Screening, Synthesis and Biochemical Characterization of SARS-CoV-2 Protease Inhibitors
Abstract
:1. Introduction
2. Results
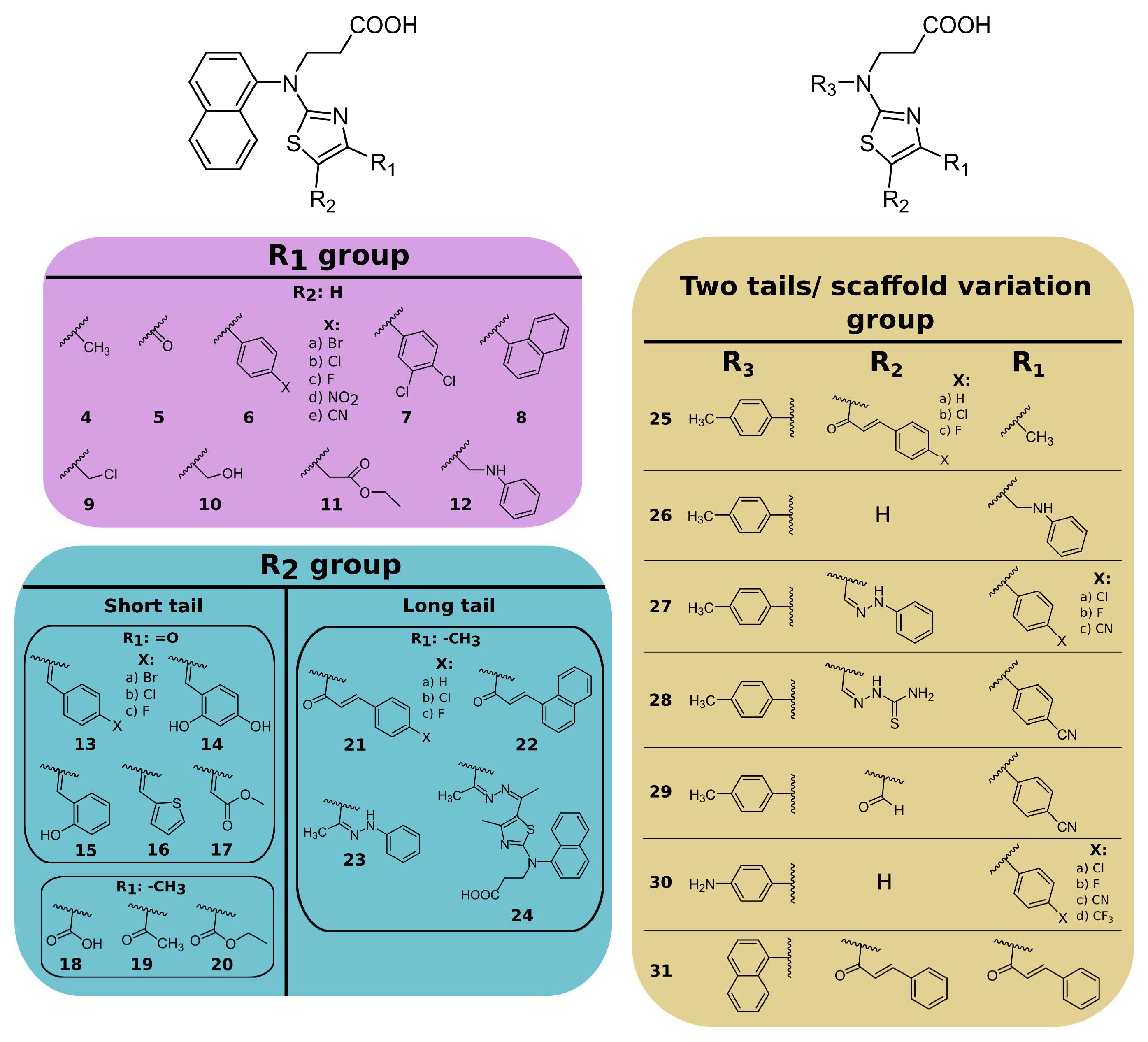
2.1. Naphthalene-Based PLpro and Mpro Inhibitors
- (i)
- substituents at the 4- position on the thiazole ring (compounds 4–12);
- (ii)
- substituents at the 4,5 positions on the thiazole ring (compounds 13–24);
- (iii)
- replacing the naphthalene group of compound 3 with the 4-methyl- or 4-aminophenyl group and introducing different substituents at the 4,5 positions of the thiazole ring (compounds 25–31).
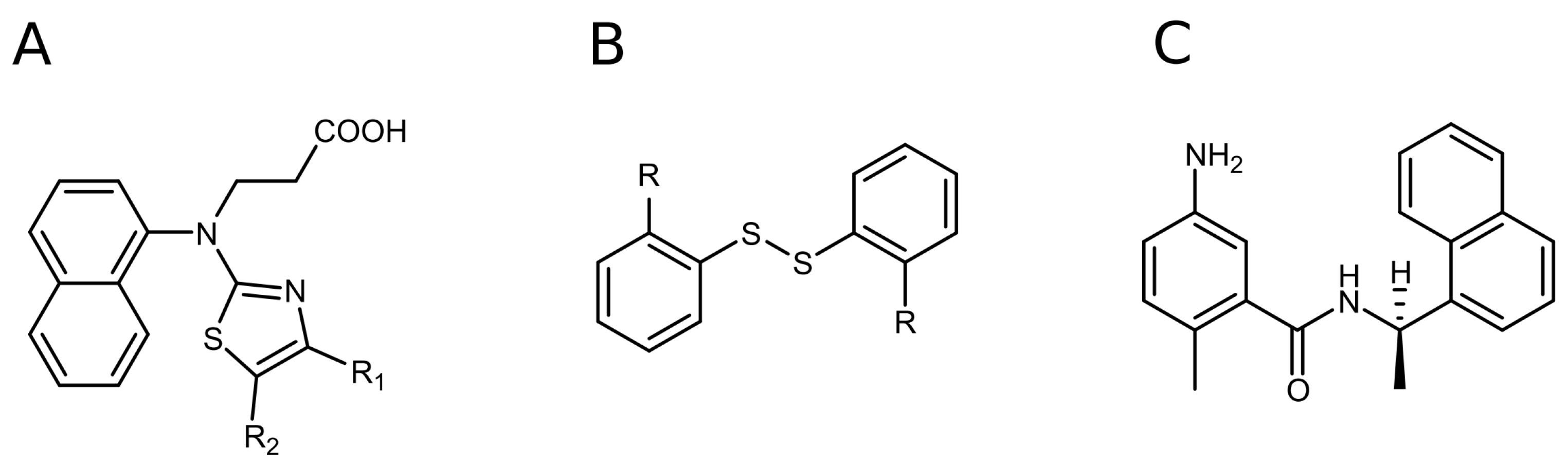
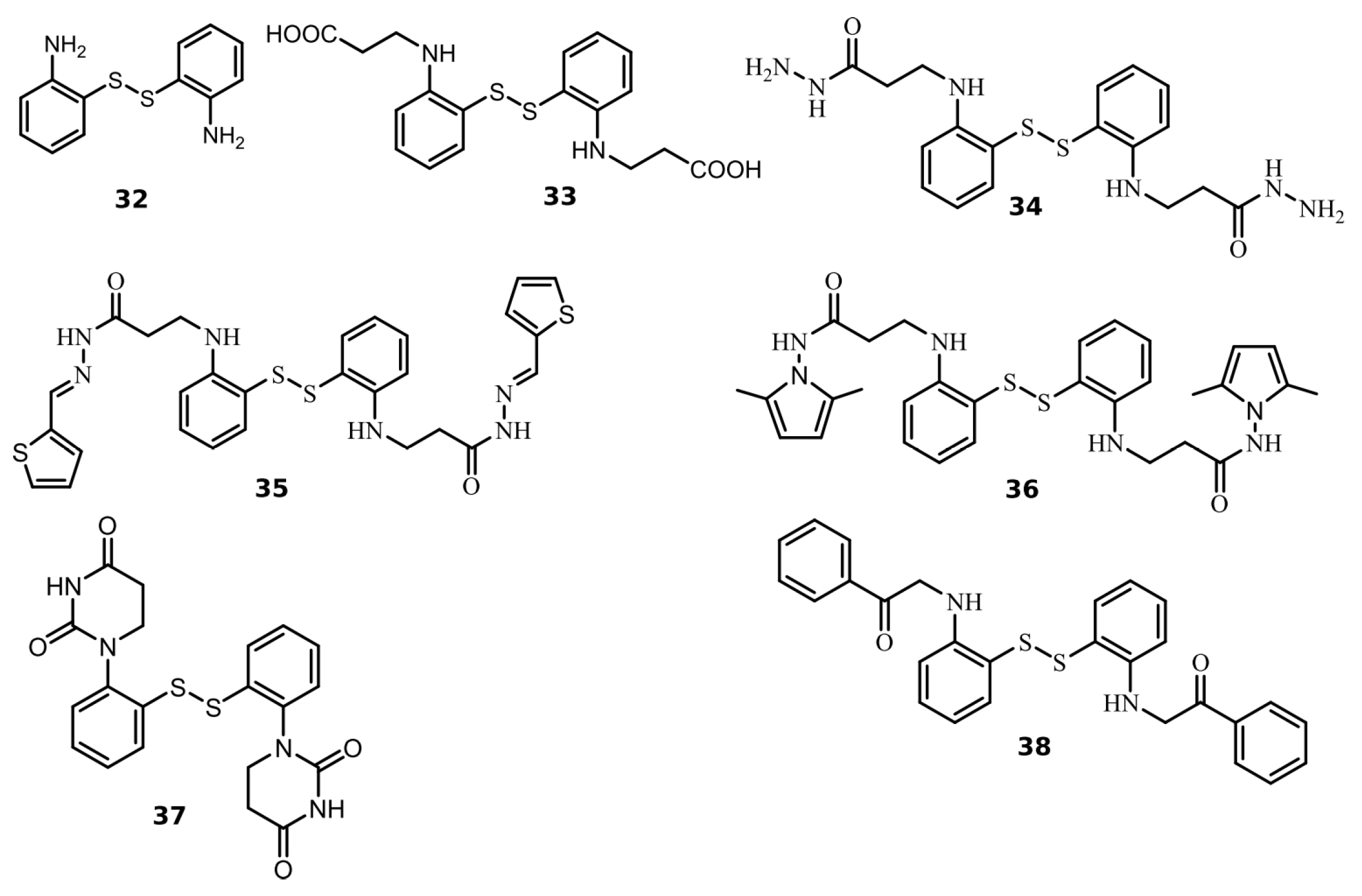
2.2. Disulfide Derivatives as PLpro and Mpro Inhibitors
3. Discussion
4. Materials and Methods
4.1. Chemistry
4.2. Protein Expression and Purification
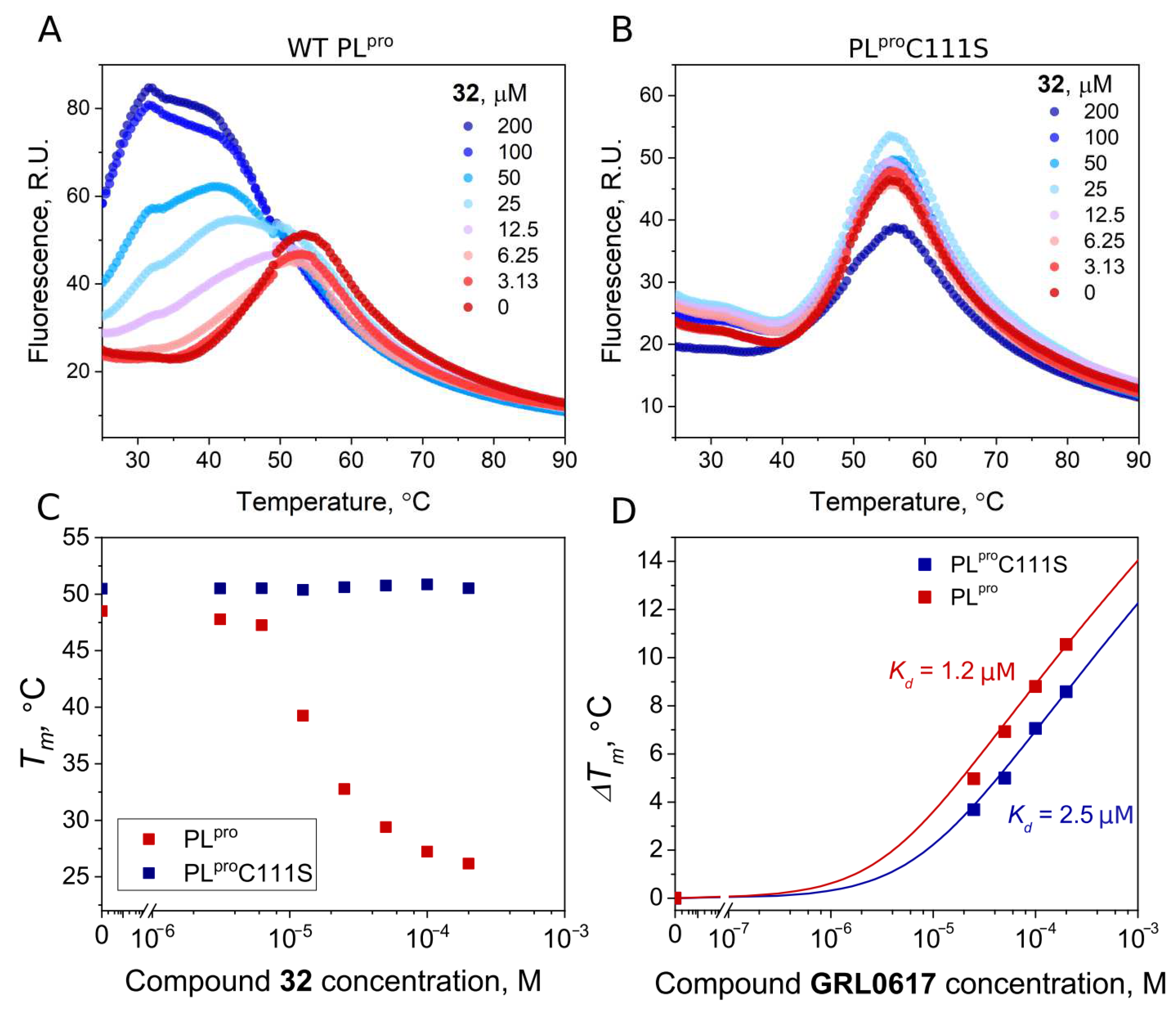
4.3. Fluorescence Thermal Shift Assay (FTSA)
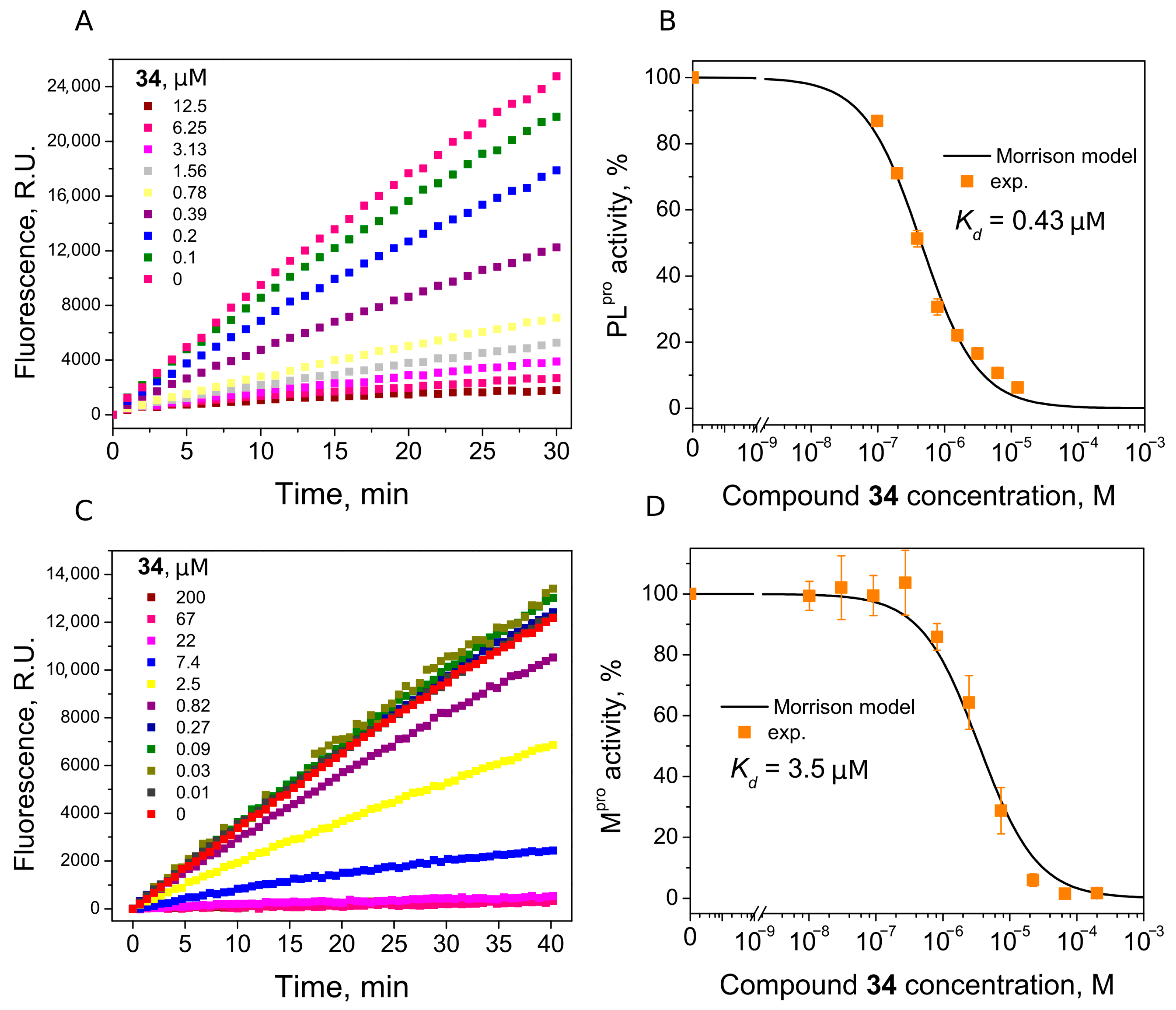
4.4. Enzymatic Inhibition Assay
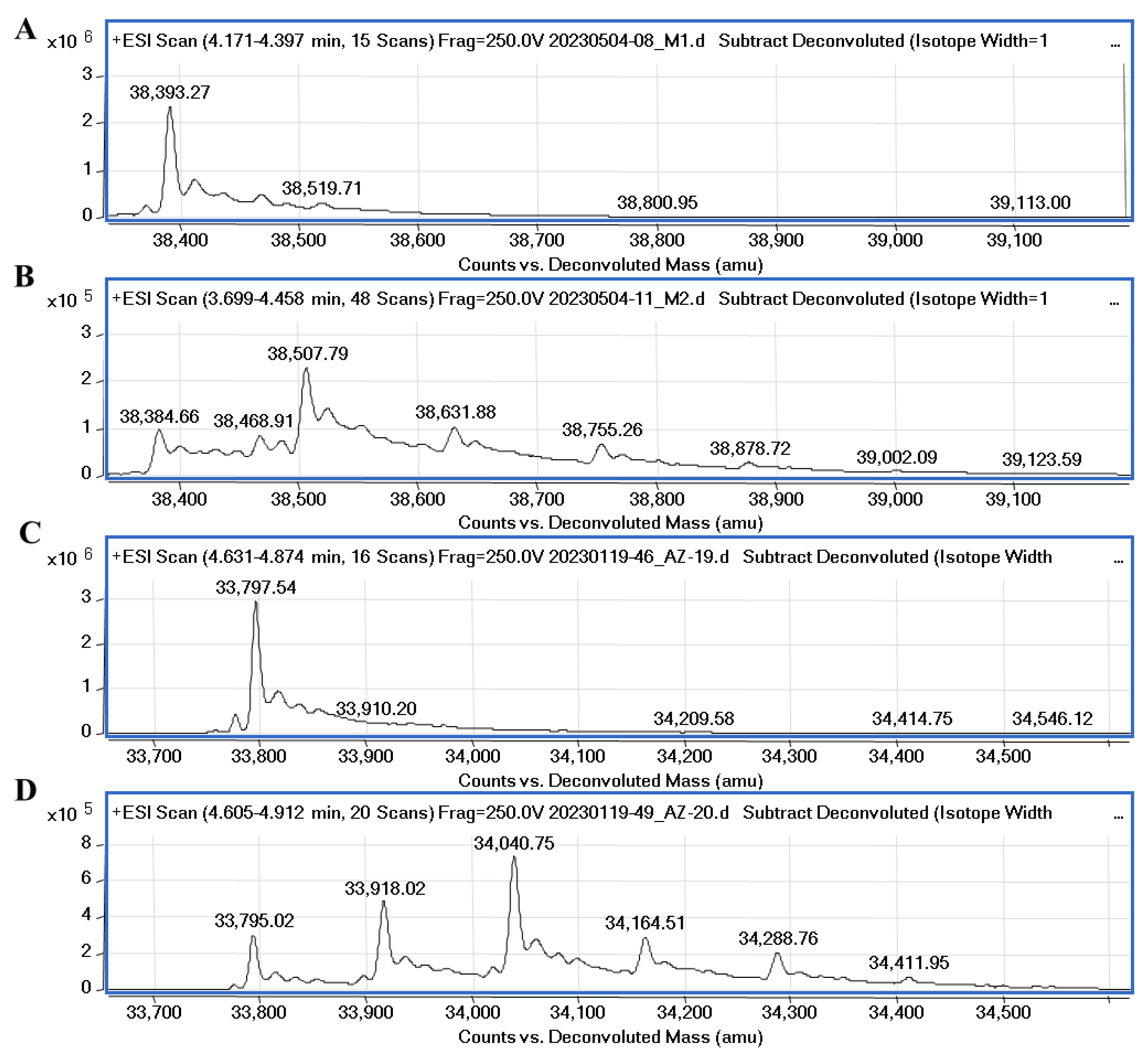
4.5. Mass Spectrometry
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- World Health Organization 2023. Available online: https://covid19.who.int/ (accessed on 18 July 2023).
- Polatoğlu, I.; Oncu-Oner, T.; Dalman, I.; Ozdogan, S. COVID-19 in Early 2023: Structure, Replication Mechanism, Variants of SARS-CoV-2, Diagnostic Tests, and Vaccine & Drug Development Studies. MedComm 2023, 4, e228. [Google Scholar] [CrossRef] [PubMed]
- Zelek, W.M.; Harrison, R.A. Complement and COVID-19: Three Years on, What We Know, What We Don’t Know, and What We Ought to Know. Immunobiology 2023, 228, 152393. [Google Scholar] [CrossRef]
- Moghadasi, S.A.; Heilmann, E.; Khalil, A.M.; Nnabuife, C.; Kearns, F.L.; Ye, C.; Moraes, S.N.; Costacurta, F.; Esler, M.A.; Aihara, H. Transmissible SARS-CoV-2 Variants with Resistance to Clinical Protease Inhibitors. Sci. Adv. 2023, 9, eade8778. [Google Scholar] [CrossRef]
- Rudan, I.; Adeloye, D.; Sheikh, A. COVID-19: Vaccines, Efficacy and Effects on Variants. Curr. Opin. Pulm. Med. 2022, 28, 180–191. [Google Scholar] [CrossRef]
- Li, G. Therapeutic Strategies for COVID-19: Progress and Lessons Learned. Nat. Rev. Drug Discov. 2023, 22, 449–475. [Google Scholar] [CrossRef]
- Yang, L.; Wang, Z. Bench-to-Bedside: Innovation of Small Molecule Anti-SARS-CoV-2 Drugs in China. Eur. J. Med. Chem. 2023, 257, 115503. [Google Scholar] [CrossRef] [PubMed]
- Săndulescu, O.; Apostolescu, C.G.; Preoțescu, L.L.; Streinu-Cercel, A.; Săndulescu, M. Therapeutic Developments for SARS-CoV-2 Infection—Molecular Mechanisms of Action of Antivirals and Strategies for Mitigating Resistance in Emerging Variants in Clinical Practice. Front. Microbiol. 2023, 14, 1132501. [Google Scholar] [CrossRef]
- Gorkhali, R.; Koirala, P.; Rijal, S.; Mainali, A.; Baral, A.; Bhattarai, H.K. Structure and Function of Major SARS-CoV-2 and SARS-CoV Proteins. Bioinform. Biol. Insights 2021, 15, 11779322211025876. [Google Scholar] [CrossRef] [PubMed]
- Moustaqil, M.; Ollivier, E.; Chiu, H.-P.; Van Tol, S.; Rudolffi-Soto, P.; Stevens, C.; Bhumkar, A.; Hunter, D.J.B.; Freiberg, A.N.; Jacques, D.; et al. SARS-CoV-2 Proteases PLpro and 3CLpro Cleave IRF3 and Critical Modulators of Inflammatory Pathways (NLRP12 and TAB1): Implications for Disease Presentation across Species. Emerg. Microbes Infect. 2021, 10, 178–195. [Google Scholar] [CrossRef] [PubMed]
- Munnur, D.; Teo, Q.; Eggermont, D.; Lee, H.H.Y.; Thery, F.; Ho, J.; van Leur, S.W.; Ng, W.W.S.; Siu, L.Y.L.; Beling, A.; et al. Altered ISGylation Drives Aberrant Macrophage-Dependent Immune Responses during SARS-CoV-2 Infection. Nat. Immunol. 2021, 22, 1416–1427. [Google Scholar] [CrossRef]
- Gao, X.; Qin, B.; Chen, P.; Zhu, K.; Hou, P.; Wojdyla, J.A.; Wang, M.; Cui, S. Crystal Structure of SARS-CoV-2 Papain-like Protease. Acta Pharm. Sin. B 2021, 11, 237–245. [Google Scholar] [CrossRef] [PubMed]
- Shin, D.; Mukherjee, R.; Grewe, D.; Bojkova, D.; Baek, K.; Bhattacharya, A.; Schulz, L.; Widera, M.; Mehdipour, A.R.; Tascher, G.; et al. Papain-like Protease Regulates SARS-CoV-2 Viral Spread and Innate Immunity. Nature 2020, 587, 657–662. [Google Scholar] [CrossRef] [PubMed]
- Hu, Q.; Xiong, Y.; Zhu, G.; Zhang, Y.; Zhang, Y.; Huang, P.; Ge, G. The SARS-CoV-2 Main Protease (M pro): Structure, Function, and Emerging Therapies for COVID-19. MedComm 2022, 3, e151. [Google Scholar] [CrossRef]
- Vidak, E.; Javoršek, U.; Vizovišek, M.; Turk, B. Cysteine Cathepsins and Their Extracellular Roles: Shaping the Microenvironment. Cells 2019, 8, 264. [Google Scholar] [CrossRef]
- Zhang, L.; Lin, D.; Sun, X.; Curth, U.; Drosten, C.; Sauerhering, L.; Becker, S.; Rox, K.; Hilgenfeld, R. Crystal Structure of SARS-CoV-2 Main Protease Provides a Basis for Design of Improved α-Ketoamide Inhibitors. Science 2020, 368, 409–412. [Google Scholar] [CrossRef] [PubMed]
- Kronenberger, T.; Laufer, S.A.; Pillaiyar, T. COVID-19 Therapeutics: Small-Molecule Drug Development Targeting SARS-CoV-2 Main Protease. Drug Discov. Today 2023, 28, 103579. [Google Scholar] [CrossRef]
- Calleja, D.J.; Lessene, G.; Komander, D. Inhibitors of SARS-CoV-2 PLpro. Front. Chem. 2022, 10, 876212. [Google Scholar] [CrossRef]
- Tan, H.; Hu, Y.; Jadhav, P.; Tan, B.; Wang, J. Progress and Challenges in Targeting the SARS-CoV-2 Papain-like Protease. J. Med. Chem. 2022, 65, 7561–7580. [Google Scholar] [CrossRef]
- Fu, Z.; Huang, B.; Tang, J.; Liu, S.; Liu, M.; Ye, Y.; Liu, Z.; Xiong, Y.; Zhu, W.; Cao, D.; et al. The Complex Structure of GRL0617 and SARS-CoV-2 PLpro Reveals a Hot Spot for Antiviral Drug Discovery. Nat. Commun. 2021, 12, 488. [Google Scholar] [CrossRef]
- Shan, H.; Liu, J.; Shen, J.; Dai, J.; Xu, G.; Lu, K.; Han, C.; Wang, Y.; Xu, X.; Tong, Y.; et al. Development of Potent and Selective Inhibitors Targeting the Papain-like Protease of SARS-CoV-2. Cell Chem. Biol. 2021, 28, 855–865.e9. [Google Scholar] [CrossRef]
- Sanders, B.C.; Pokhrel, S.; Labbe, A.D.; Mathews, I.I.; Cooper, C.J.; Davidson, R.B.; Phillips, G.; Weiss, K.L.; Zhang, Q.; O’Neill, H.; et al. Potent and Selective Covalent Inhibition of the Papain-like Protease from SARS-CoV-2. Nat. Commun. 2023, 14, 1733. [Google Scholar] [CrossRef] [PubMed]
- Fischer, C.; Feys, J.R. SARS-CoV-2 Mpro Inhibitors: Achieved Diversity, Developing Resistance and Future Strategies. Future Pharmacol. 2023, 3, 80–107. [Google Scholar] [CrossRef]
- Huang, C.; Shuai, H.; Qiao, J.; Hou, Y.; Zeng, R.; Xia, A.; Xie, L.; Fang, Z.; Li, Y.; Yoon, C.; et al. A New Generation Mpro Inhibitor with Potent Activity against SARS-CoV-2 Omicron Variants. Signal Transduct. Target. Ther. 2023, 8, 128. [Google Scholar] [CrossRef] [PubMed]
- Anjani; Kumar, S.; Rathi, B.; Poonam. Recent Updates on the Biological Efficacy of Approved Drugs and Potent Synthetic Compounds against SARS-CoV-2. RSC Adv. 2023, 13, 3677–3687. [Google Scholar] [CrossRef]
- Narayanan, A.; Narwal, M.; Majowicz, S.A.; Varricchio, C.; Toner, S.A.; Ballatore, C.; Brancale, A.; Murakami, K.S.; Jose, J. Identification of SARS-CoV-2 Inhibitors Targeting Mpro and PLpro Using in-Cell-Protease Assay. Commun. Biol. 2022, 5, 169. [Google Scholar] [CrossRef]
- Duveau, D.Y.; Thomas, C.J. The Remarkable Selectivity of Nirmatrelvir. ACS Pharmacol. Transl. Sci. 2022, 5, 445–447. [Google Scholar] [CrossRef] [PubMed]
- Unoh, Y.; Uehara, S.; Nakahara, K.; Nobori, H.; Yamatsu, Y.; Yamamoto, S.; Maruyama, Y.; Taoda, Y.; Kasamatsu, K.; Suto, T.; et al. Discovery of S-217622, a Noncovalent Oral SARS-CoV-2 3CL Protease Inhibitor Clinical Candidate for Treating COVID-19. J. Med. Chem. 2022, 65, 6499–6512. [Google Scholar] [CrossRef] [PubMed]
- Mukae, H.; Yotsuyanagi, H.; Ohmagari, N.; Doi, Y.; Sakaguchi, H.; Sonoyama, T.; Ichihashi, G.; Sanaki, T.; Baba, K.; Tsuge, Y.; et al. Efficacy and Safety of Ensitrelvir in Patients With Mild-to-Moderate Coronavirus Disease 2019: The Phase 2b Part of a Randomized, Placebo-Controlled, Phase 2/3 Study. Clin. Infect. Dis. 2023, 76, 1403–1411. [Google Scholar] [CrossRef]
- Di Sarno, V.; Lauro, G.; Musella, S.; Ciaglia, T.; Vestuto, V.; Sala, M.; Scala, M.C.; Smaldone, G.; Di Matteo, F.; Novi, S.; et al. Identification of a Dual Acting SARS-CoV-2 Proteases Inhibitor through in Silico Design and Step-by-Step Biological Characterization. Eur. J. Med. Chem. 2021, 226, 113863. [Google Scholar] [CrossRef]
- Meewan, I.; Kattoula, J.; Kattoula, J.Y.; Skinner, D.; Fajtová, P.; Giardini, M.A.; Woodworth, B.; McKerrow, J.H.; Lage de Siqueira-Neto, J.; O’Donoghue, A.J.; et al. Discovery of Triple Inhibitors of Both SARS-CoV-2 Proteases and Human Cathepsin L. Pharmaceuticals 2022, 15, 744. [Google Scholar] [CrossRef] [PubMed]
- Gedgaudas, M.; Baronas, D.; Kazlauskas, E.; Petrauskas, V.; Matulis, D. Thermott: A Comprehensive Online Tool for Protein–Ligand Binding Constant Determination. Drug Discov. Today 2022, 27, 2076–2079. [Google Scholar] [CrossRef]
- Skrickus, K.; Šiugždaitė, J.; Lelešiu, R.; Anusevičius, R.; Grybaitė, B.; Vaickelionienė, R.; Mickevičius, V. Synthesis, Characterization and Antibacterial Assays of Novel N,N1-Disubstituted 2,2′-Dithiodianiline Derivatives. ChemistrySelect 2023, 8, e202300332. [Google Scholar] [CrossRef]
- Ma, C.; Hu, Y.; Townsend, J.A.; Lagarias, P.I.; Marty, M.T.; Kolocouris, A.; Wang, J. Ebselen, Disulfiram, Carmofur, PX-12, Tideglusib, and Shikonin Are Nonspecific Promiscuous SARS-CoV-2 Main Protease Inhibitors. ACS Pharmacol. Transl. Sci. 2020, 3, 1265–1277. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Bao, B.-B.; Song, G.-Q.; Chen, C.; Zhang, X.-M.; Lu, W.; Wang, Z.; Cai, Y.; Li, S.; Fu, S.; et al. Discovery of Unsymmetrical Aromatic Disulfides as Novel Inhibitors of SARS-CoV Main Protease: Chemical Synthesis, Biological Evaluation, Molecular Docking and 3D-QSAR Study. Eur. J. Med. Chem. 2017, 137, 450–461. [Google Scholar] [CrossRef] [PubMed]
- Gao, K.; Oerlemans, R.; Groves, M.R. Theory and Applications of Differential Scanning Fluorimetry in Early-Stage Drug Discovery. Biophys. Rev. 2020, 12, 85–104. [Google Scholar] [CrossRef]
- Waldron, T.T.; Murphy, K.P. Stabilization of Proteins by Ligand Binding: Application to Drug Screening and Determination of Unfolding Energetics. Biochemistry 2003, 42, 5058–5064. [Google Scholar] [CrossRef]
- Llowarch, P.; Usselmann, L.; Ivanov, D.; Holdgate, G.A. Thermal Unfolding Methods in Drug Discovery. Biophys. Rev. 2023, 4, 021305. [Google Scholar] [CrossRef]
- Dai, R.; Wilson, D.J.; Geders, T.W.; Aldrich, C.C.; Finzel, B.C. Inhibition of Mycobacterium tuberculosis Transaminase BioA by Aryl Hydrazines and Hydrazides. ChemBioChem 2014, 15, 575–586. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.; Sacco, M.D.; Xia, Z.; Lambrinidis, G.; Townsend, J.A.; Hu, Y.; Meng, X.; Szeto, T.; Ba, M.; Zhang, X.; et al. Discovery of SARS-CoV-2 Papain-like Protease Inhibitors through a Combination of High-Throughput Screening and a FlipGFP-Based Reporter Assay. ACS Cent. Sci. 2021, 7, 1245–1260. [Google Scholar] [CrossRef] [PubMed]
- Grybaitė, B.; Jonuškienė, I.; Vaickelionienė, R.; Mickevičius, V. Synthesis, transformation and antibacterial activity of new N,N-disubstituted 2-aminothiazole derivatives. Chemija 2017, 28, 64–73. [Google Scholar]
- Grybaitė, B.; Vaickelionienė, R.; Stasevych, M.; Komarovska-Porokhnyavets, O.; Novikov, V.; Mickevičius, V. Synthesis, Transformation of 3-[(4-Arylthiazol-2-yl)(p-tolyl)amino]propanoic Acids, Bis(thiazol-5-yl)phenyl-, Bis(thiazol-5-yl)methane Derivatives, and Their Antimicrobial Activity. Heterocycles 2018, 96, 86. [Google Scholar] [CrossRef]
- Tumosienė, I.; Jakienė, E.; Beresnevičius, Z.J.; Mikulskienė, G. Synthesis and properties of dihydrazides of N-phenyl-and N-(4-methylphenyl)-N-carboxyethyl-β-alanines. Cheminė Technol. 2006, 3, 58–64. [Google Scholar]
- Grybaitė, B.; Vaickelionienė, R.; Stasevych, M.; Komarovska-Porokhnyavets, O.; Kantminienė, K.; Novikov, V.; Mickevičius, V. Synthesis and Antimicrobial Activity of Novel Thiazoles with Reactive Functional Groups. ChemistrySelect 2019, 4, 6965–6970. [Google Scholar] [CrossRef]
- Sapijanskaitė-Banevič, B.; Grybaitė, B.; Vaickelionienė, R.; Bružaitė, I. Synthesis, transformation and preliminary bioassay of 3-(thiazol-2-yl(p-tolyl)amino)propanoic acid derivatives. Chemija 2023, 34, 57–69. [Google Scholar] [CrossRef]
- Minickaitė, R.; Grybaitė, B.; Vaickelionienė, R.; Kavaliauskas, P.; Petraitis, V.; Petraitienė, R.; Tumosienė, I.; Jonuškienė, I.; Mickevičius, V. Synthesis of Novel Aminothiazole Derivatives as Promising Antiviral, Antioxidant and Antibacterial Candidates. Int. J. Mol. Sci. 2022, 23, 7688. [Google Scholar] [CrossRef]

| Compound | Kd, µM | |||
|---|---|---|---|---|
| PLpro | Mpro | |||
| 1 | 68 | >200 | ||
| 2 | 29 | >200 | ||
| 3 | 14 | 25 | ||
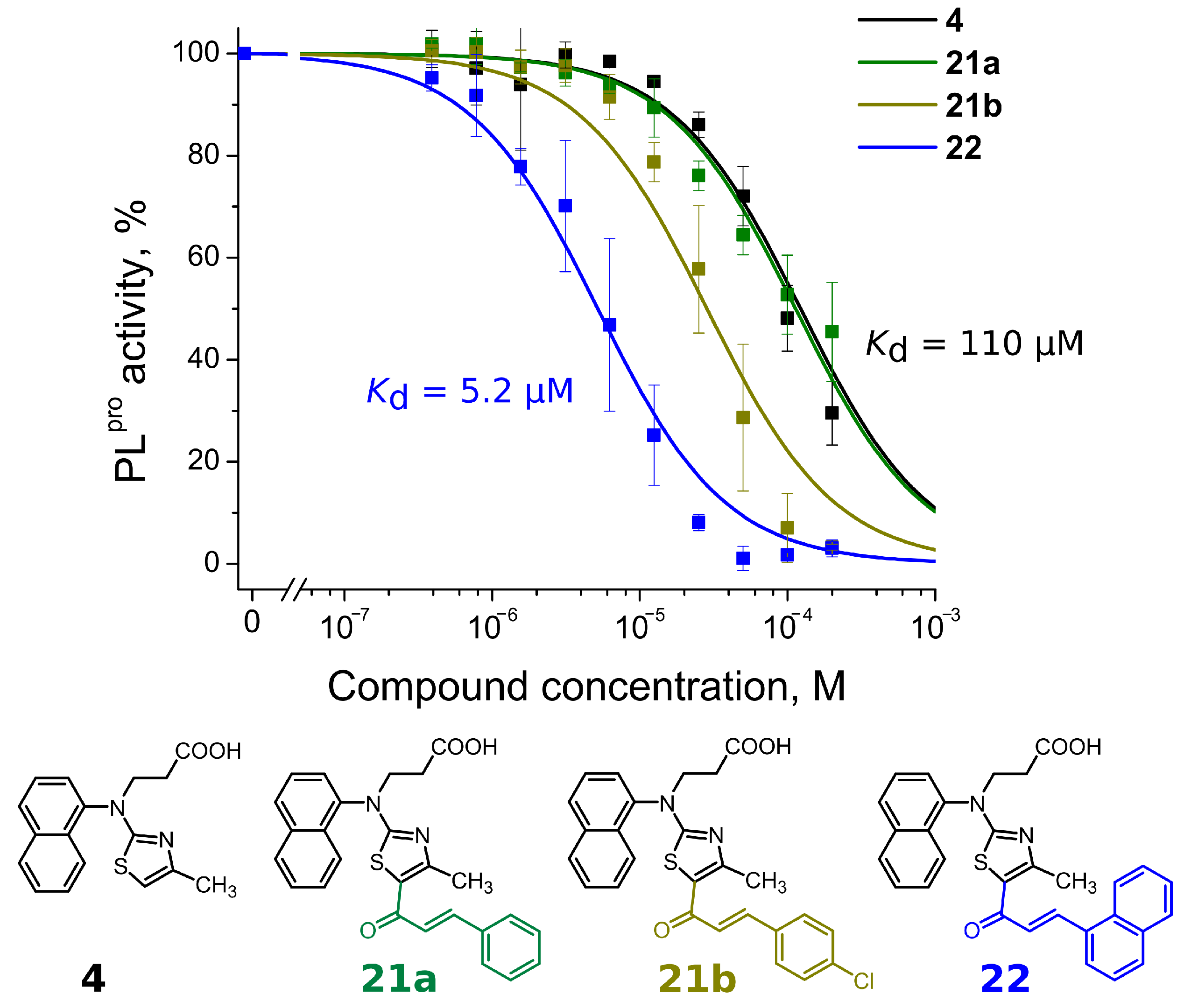
| R1 group | 4 | 110 | >200 | |
| 5 | >200 | >200 | ||
| 6a | 50 | >200 | ||
| 6b | 50 | >200 | ||
| 6c | 50 | >200 | ||
| 6d | 25 | 26 | ||
| 6e | 20 | >200 | ||
| 7 | 50 | >200 | ||
| 8 | 35 | >200 | ||
| 9 | 50 | >200 | ||
| 10 | 35 | >200 | ||
| 11 | >200 | >200 | ||
| 12 | 29 | >200 | ||
| R2 group | Short tail | 13a | 24 | 6.5 |
| 13b | 50 | >200 | ||
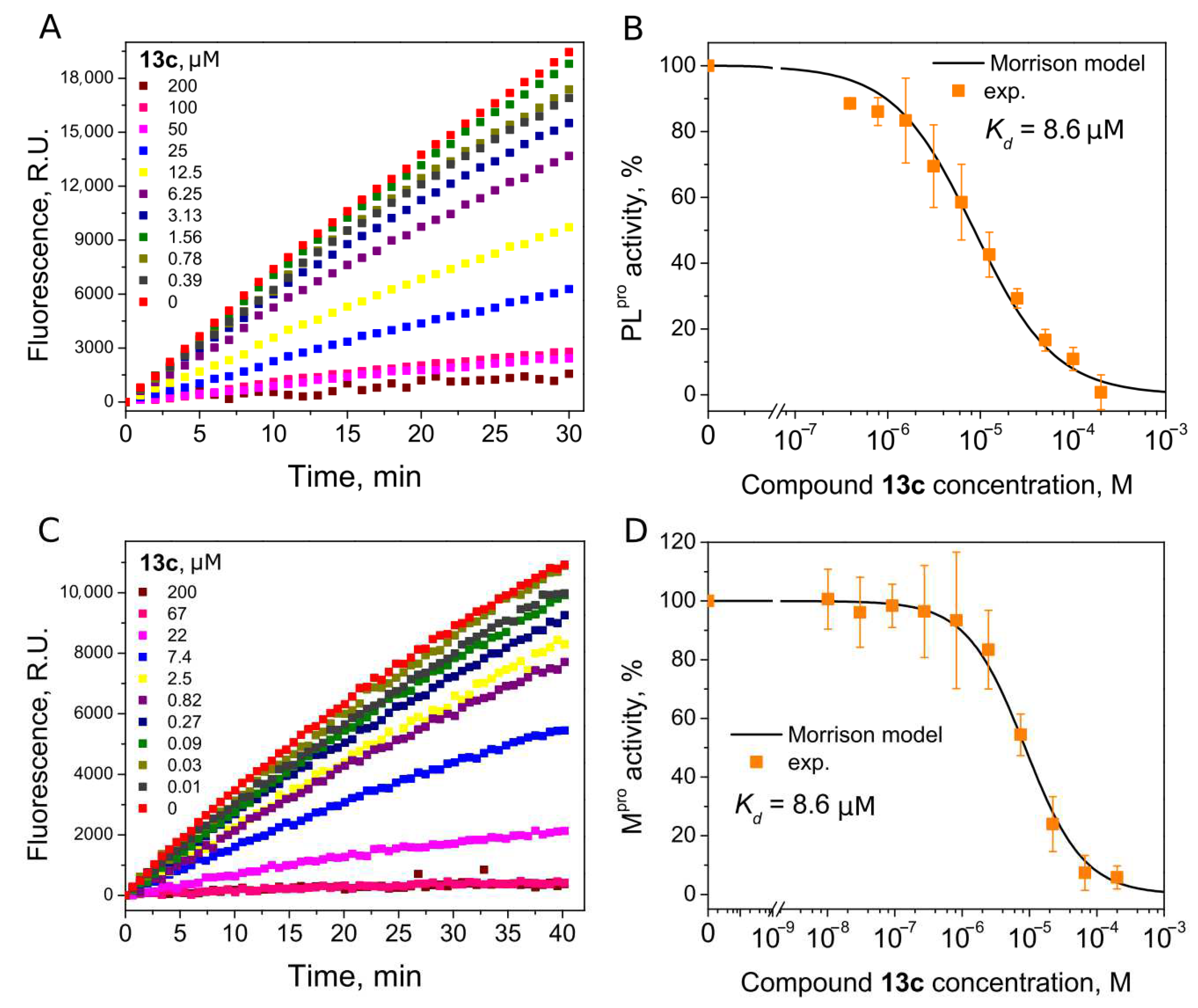
| 13c | 8.6 | 8.6 | ||
| 14 | 9.2 | >200 | ||
| 15 | 50 | >200 | ||
| 16 | 50 | >200 | ||
| 17 | >200 | >200 | ||
| 18 | 50 | >200 | ||
| 19 | 50 | >200 | ||
| 20 | 29 | >200 | ||
| Long tail | 21a | 110 | >200 | |
| 21b | 29 | 17 | ||
| 21c | 52 | >200 | ||
| 22 | 5.2 | 10 | ||
| 23 | 75 | >200 | ||
| 24 | 8.0 | >200 | ||
| Two tails/ scaffold variation group | 25a | 83 | >200 | |
| 25b | 66 | >200 | ||
| 25c | 58 | >200 | ||
| 26 | 16 | >200 | ||
| 27a | 35 | >200 | ||
| 27b | 50 | >200 | ||
| 27c | 15 | >200 | ||
| 28 | 8.8 | 50 | ||
| 29 | >200 | >200 | ||
| 30a | 50 | >200 | ||
| 30b | 50 | >200 | ||
| 30c | 32 | >200 | ||
| 30d | 50 | >200 | ||
| 31 | 50 | 50 | ||
| GRL0617 | 1.6 | >200 | ||
| Compound | Kd, µM | |
|---|---|---|
| PLpro | Mpro | |
| 32 | 0.54 | 2.8 |
| 33 | 2.0 | >200 |
| 34 | 0.43 | 3.5 |
| 35 | 2.5 | >200 |
| 36 | 1.9 | >200 |
| 37 | 0.63 | >200 |
| 38 | 1.9 | >200 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bagdonas, M.; Čerepenkaitė, K.; Mickevičiūtė, A.; Kananavičiūtė, R.; Grybaitė, B.; Anusevičius, K.; Rukšėnaitė, A.; Kojis, T.; Gedgaudas, M.; Mickevičius, V.; et al. Screening, Synthesis and Biochemical Characterization of SARS-CoV-2 Protease Inhibitors. Int. J. Mol. Sci. 2023, 24, 13491. https://doi.org/10.3390/ijms241713491
Bagdonas M, Čerepenkaitė K, Mickevičiūtė A, Kananavičiūtė R, Grybaitė B, Anusevičius K, Rukšėnaitė A, Kojis T, Gedgaudas M, Mickevičius V, et al. Screening, Synthesis and Biochemical Characterization of SARS-CoV-2 Protease Inhibitors. International Journal of Molecular Sciences. 2023; 24(17):13491. https://doi.org/10.3390/ijms241713491
Chicago/Turabian StyleBagdonas, Martynas, Kamilė Čerepenkaitė, Aurelija Mickevičiūtė, Rūta Kananavičiūtė, Birutė Grybaitė, Kazimieras Anusevičius, Audronė Rukšėnaitė, Tautvydas Kojis, Marius Gedgaudas, Vytautas Mickevičius, and et al. 2023. "Screening, Synthesis and Biochemical Characterization of SARS-CoV-2 Protease Inhibitors" International Journal of Molecular Sciences 24, no. 17: 13491. https://doi.org/10.3390/ijms241713491
APA StyleBagdonas, M., Čerepenkaitė, K., Mickevičiūtė, A., Kananavičiūtė, R., Grybaitė, B., Anusevičius, K., Rukšėnaitė, A., Kojis, T., Gedgaudas, M., Mickevičius, V., Matulis, D., Zubrienė, A., & Matulienė, J. (2023). Screening, Synthesis and Biochemical Characterization of SARS-CoV-2 Protease Inhibitors. International Journal of Molecular Sciences, 24(17), 13491. https://doi.org/10.3390/ijms241713491










