Bouzayen, M.; Zouine, M. Auxin Response Factors (ARFs) are potential mediators of auxin action in tomato response to biotic and abiotic stress (Solanum lycopersicum). PLoS ONE 2018, 13, e0193517. [Google Scholar] [CrossRef]
- Kang, C.; He, S.; Zhai, H.; Li, R.; Zhao, N.; Liu, Q. A Sweetpotato Auxin Response Factor Gene (IbARF5) Is Involved in Carotenoid Biosynthesis and Salt and Drought Tolerance in Transgenic Arabidopsis. Front. Plant Sci. 2018, 9, 1307. [Google Scholar] [CrossRef] [PubMed]
- Lim, J.; Lim, C.W.; Lee, S.C. Pepper Novel Pseudo Response Regulator Protein CaPRR2 Modulates Drought and High Salt Tolerance. Front. Plant Sci. 2021, 12, 736421. [Google Scholar] [CrossRef]
- Ambawat, S.; Sharma, P.; Yadav, N.R.; Yadav, R.C. MYB transcription factor genes as regulators for plant responses: An overview. Physiol. Mol. Biol. Plants 2013, 19, 307–321. [Google Scholar] [CrossRef]
- Baldoni, E.; Genga, A.; Cominelli, E. Plant MYB Transcription Factors: Their Role in Drought Response Mechanisms. Int. J. Mol. Sci. 2015, 16, 15811–15851. [Google Scholar] [CrossRef]
- Zhao, Y.; Xing, L.; Wang, X.; Hou, Y.J.; Gao, J.; Wang, P.; Duan, C.G.; Zhu, X.; Zhu, J.K. The ABA receptor PYL8 promotes lateral root growth by enhancing MYB77-dependent transcription of auxin-responsive genes. Sci. Signal. 2014, 7, ra53. [Google Scholar] [CrossRef] [PubMed]
- Wan, J.; Wang, R.; Zhang, P.; Sun, L.; Ju, Q.; Huang, H.; Lü, S.; Tran, L.S.; Xu, J. MYB70 modulates seed germination and root system development in Arabidopsis. iScience 2021, 24, 103228. [Google Scholar] [CrossRef] [PubMed]
- Jung, C.; Seo, J.S.; Han, S.W.; Koo, Y.J.; Kim, C.H.; Song, S.I.; Nahm, B.H.; Choi, Y.D.; Cheong, J.J. Overexpression of AtMYB44 enhances stomatal closure to confer abiotic stress tolerance in transgenic Arabidopsis. Plant Physiol. 2008, 146, 623–635. [Google Scholar] [CrossRef] [PubMed]
- Cruz de Carvalho, M.H. Drought stress and reactive oxygen species: Production, scavenging and signaling. Plant Signal. Behav. 2008, 3, 156–165. [Google Scholar] [CrossRef] [PubMed]
- Gill, S.S.; Tuteja, N. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 2010, 48, 909–930. [Google Scholar] [CrossRef] [PubMed]
- Ali, S.S.; Ahsan, H.; Zia, M.K.; Siddiqui, T.; Khan, F.H. Understanding oxidants and antioxidants: Classical team with new players. J. Food Biochem. 2020, 44, e13145. [Google Scholar] [CrossRef]
- Yin, Z.; Zhou, F.; Chen, Y.; Wu, H.; Yin, T. Genome-Wide Analysis of the Expansin Gene Family in Populus and Characterization of Expression Changes in Response to Phytohormone (Abscisic Acid) and Abiotic (Low-Temperature) Stresses. Int. J. Mol. Sci. 2023, 24, 7759. [Google Scholar] [CrossRef]
- Fang, Z.; Ji, Y.; Hu, J.; Guo, R.; Sun, S.; Wang, X. Strigolactones and Brassinosteroids Antagonistically Regulate the Stability of the D53-OsBZR1 Complex to Determine FC1 Expression in Rice Tillering. Mol. Plant 2020, 13, 586–597. [Google Scholar] [CrossRef]
- Jiang, L.; Liu, X.; Xiong, G.; Liu, H.; Chen, F.; Wang, L.; Meng, X.; Liu, G.; Yu, H.; Yuan, Y.; et al. DWARF 53 acts as a repressor of strigolactone signalling in rice. Nature 2013, 504, 401–405. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Mughal, I.; Shah, Y.; Tahir, S.; Haider, W.; Fayyaz, M.; Yasmin, T.; Ilyas, M.; Farrakh, S. Protein quantification and enzyme activity estimation of Pakistani wheat landraces. PLoS ONE 2020, 15, e0239375. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, X.; Wang, H.; Chen, Y.; Huang, M.; Zhu, S. The Over-Expression of Two R2R3-MYB Genes, PdMYB2R089 and PdMYB2R151, Increases the Drought-Resistant Capacity of Transgenic Arabidopsis. Int. J. Mol. Sci. 2023, 24, 13466. https://doi.org/10.3390/ijms241713466
Zhang X, Wang H, Chen Y, Huang M, Zhu S. The Over-Expression of Two R2R3-MYB Genes, PdMYB2R089 and PdMYB2R151, Increases the Drought-Resistant Capacity of Transgenic Arabidopsis. International Journal of Molecular Sciences. 2023; 24(17):13466. https://doi.org/10.3390/ijms241713466
Chicago/Turabian StyleZhang, Xueli, Haoran Wang, Ying Chen, Minren Huang, and Sheng Zhu. 2023. "The Over-Expression of Two R2R3-MYB Genes, PdMYB2R089 and PdMYB2R151, Increases the Drought-Resistant Capacity of Transgenic Arabidopsis" International Journal of Molecular Sciences 24, no. 17: 13466. https://doi.org/10.3390/ijms241713466




