Identification of a Non-Invasive Urinary Exosomal Biomarker for Diabetic Nephropathy Using Data-Independent Acquisition Proteomics
Abstract
:1. Introduction
2. Results
2.1. Characterization of the Exosomes
2.2. Proteomic Analysis
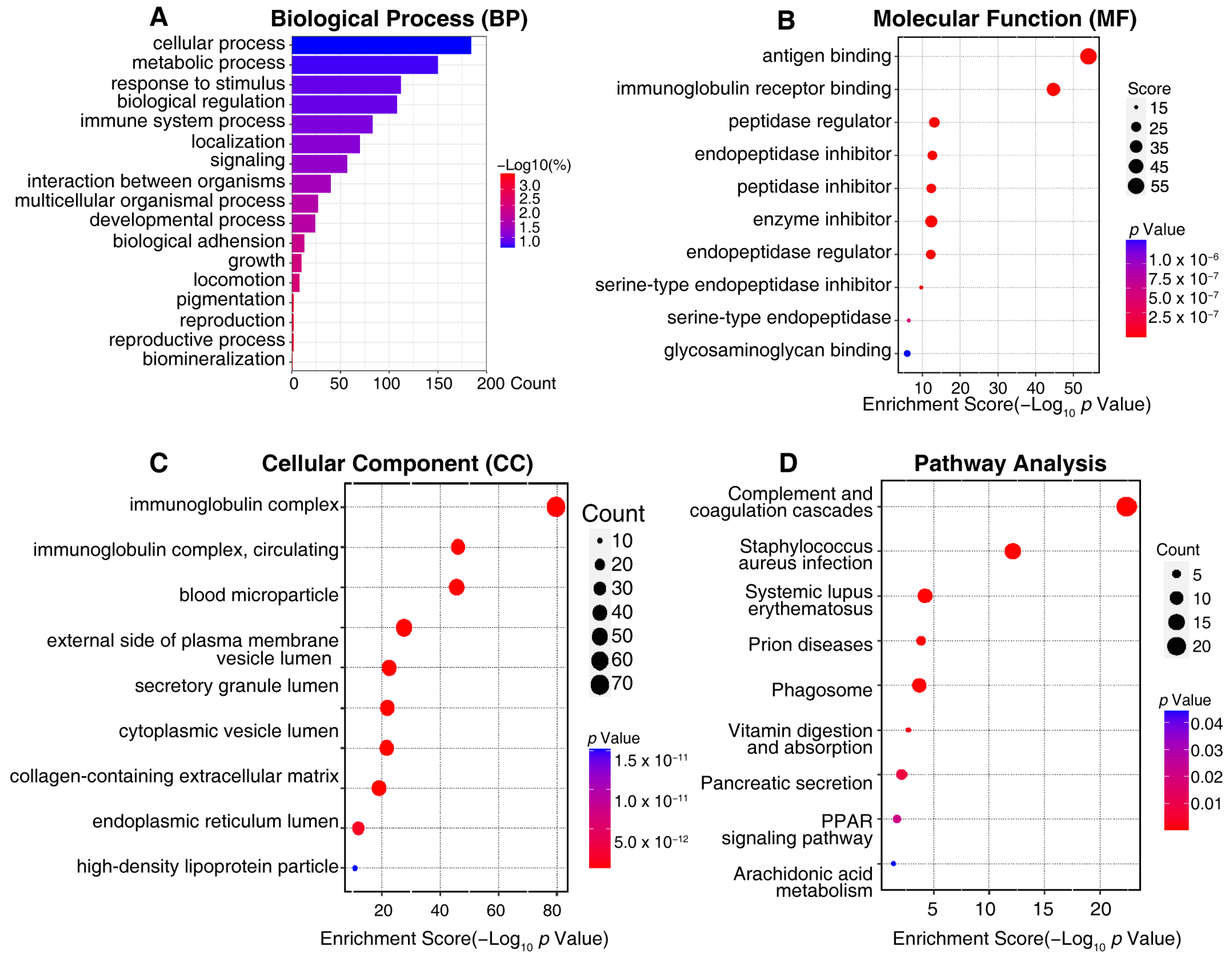
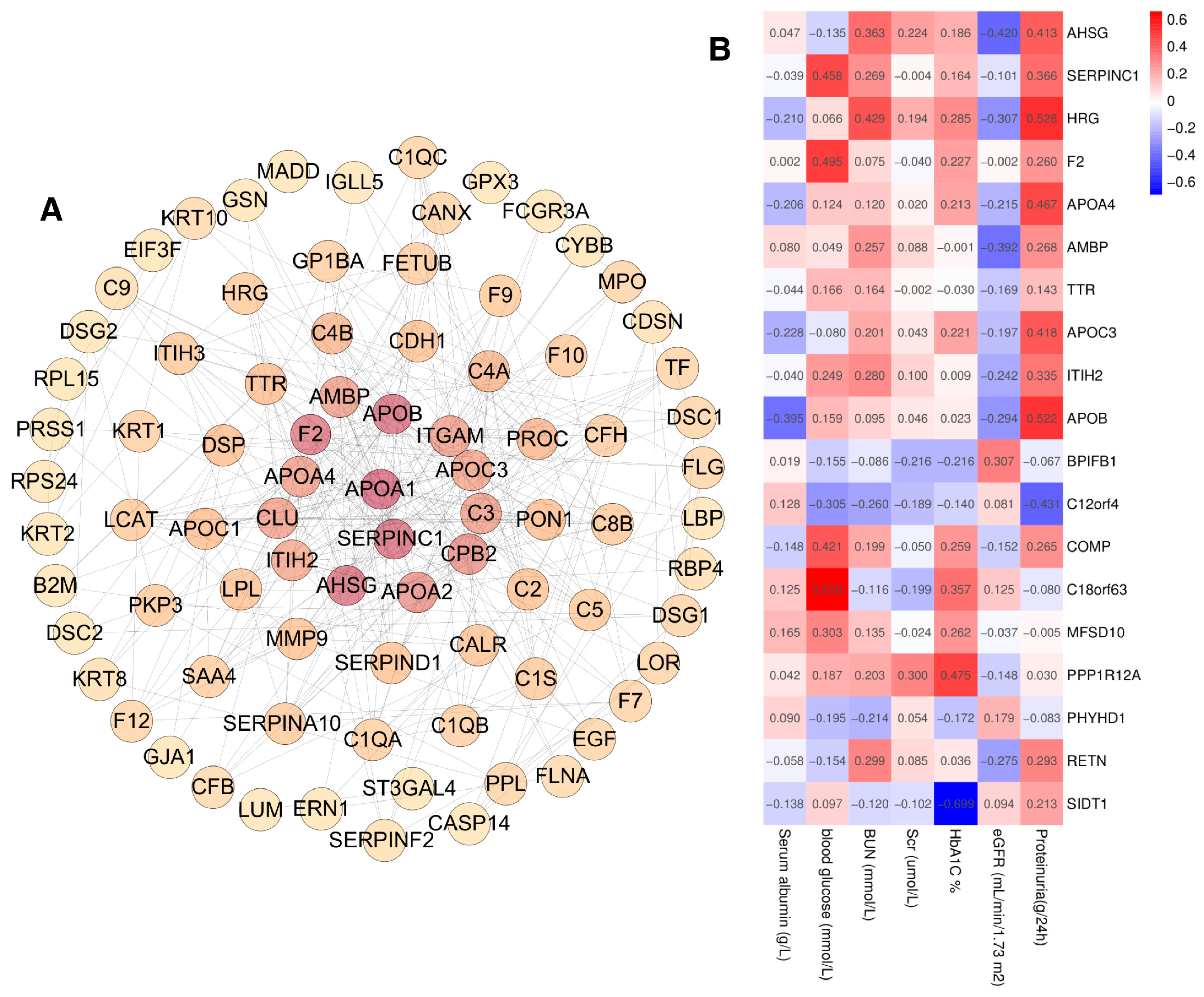
2.3. Bioinformatic Analysis of Differentially Expressed Proteins
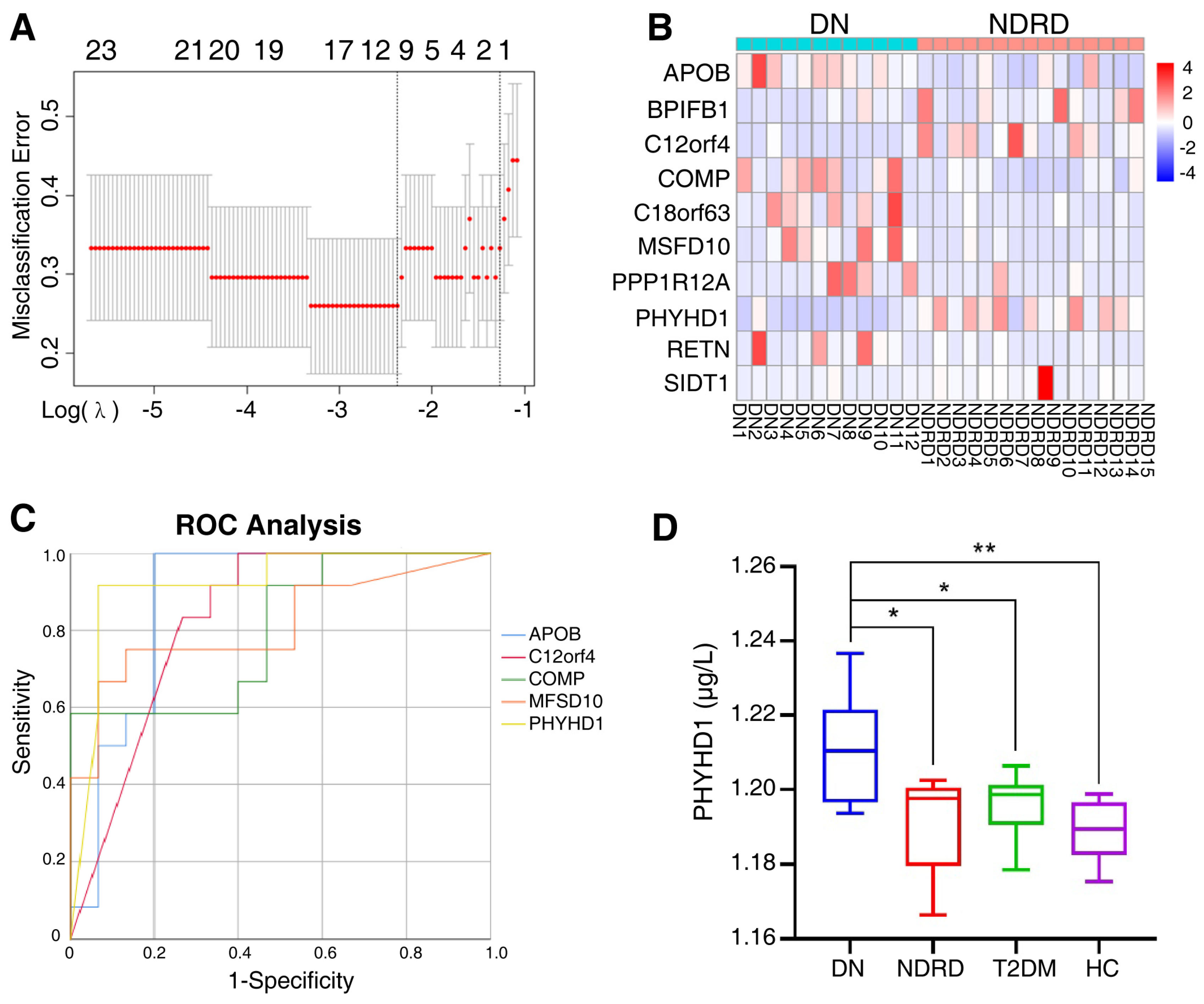
2.4. Screening for Potential Biomarkers
2.5. Validation of Biomarker
2.6. Clinical Correlation
3. Discussion
Study Limitations
4. Materials and Methods
4.1. Description of the Cohort
4.2. Urine Exosomes Isolation
4.3. Characterization of the Exosomes
4.4. Proteomic Analyses
4.5. Bioinformatics Analyses and Statistical Rationale
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ACN | Acetonitrile |
| ADA | American Diabetes Association |
| AMBP | Alpha-1-microglobulin/bikunin precursor |
| AUC | Area under the curve |
| DIA | Data-independent acquisition |
| DN | Diabetic nephropathy |
| DPP-4 | Dipeptidyl peptidase-4 |
| eGFR | Estimated glomerular filtration rate |
| ESRD | End-stage renal disease |
| FA | Formic acid |
| FDR | False discovery rate |
| FSGS | Focal segmental glomerulosclerosis |
| GO | Gene ontology |
| HC | Healthy control |
| IAM | Iodoacetamide |
| IgAN | IgA nephropathy |
| IL-6 | Interleukin-6 |
| ISEV | International Society for Extracellular Vesicles |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| LASSO | Least absolute shrinkage and selection operator |
| MN | Membranous nephropathy |
| MS | Mass spectrometry |
| NDRD | Non-diabetic renal disease |
| NTA | Nanoparticle tracking analysis |
| PBS | Phosphate-buffered saline |
| PHYHD1 | Phytanoyl-CoA dioxygenase domain containing 1 |
| PLA | People’s Liberation Army |
| PPI | Protein–protein interaction networks |
| ROC | Receiver operating characteristic curve |
| T1DM | Type 1 diabetes mellitus |
| T2DM | Type 2 diabetes mellitus |
| TEM | Transmission electron microscope |
| TGF-β | Transforming growth factor-β |
| UACR | Urinary albumin-to-creatinine ratio |
References
- Saeedi, P.; Petersohn, I.; Salpea, P.; Malanda, B.; Karuranga, S.; Unwin, N.; Colagiuri, S.; Guariguata, L.; Motala, A.A.; Ogurtsova, K.; et al. Global and regional diabetes prevalence estimates for 2019 and projections for 2030 and 2045: Results from the International Diabetes Federation Diabetes Atlas, 9(th) edition. Diabetes Res. Clin. Pract. 2019, 157, 107843. [Google Scholar] [CrossRef]
- American Diabetes Association Professional Practice Committee. 11. Chronic Kidney Disease and Risk Management: Standards of Medical Care in Diabetes—2022. Diabetes Care 2021, 45, S175–S184. [Google Scholar]
- Cockwell, P.; Fisher, L.A. The global burden of chronic kidney disease. Lancet 2020, 395, 662–664. [Google Scholar] [CrossRef]
- Sharma, S.G.; Bomback, A.S.; Radhakrishnan, J.; Herlitz, L.C.; Stokes, M.B.; Markowitz, G.S.; D’Agati, V.D. The modern spectrum of renal biopsy findings in patients with diabetes. Clin. J. Am. Soc. Nephrol. 2013, 8, 1718–1724. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Liu, Y.; Liu, S.; Gao, M.; Wang, W.; Chen, K.; Huang, L.; Liu, Y. Diabetic vascular diseases: Molecular mechanisms and therapeutic strategies. Signal. Transduct. Target. Ther. 2023, 8, 152. [Google Scholar]
- Bermejo, S.; Pascual, J.; Soler, M.J. The current role of renal biopsy in diabetic patients. Minerva Med. 2018, 109, 116–125. [Google Scholar] [CrossRef]
- Santucci, L.; Candiano, G.; Petretto, A.; Bruschi, M.; Lavarello, C.; Inglese, E.; Righetti, P.G.; Ghiggeri, G.M. From hundreds to thousands: Widening the normal human Urinome (1). J. Proteom. 2015, 112, 53–62. [Google Scholar] [CrossRef]
- Erdbrügger, U.; Blijdorp, C.J.; Bijnsdorp, I.V.; Borràs, F.E.; Burger, D.; Bussolati, B.; Byrd, J.B.; Clayton, A.; Dear, J.W.; Falcón-Pérez, J.M.; et al. Urinary extracellular vesicles: A position paper by the Urine Task Force of the International Society for Extracellular Vesicles. J. Extracell. Vesicles 2021, 10, e12093. [Google Scholar] [CrossRef]
- Vitorino, R.; Ferreira, R.; Guedes, S.; Amado, F.; Thongboonkerd, V. What can urinary exosomes tell us? Cell. Mol. Life Sci. 2021, 78, 3265–3283. [Google Scholar] [CrossRef]
- Meldolesi, J. Exosomes and Ectosomes in Intercellular Communication. Curr. Biol. 2018, 28, R435–R444. [Google Scholar] [PubMed]
- Yuan, Y.; Mei, Z.; Qu, Z.; Li, G.; Yu, S.; Liu, Y.; Liu, K.; Shen, Z.; Pu, J.; Wang, Y.; et al. Exosomes secreted from cardiomyocytes suppress the sensitivity of tumor ferroptosis in ischemic heart failure. Signal. Transduct. Target. Ther. 2023, 8, 121. [Google Scholar] [CrossRef]
- Xing, C.; Li, H.; Li, R.J.; Yin, L.; Zhang, H.F.; Huang, Z.N.; Cheng, Z.; Li, J.; Wang, Z.H.; Peng, H.L. The roles of exosomal immune checkpoint proteins in tumors. Mil. Med. Res. 2021, 8, 56. [Google Scholar] [CrossRef]
- Paskeh, M.D.A.; Entezari, M.; Mirzaei, S.; Zabolian, A.; Saleki, H.; Naghdi, M.J.; Sabet, S.; Khoshbakht, M.A.; Hashemi, M.; Hushmandi, K.; et al. Emerging role of exosomes in cancer progression and tumor microenvironment remodeling. J. Hematol. Oncol. 2022, 15, 83. [Google Scholar] [CrossRef] [PubMed]
- Vergani, E.; Daveri, E.; Vallacchi, V.; Bergamaschi, L.; Lalli, L.; Castelli, C.; Rodolfo, M.; Rivoltini, L.; Huber, V. Extracellular vesicles in anti-tumor immunity. Semin. Cancer Biol. 2022, 86, 64–79. [Google Scholar] [CrossRef] [PubMed]
- Kalluri, R.; LeBleu, V.S. The biology, function, and biomedical applications of exosomes. Science 2020, 367, eaau6977. [Google Scholar] [CrossRef]
- Zubiri, I.; Posada-Ayala, M.; Sanz-Maroto, A.; Calvo, E.; Martin-Lorenzo, M.; Gonzalez-Calero, L.; de la Cuesta, F.; Lopez, J.A.; Fernandez-Fernandez, B.; Ortiz, A.; et al. Diabetic nephropathy induces changes in the proteome of human urinary exosomes as revealed by label-free comparative analysis. J. Proteom. 2014, 96, 92–102. [Google Scholar] [CrossRef]
- Kamińska, A.; Platt, M.; Kasprzyk, J.; Kuśnierz-Cabala, B.; Gala-Błądzińska, A.; Woźnicka, O.; Jany, B.R.; Krok, F.; Piekoszewski, W.; Kuźniewski, M.; et al. Urinary Extracellular Vesicles: Potential Biomarkers of Renal Function in Diabetic Patients. J. Diabetes Res. 2016, 2016, 5741518. [Google Scholar] [CrossRef]
- De, S.; Kuwahara, S.; Hosojima, M.; Ishikawa, T.; Kaseda, R.; Sarkar, P.; Yoshioka, Y.; Kabasawa, H.; Iida, T.; Goto, S.; et al. Exocytosis-Mediated Urinary Full-Length Megalin Excretion Is Linked with the Pathogenesis of Diabetic Nephropathy. Diabetes 2017, 66, 1391–1404. [Google Scholar] [CrossRef] [PubMed]
- Sun, A.-L.; Deng, J.-T.; Guan, G.-J.; Chen, S.-H.; Liu, Y.-T.; Cheng, J.; Li, Z.-W.; Zhuang, X.-H.; Sun, F.-D.; Deng, H.-P. Dipeptidyl peptidase-IV is a potential molecular biomarker in diabetic kidney disease. Diabetes Vasc. Dis. Res. 2012, 9, 301–308. [Google Scholar] [CrossRef] [PubMed]
- Musante, L.; Tataruch, D.; Gu, D.; Liu, X.; Forsblom, C.; Groop, P.-H.; Holthofer, H. Proteases and Protease Inhibitors of Urinary Extracellular Vesicles in Diabetic Nephropathy. J. Diabetes Res. 2015, 2015, 289734. [Google Scholar] [CrossRef]
- Ding, X.; Wang, X.; Du, J.; Han, Q.; Zhang, D.; Zhu, H. A systematic review and Meta-analysis of urinary extracellular vesicles proteome in diabetic nephropathy. Front. Endocrinol. 2022, 13, 866252. [Google Scholar] [CrossRef] [PubMed]
- Fonseka, P.; Pathan, M.; Chitti, S.V.; Kang, T.; Mathivanan, S. FunRich enables enrichment analysis of OMICs datasets. J. Mol. Biol. 2021, 433, 166747. [Google Scholar] [CrossRef] [PubMed]
- Gonzales, P.A.; Pisitkun, T.; Hoffert, J.D.; Tchapyjnikov, D.; Star, R.A.; Kleta, R.; Wang, N.S.; Knepper, M.A. Large-scale proteomics and phosphoproteomics of urinary exosomes. J. Am. Soc. Nephrol. 2009, 20, 363–379. [Google Scholar] [CrossRef] [PubMed]
- Pisitkun, T.; Shen, R.F.; Knepper, M.A. Identification and proteomic profiling of exosomes in human urine. Proc. Natl. Acad. Sci. USA 2004, 101, 13368–13373. [Google Scholar] [CrossRef]
- Teng, F.; Fussenegger, M. Shedding Light on Extracellular Vesicle Biogenesis and Bioengineering. Adv. Sci. 2020, 8, 2003505. [Google Scholar] [CrossRef]
- van Niel, G.; D’Angelo, G.; Raposo, G. Shedding light on the cell biology of extracellular vesicles. Nat. Rev. Mol. Cell Biol. 2018, 19, 213–228. [Google Scholar] [CrossRef]
- Matsuzaka, Y.; Yashiro, R. Advances in Purification, Modification, and Application of Extracellular Vesicles for Novel Clinical Treatments. Membranes 2022, 12, 1244. [Google Scholar] [CrossRef]
- Jeppesen, D.K.; Fenix, A.M.; Franklin, J.L.; Higginbotham, J.N.; Zhang, Q.; Zimmerman, L.J.; Liebler, D.C.; Ping, J.; Liu, Q.; Evans, R.; et al. Reassessment of Exosome Composition. Cell 2019, 177, 428–445.e418. [Google Scholar] [CrossRef]
- Sidhom, K.; Obi, P.O.; Saleem, A. A Review of Exosomal Isolation Methods: Is Size Exclusion Chromatography the Best Option? Int. J. Mol. Sci. 2020, 21, 6466. [Google Scholar] [CrossRef]
- Barranco, I.; Sanchez-López, C.M.; Bucci, D.; Alvarez-Barrientos, A.; Rodriguez-Martinez, H.; Marcilla, A.; Roca, J. The Proteome of Large or Small Extracellular Vesicles in Pig Seminal Plasma Differs, Defining Sources and Biological Functions. Mol. Cell. Proteom. 2023, 22, 100514. [Google Scholar] [CrossRef]
- Merchant, M.L.; Rood, I.M.; Deegens, J.K.J.; Klein, J.B. Isolation and characterization of urinary extracellular vesicles: Implications for biomarker discovery. Nat. Rev. Nephrol. 2017, 13, 731–749. [Google Scholar] [CrossRef] [PubMed]
- Weiss, G.L.; Stanisich, J.J.; Sauer, M.M.; Lin, C.W.; Eras, J.; Zyla, D.S.; Trück, J.; Devuyst, O.; Aebi, M.; Pilhofer, M.; et al. Architecture and function of human uromodulin filaments in urinary tract infections. Science 2020, 369, 1005–1010. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Llama, P.; Khositseth, S.; Gonzales, P.A.; Star, R.A.; Pisitkun, T.; Knepper, M.A. Tamm-Horsfall protein and urinary exosome isolation. Kidney Int. 2010, 77, 736–742. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Kochan, G.T.; Ng, S.S.; Kavanagh, K.L.; Oppermann, U.; Schofield, C.J.; McDonough, M.A. Crystal structure of PHYHD1A, a 2OG oxygenase related to phytanoyl-CoA hydroxylase. Biochem. Biophys. Res. Commun. 2011, 408, 553–558. [Google Scholar] [CrossRef] [PubMed]
- Tang, D.; Kroemer, G. Peroxisome: The new player in ferroptosis. Signal. Transduct. Target. Ther. 2020, 5, 273. [Google Scholar] [CrossRef] [PubMed]
- Zou, Y.; Henry, W.S.; Ricq, E.L.; Graham, E.T.; Phadnis, V.V.; Maretich, P.; Paradkar, S.; Boehnke, N.; Deik, A.A.; Reinhardt, F.; et al. Plasticity of ether lipids promotes ferroptosis susceptibility and evasion. Nature 2020, 585, 603–608. [Google Scholar] [CrossRef]
- Brown, C.W.; Amante, J.J.; Chhoy, P.; Elaimy, A.L.; Liu, H.; Zhu, L.J.; Baer, C.E.; Dixon, S.J.; Mercurio, A.M. Prominin2 Drives Ferroptosis Resistance by Stimulating Iron Export. Dev. Cell 2019, 51, 575–586.e574. [Google Scholar] [CrossRef]
- Strzyz, P. Iron expulsion by exosomes drives ferroptosis resistance. Nat. Rev. Mol. Cell Biol. 2020, 21, 4–5. [Google Scholar] [CrossRef]
- Kim, S.; Kang, S.W.; Joo, J.; Han, S.H.; Shin, H.; Nam, B.Y.; Park, J.; Yoo, T.H.; Kim, G.; Lee, P.; et al. Characterization of ferroptosis in kidney tubular cell death under diabetic conditions. Cell Death Dis. 2021, 12, 160. [Google Scholar] [CrossRef]
- Furusawa, Y.; Kubo, T.; Fukazawa, T. Phyhd1, an XPhyH-like homologue, is induced in mouse T cells upon T cell stimulation. Biochem. Biophys. Res. Commun. 2016, 472, 551–556. [Google Scholar] [CrossRef]
- Vandewalle, C.; Van Roy, F.; Berx, G. The role of the ZEB family of transcription factors in development and disease. Cell. Mol. Life Sci. 2009, 66, 773–787. [Google Scholar] [CrossRef] [PubMed]
- Krupa, A.; Jenkins, R.; Luo, D.D.; Lewis, A.; Phillips, A.; Fraser, D. Loss of MicroRNA-192 promotes fibrogenesis in diabetic nephropathy. J. Am. Soc. Nephrol. 2010, 21, 438–447. [Google Scholar] [CrossRef] [PubMed]
- Putta, S.; Lanting, L.; Sun, G.; Lawson, G.; Kato, M.; Natarajan, R. Inhibiting microRNA-192 ameliorates renal fibrosis in diabetic nephropathy. J. Am. Soc. Nephrol. 2012, 23, 458–469. [Google Scholar] [CrossRef] [PubMed]
- Hotta, O.; Sugai, H.; Kitamura, H.; Yusa, N.; Taguma, Y. Predictive value of urinary micro-cholesterol (mCHO) levels in patients with progressive glomerular disease. Kidney Int. 2004, 66, 2374–2381. [Google Scholar] [CrossRef] [PubMed]
- Andreu, Z.; Otta Oshiro, R.; Redruello, A.; López-Martín, S.; Gutiérrez-Vázquez, C.; Morato, E.; Marina, A.I.; Olivier Gómez, C.; Yáñez-Mó, M. Extracellular vesicles as a source for non-invasive biomarkers in bladder cancer progression. Eur. J. Pharm. Sci. 2017, 98, 70–79. [Google Scholar] [CrossRef] [PubMed]
- Morell, M.; Varela, N.; Castillejo-López, C.; Coppard, C.; Luque, M.J.; Wu, Y.Y.; Martín-Morales, N.; Pérez-Cózar, F.; Gómez-Hernández, G.; Kumar, R.; et al. SIDT1 plays a key role in type I IFN responses to nucleic acids in plasmacytoid dendritic cells and mediates the pathogenesis of an imiquimod-induced psoriasis model. EBioMedicine 2022, 76, 103808. [Google Scholar] [CrossRef]
- Nguyen, T.A.; Smith, B.R.C.; Elgass, K.D.; Creed, S.J.; Cheung, S.; Tate, M.D.; Belz, G.T.; Wicks, I.P.; Masters, S.L.; Pang, K.C. SIDT1 Localizes to Endolysosomes and Mediates Double-Stranded RNA Transport into the Cytoplasm. J. Immunol. 2019, 202, 3483–3492. [Google Scholar] [CrossRef]
- Stefan, N.; Hennige, A.M.; Staiger, H.; Machann, J.; Schick, F.; Kröber, S.M.; Machicao, F.; Fritsche, A.; Häring, H.U. Alpha2-Heremans-Schmid glycoprotein/fetuin-A is associated with insulin resistance and fat accumulation in the liver in humans. Diabetes Care 2006, 29, 853–857. [Google Scholar] [CrossRef]
- Heo, J.I.; Yoon, D.W.; Yu, J.H.; Kim, N.H.; Yoo, H.J.; Seo, J.A.; Kim, S.G.; Choi, K.M.; Baik, S.H.; Choi, D.S.; et al. Melatonin improves insulin resistance and hepatic steatosis through attenuation of alpha-2-HS-glycoprotein. J. Pineal Res. 2018, 65, e12493. [Google Scholar] [CrossRef]
- Gross, B.; Pawlak, M.; Lefebvre, P.; Staels, B. PPARs in obesity-induced T2DM, dyslipidaemia and NAFLD. Nat. Rev. Endocrinol. 2017, 13, 36–49. [Google Scholar] [CrossRef]
- Lodhi, I.J.; Semenkovich, C.F. Peroxisomes: A nexus for lipid metabolism and cellular signaling. Cell Metab. 2014, 19, 380–392. [Google Scholar] [CrossRef]
- Bansal, G.; Thanikachalam, P.V.; Maurya, R.K.; Chawla, P.; Ramamurthy, S. An overview on medicinal perspective of thiazolidine-2,4-dione: A remarkable scaffold in the treatment of type 2 diabetes. J. Adv. Res. 2020, 23, 163–205. [Google Scholar] [CrossRef] [PubMed]
- American Diabetes Association. 2. Classification and Diagnosis of Diabetes: Standards of Medical Care in Diabetes-2020. Diabetes Care 2020, 43, S14–S31. [Google Scholar] [CrossRef] [PubMed]
- Tervaert, T.W.; Mooyaart, A.L.; Amann, K.; Cohen, A.H.; Cook, H.T.; Drachenberg, C.B.; Ferrario, F.; Fogo, A.B.; Haas, M.; de Heer, E.; et al. Pathologic classification of diabetic nephropathy. J. Am. Soc. Nephrol. 2010, 21, 556–563. [Google Scholar] [CrossRef]
- Han, Q.; Zhang, Y.; Jiao, T.; Li, Q.; Ding, X.; Zhang, D.; Cai, G.; Zhu, H. Urinary sediment microRNAs can be used as potential noninvasive biomarkers for diagnosis, reflecting the severity and prognosis of diabetic nephropathy. Nutr. Diabetes 2021, 11, 24. [Google Scholar] [CrossRef] [PubMed]
- Bachurski, D.; Schuldner, M.; Nguyen, P.H.; Malz, A.; Reiners, K.S.; Grenzi, P.C.; Babatz, F.; Schauss, A.C.; Hansen, H.P.; Hallek, M.; et al. Extracellular vesicle measurements with nanoparticle tracking analysis—An accuracy and repeatability comparison between NanoSight NS300 and ZetaView. J. Extracell. Vesicles 2019, 8, 1596016. [Google Scholar] [CrossRef]
- Théry, C.; Amigorena, S.; Raposo, G.; Clayton, A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr. Protoc. Cell Biol. 2006, 30, 3.22.1–3.22.29. [Google Scholar] [CrossRef]
- Botha, J.; Handberg, A.; Simonsen, J.B. Lipid-based strategies used to identify extracellular vesicles in flow cytometry can be confounded by lipoproteins: Evaluations of annexin V, lactadherin, and detergent lysis. J. Extracell. Vesicles 2022, 11, e12200. [Google Scholar] [CrossRef]
- Théry, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef]
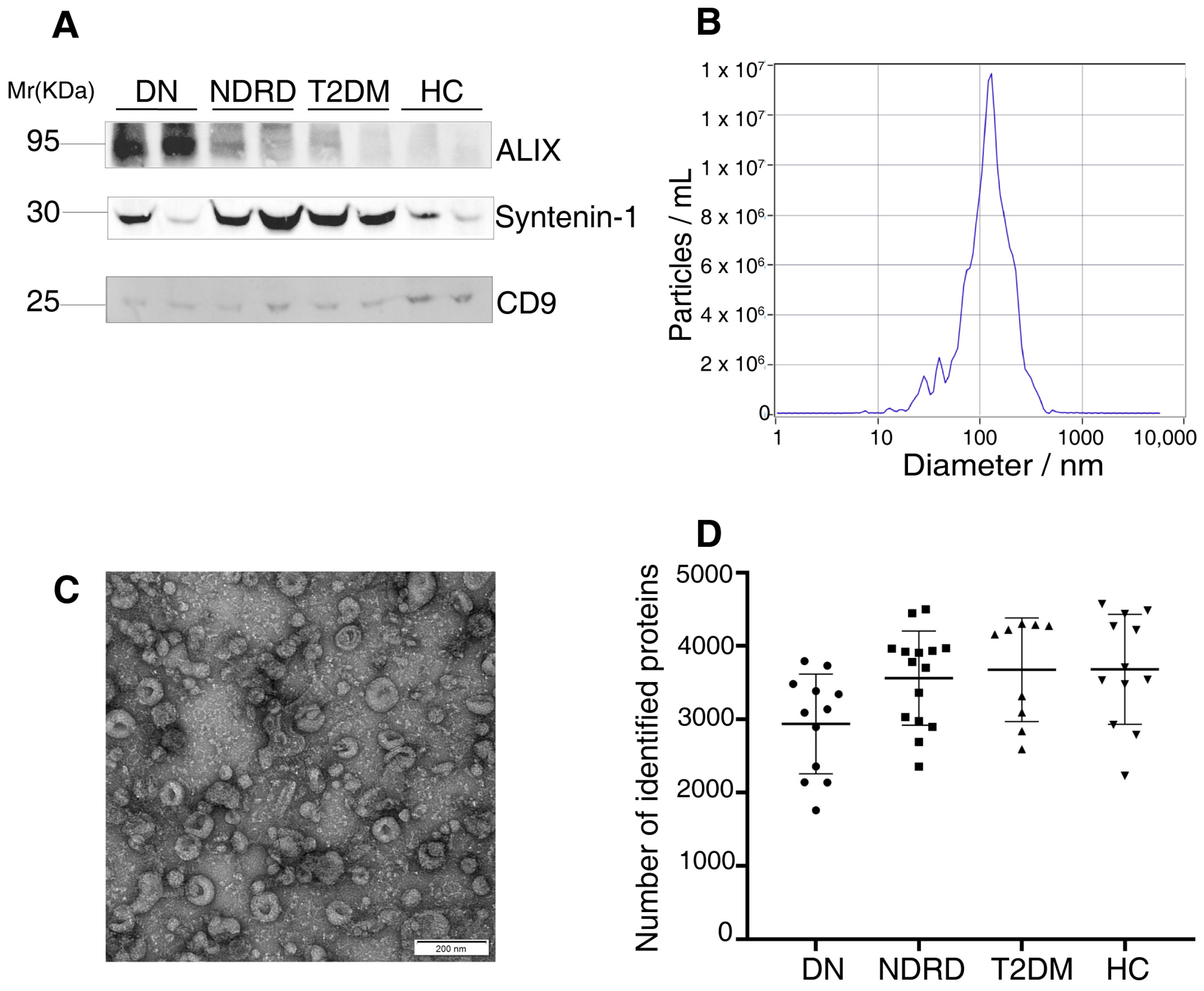
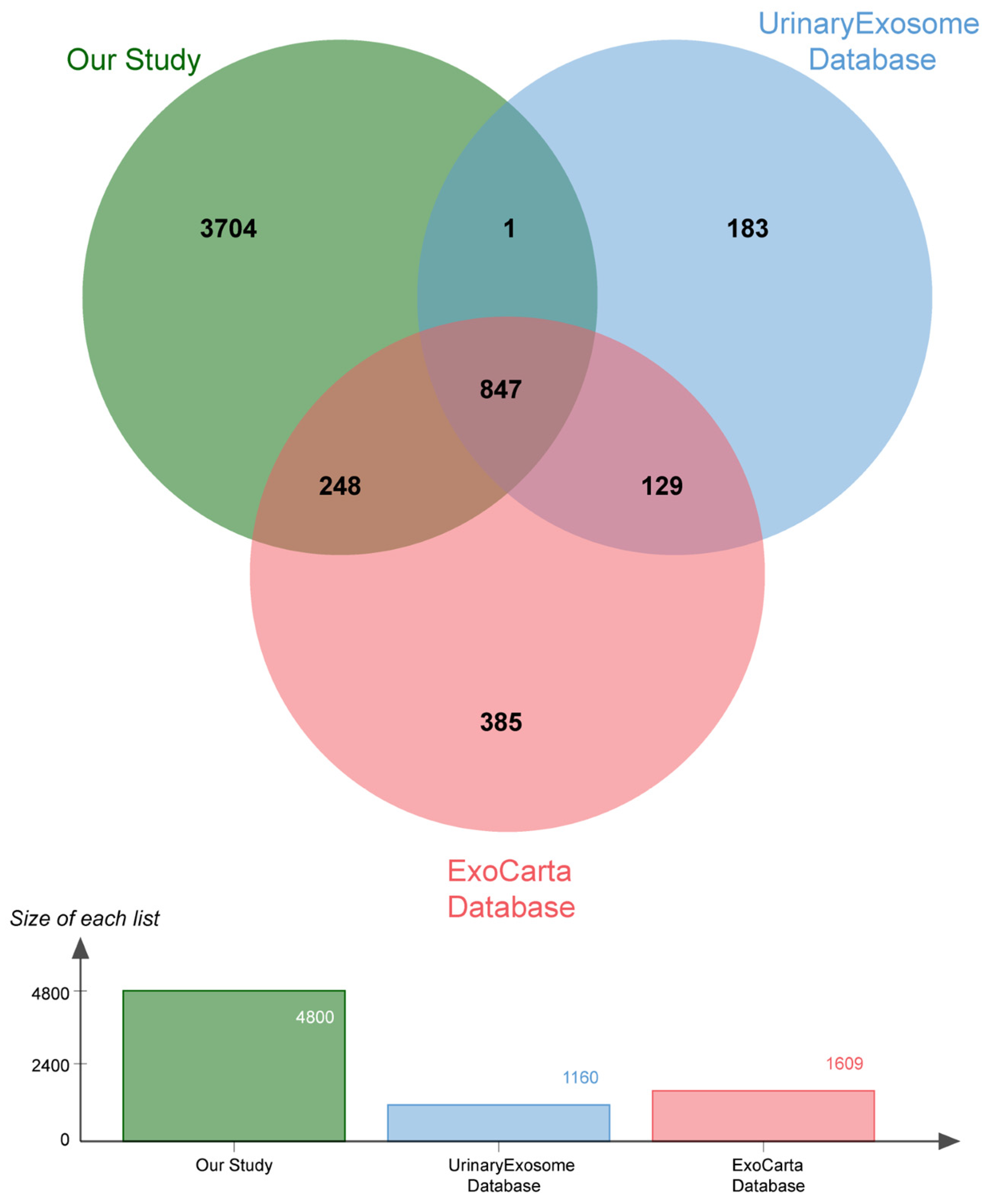
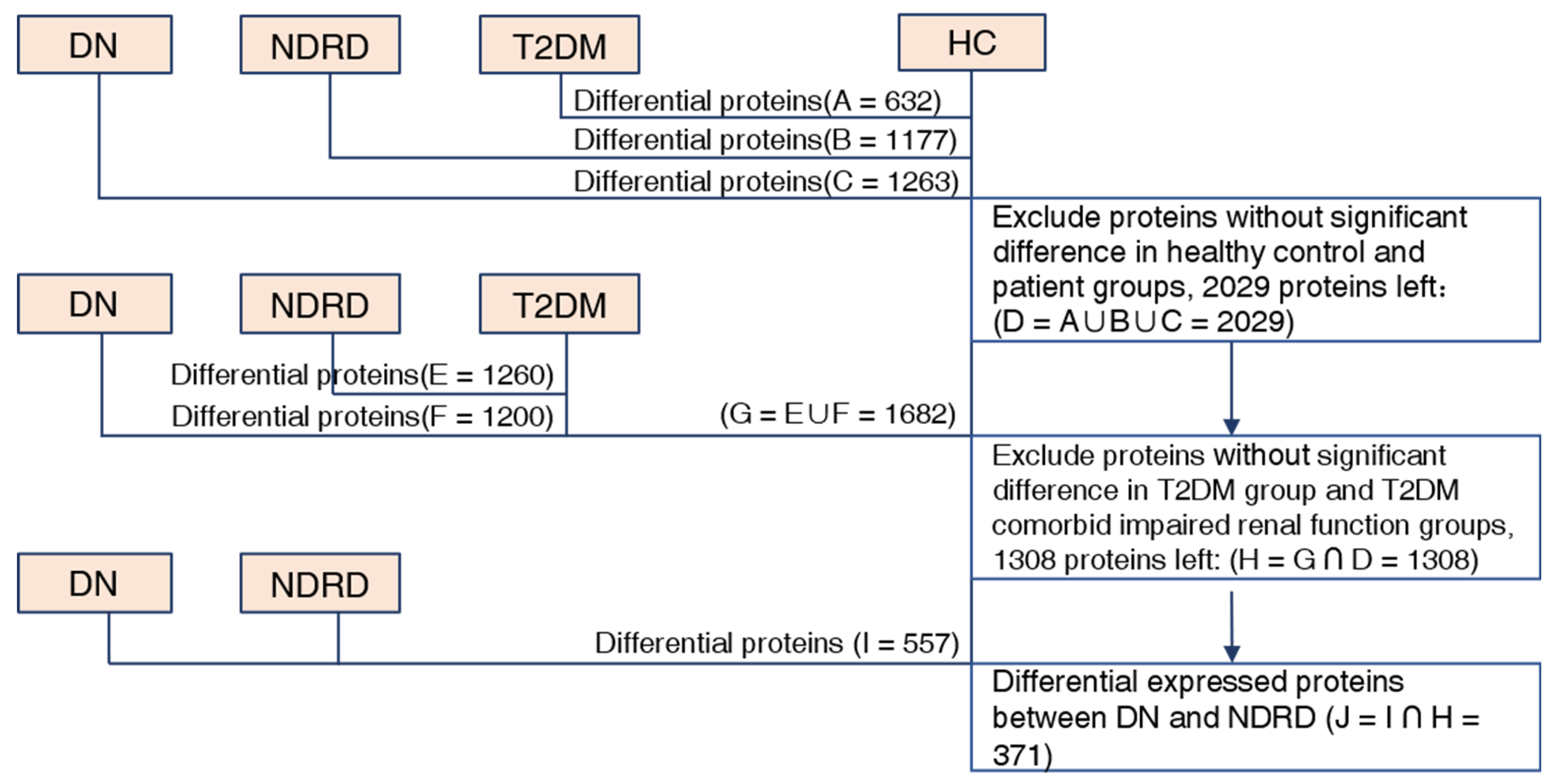
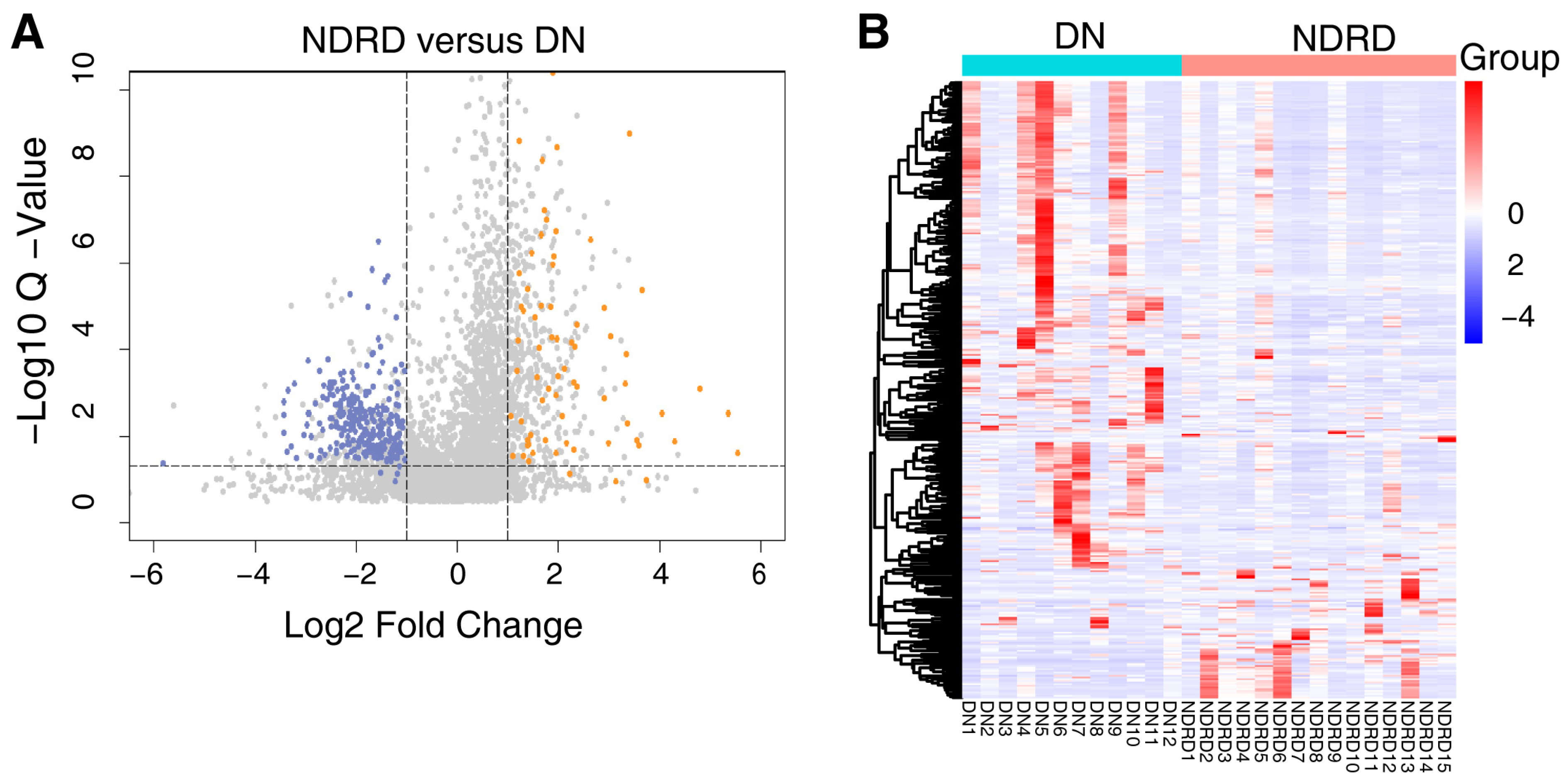

| DN (n = 12) | NDRD (n = 15) | T2DM (n = 9) | HC (n = 12) | |
|---|---|---|---|---|
| Age (years) | 49.08 ± 3.21 | 51.29 ± 2.50 | 60.11 ± 3.09 a | 47.83 ± 1.90 |
| Sex (male/female) | (6/6) | (12/3) | (4/5) | (8/4) |
| Serum albumin (g/L) | 32.24 ± 2.75 ab | 31.16 ± 3.82 a | 42.43 ± 1.04 | 44.35 ± 1.06 |
| HbA1c (%) | 7.54 ± 0.53 a | 6.3 ± 0.27 b | 9.56 ± 0.62 a | 5.44 ± 0.069 |
| Plasma glucose (mmol/L) | 8.20 ± 1.63 a | 5.42 ± 0.29 | 10.33 ± 1.57 a | 5.07 ± 0.14 |
| Urinary albumin (g/24 h) | 2.42 ± 0.59 b | 1.77 ± 0.50 b | 0.04 ± 0.03 | |
| Serum creatinine (μmol/L) | 169.10 ± 16.73 ab | 107.53 ± 18.32 ab | 69.01 ± 7.01 | 64.77 ± 3.64 |
| BUN (mmol/L) | 10.08 ± 1.47 ab | 6.15 ± 0.66 a | 5.18 ± 0.58 | 4.38 ± 0.17 |
| eGFR (mL min−1 1.73 m−2) | 32.23 ± 3.46 b | 68.41 ± 14.44 | 120.34 ± 10.96 |
| Number of Differential Proteins | Upregulated Proteins | Downregulated Proteins | |
|---|---|---|---|
| HC vs. DN | 1263 | 497 | 766 |
| HC vs. NDRD | 1177 | 508 | 669 |
| HC vs. T2DM | 632 | 462 | 170 |
| T2DM vs. DN | 1200 | 337 | 863 |
| T2DM vs. NDRD | 1260 | 347 | 913 |
| NDRD vs. DN | 557 | 156 | 401 |
| Protein Names | Protein Descriptions | DN vs. NDRD | ||
|---|---|---|---|---|
| Genes | AVG Log2 Ratio | Q-Value | ||
| PEPC | Gastricsin | PGC | 5.45 | 0.01 |
| PLCL1 | Inactive phospholipase C-like protein 1 | PLCL1 | 5.20 | 0.01 |
| PIP | Prolactin-inducible protein | PIP | 4.84 | <0.01 |
| SEMG2 | Semenogelin-2 | SEMG2 | 4.15 | 0.01 |
| SEMG1 | Semenogelin-1 | SEMG1 | 3.39 | <0.01 |
| GTR14 | Solute carrier family 2, facilitated glucose transporter member 14 | SLC2A14 | 3.32 | 0.01 |
| PLA1A | Phospholipase A1 member A | PLA1A | 3.13 | 0.03 |
| ILDR1 | Immunoglobulin-like domain-containing receptor 1 | ILDR1 | 2.65 | 0.03 |
| CRCT1 | Cysteine-rich C-terminal protein 1 | CRCT1 | 2.35 | <0.01 |
| SIDT1 | SID1 transmembrane family member 1 | SIDT1 | 2.29 | <0.01 |
| PERM | Myeloperoxidase | MPO | −3.50 | 0.02 |
| FIG4 | Polyphosphoinositide phosphatase | FIG4 | −3.38 | <0.01 |
| M4K4 | Mitogen-activated protein kinase kinase 4 | MAP4K4 | −3.34 | 0.03 |
| TETN | Tetranectin | CLEC3B | −3.26 | 0.02 |
| LV746 | Immunoglobulin lambda variable 7-46 | IGLV7-46 | −3.22 | <0.01 |
| PKP3 | Plakophilin-3 | PKP3 | −3.13 | 0.02 |
| GNTK | Probable glucokinase | IDNK | −3.09 | 0.04 |
| H3PS2 | Histone HIST2H3PS2 | H3-2 | −3.06 | 0.04 |
| PERE | Eosinophil peroxidase | EPX | −3.04 | 0.02 |
| SP100 | Nuclear autoantigen Sp-100 | SP100 | −3.03 | 0.01 |
| Variable | DN (n = 8) | NDRD (n = 5) | T2DM (n = 7) | HC (n = 9) |
|---|---|---|---|---|
| Age (years) | 57.50 ± 7.71 | 58.00 ± 5.00 | 54.29 ± 11.64 | 47.67 ± 6.00 |
| Sex (male/female) | (5/3) | (3/2) | (4/3) | (5/4) |
| Serum albumin (g/L) | 34.20 ± 7.47 a | 34.92 ± 12.43 | 42.49 ± 4.20 | 45.03 ± 2.55 |
| HbA1c (%) | 7.39 ± 1.60 a | 6.62 ± 0.99 | 9.57 ± 2.32 a | 5.43 ± 0.24 |
| Plasma glucose (mmol/L) | 6.99 ± 2.21 | 6.50 ± 2.88 | 11.21 ± 5.24 a | 5.09 ± 0.36 |
| Urinary albumin (g/24 h) | 2.71 ± 1.98 ab | 2.83 ± 4.11 a | 0.02 ± 0.05 | |
| Serum creatinine (μmol/L) | 150.44 ± 53.27 ab | 114.40 ± 35.06 | 71.94 ± 23.20 | 65.89 ± 10.67 |
| BUN (mmol/L) | 9.51 ± 2.53 ab | 7.97 ± 1.89 | 4.90 ± 1.80 | 4.67 ± 0.96 |
| eGFR (mL min−1 1.73 m−2) | 39.85 ± 17.76 b | 50.48 ± 17.91 | 97.55 ± 25.17 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ding, X.; Zhang, D.; Ren, Q.; Hu, Y.; Wang, J.; Hao, J.; Wang, H.; Zhao, X.; Wang, X.; Song, C.; et al. Identification of a Non-Invasive Urinary Exosomal Biomarker for Diabetic Nephropathy Using Data-Independent Acquisition Proteomics. Int. J. Mol. Sci. 2023, 24, 13560. https://doi.org/10.3390/ijms241713560
Ding X, Zhang D, Ren Q, Hu Y, Wang J, Hao J, Wang H, Zhao X, Wang X, Song C, et al. Identification of a Non-Invasive Urinary Exosomal Biomarker for Diabetic Nephropathy Using Data-Independent Acquisition Proteomics. International Journal of Molecular Sciences. 2023; 24(17):13560. https://doi.org/10.3390/ijms241713560
Chicago/Turabian StyleDing, Xiaonan, Dong Zhang, Qinqin Ren, Yilan Hu, Jifeng Wang, Jing Hao, Haoran Wang, Xiaolin Zhao, Xiaochen Wang, Chenwen Song, and et al. 2023. "Identification of a Non-Invasive Urinary Exosomal Biomarker for Diabetic Nephropathy Using Data-Independent Acquisition Proteomics" International Journal of Molecular Sciences 24, no. 17: 13560. https://doi.org/10.3390/ijms241713560
APA StyleDing, X., Zhang, D., Ren, Q., Hu, Y., Wang, J., Hao, J., Wang, H., Zhao, X., Wang, X., Song, C., Du, J., Yang, F., & Zhu, H. (2023). Identification of a Non-Invasive Urinary Exosomal Biomarker for Diabetic Nephropathy Using Data-Independent Acquisition Proteomics. International Journal of Molecular Sciences, 24(17), 13560. https://doi.org/10.3390/ijms241713560







