Copy Number Variations in Neuropsychiatric Disorders
Abstract
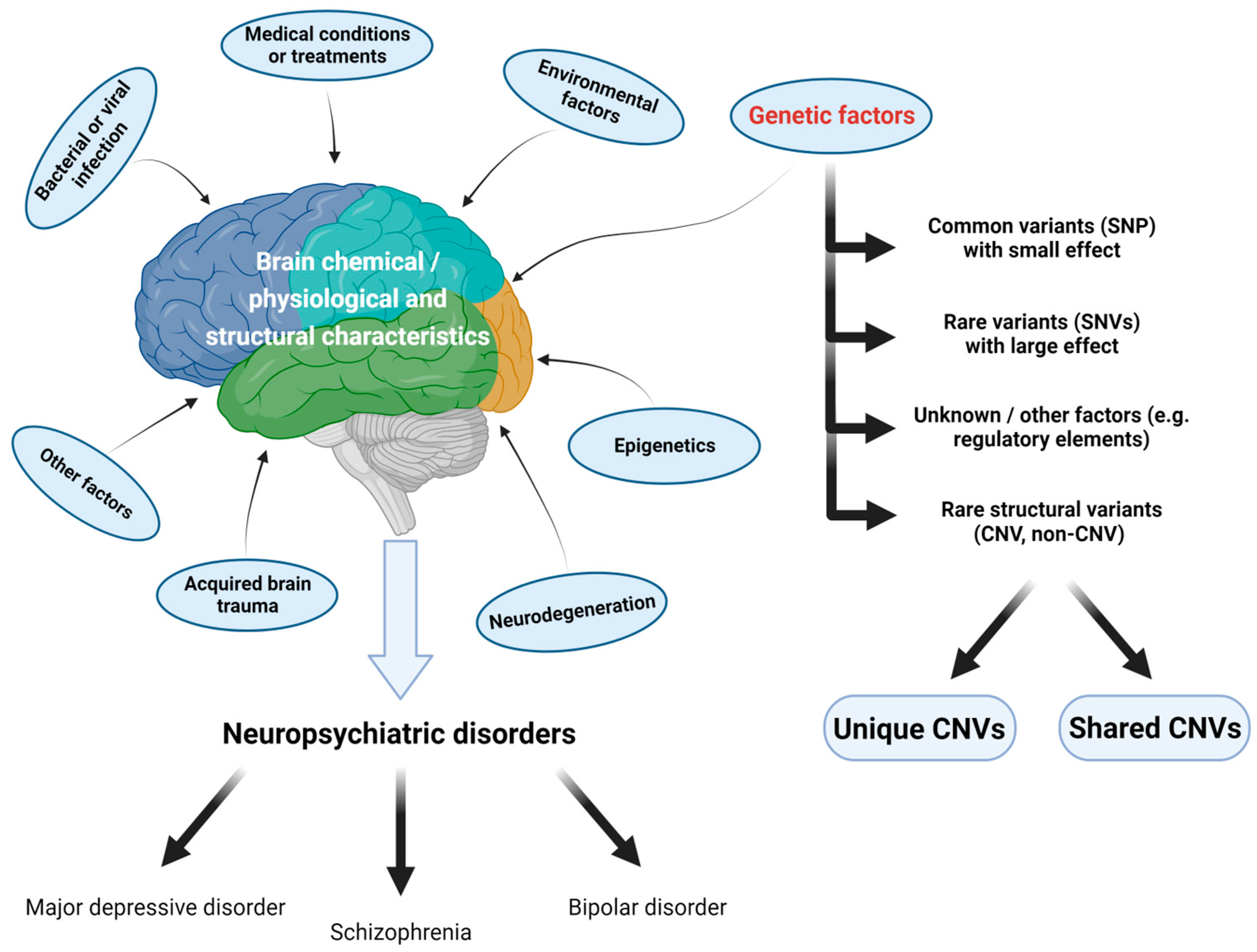
1. Introduction
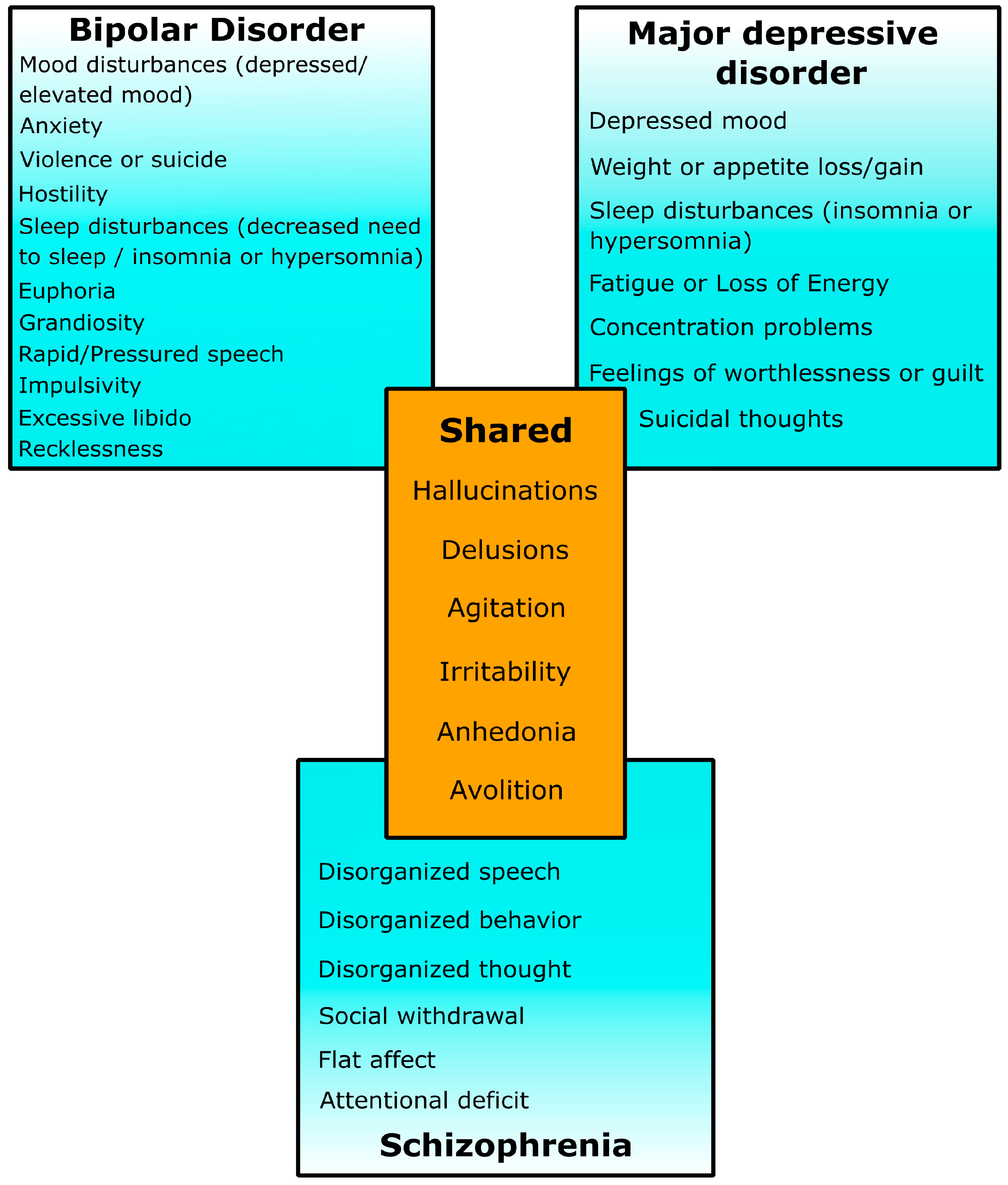
2. CNVs in Schizophrenia
3. CNVs in Bipolar Disorder
4. CNVs in Major Depressive Disorder
5. Tools for Detection and Investigation of the Biological Significance of CNVs
6. Biological Impacts of CNVs
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bray, N.J.; O’Donovan, M.C. The genetics of neuropsychiatric disorders. Brain Neurosci. Adv. 2019, 2, 2398212818799271. [Google Scholar] [CrossRef] [PubMed]
- McGuffin, P.; Rijsdijk, F.; Andrew, M.; Sham, P.; Katz, R.; Cardno, A. The heritability of bipolar affective disorder and the genetic relationship to unipolar depression. Arch. Gen. Psychiatry 2003, 60, 497–502. [Google Scholar] [CrossRef]
- Rietveld, M.J.; Hudziak, J.J.; Bartels, M.; van Beijsterveldt, C.E.; Boomsma, D.I. Heritability of attention problems in children: I. cross-sectional results from a study of twins, age 3-12 years. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2003, 117B, 102–113. [Google Scholar] [CrossRef]
- Sullivan, P.F.; Kendler, K.S.; Neale, M.C. Schizophrenia as a complex trait: Evidence from a meta-analysis of twin studies. Arch. Gen. Psychiatry 2003, 60, 1187–1192. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, P.F.; Neale, M.C.; Kendler, K.S. Genetic epidemiology of major depression: Review and meta-analysis. Am. J. Psychiatry 2000, 157, 1552–1562. [Google Scholar] [CrossRef]
- Schizophrenia Working Group of the Psychiatric Genomics C. Biological insights from 108 schizophrenia-associated genetic loci. Nature 2014, 511, 421–427. [Google Scholar] [CrossRef]
- Pardinas, A.F.; Holmans, P.; Pocklington, A.J.; Escott-Price, V.; Ripke, S.; Carrera, N.; Legge, S.E.; Bishop, S.; Cameron, D.; Hamshere, M.L.; et al. Common schizophrenia alleles are enriched in mutation-intolerant genes and in regions under strong background selection. Nat. Genet. 2018, 50, 381–389. [Google Scholar] [CrossRef] [PubMed]
- Mokhtari, M.A.; Sargazi, S.; Saravani, R.; Heidari Nia, M.; Mirinejad, S.; Hadzsiev, K.; Bene, J.; Shakiba, M. Genetic Polymorphisms in miR-137 and Its Target Genes, TCF4 and CACNA1C, Contribute to the Risk of Bipolar Disorder: A Preliminary Case-Control Study and Bioinformatics Analysis. Dis. Markers 2022, 2022, 1886658. [Google Scholar] [CrossRef]
- Doherty, J.L.; Owen, M.J. Genomic insights into the overlap between psychiatric disorders: Implications for research and clinical practice. Genome Med. 2014, 6, 29. [Google Scholar] [CrossRef]
- Yuen, R.K.C.; Merico, D.; Bookman, M.; LHowe, J.; Thiruvahindrapuram, B.; Patel, R.V.; Whitney, J.; Deflaux, N.; Bingham, J.; Wang, Z.; et al. Whole genome sequencing resource identifies 18 new candidate genes for autism spectrum disorder. Nat. Neurosci. 2017, 20, 602–611. [Google Scholar] [CrossRef]
- Marshall, C.R.; Howrigan, D.P.; Merico, D.; Thiruvahindrapuram, B.; Wu, W.; Greer, D.S.; Antaki, D.; Shetty, A.; Holmans, P.A.; Pinto, D.; et al. Contribution of copy number variants to schizophrenia from a genome-wide study of 41,321 subjects. Nat. Genet. 2017, 49, 27–35. [Google Scholar] [CrossRef]
- Feuk, L.; Carson, A.R.; Scherer, S.W. Structural variation in the human genome. Nat. Rev. Genet. 2006, 7, 85–97. [Google Scholar] [CrossRef] [PubMed]
- Rees, E.; Kirov, G. Copy number variation and neuropsychiatric illness. Curr. Opin. Genet. Dev. 2021, 68, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Vacic, V.; McCarthy, S.; Malhotra, D.; Murray, F.; Chou, H.H.; Peoples, A.; Makarov, V.; Yoon, S.; Bhandari, A.; Corominas, R.; et al. Duplications of the neuropeptide receptor gene VIPR2 confer significant risk for schizophrenia. Nature 2011, 471, 499–503. [Google Scholar] [CrossRef]
- van Ommen, G.J. Frequency of new copy number variation in humans. Nat. Genet. 2005, 37, 333–334. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.A.; Carvalho, C.M.; Lupski, J.R. A DNA replication mechanism for generating nonrecurrent rearrangements associated with genomic disorders. Cell 2007, 131, 1235–1247. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Gu, W.; Hurles, M.E.; Lupski, J.R. Copy number variation in human health, disease, and evolution. Annu. Rev. Genom. Hum. Genet. 2009, 10, 451–481. [Google Scholar] [CrossRef]
- Harel, T.; Lupski, J.R. Genomic disorders 20 years on-mechanisms for clinical manifestations. Clin. Genet. 2018, 93, 439–449. [Google Scholar] [CrossRef]
- Szecowka, K.; Misiak, B.; Laczmanska, I.; Frydecka, D.; Moustafa, A.A. Copy Number Variations and Schizophrenia. Mol. Neurobiol. 2023, 60, 1854–1864. [Google Scholar] [CrossRef]
- Itsara, A.; Cooper, G.M.; Baker, C.; Girirajan, S.; Li, J.; Absher, D.; Krauss, R.M.; Myers, R.M.; Ridker, P.M.; Chasman, D.I.; et al. Population analysis of large copy number variants and hotspots of human genetic disease. Am. J. Hum. Genet. 2009, 84, 148–161. [Google Scholar] [CrossRef]
- Rice, A.M.; McLysaght, A. Dosage sensitivity is a major determinant of human copy number variant pathogenicity. Nat. Commun. 2017, 8, 14366. [Google Scholar] [CrossRef]
- Manolio, T.A.; Collins, F.S.; Cox, N.J.; Goldstein, D.B.; Hindorff, L.A.; Hunter, D.J.; McCarthy, M.I.; Ramos, E.M.; Cardon, L.R.; Chakravarti, A.; et al. Finding the missing heritability of complex diseases. Nature 2009, 461, 747–753. [Google Scholar] [CrossRef] [PubMed]
- Nakatochi, M.; Kushima, I.; Ozaki, N. Implications of germline copy-number variations in psychiatric disorders: Review of large-scale genetic studies. J. Hum. Genet. 2021, 66, 25–37. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Wang, Y.; Zheng, X.; Sheng, J.; Guo, H.; Long, W.; Xu, Y. Quantitative evaluation of PTPN22 copy number variation by digital droplet PCR and association with type 2 diabetes risk. Endocr. J. 2021, 68, 153–162. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Smith, C.E.; Parnell, L.D.; Lee, Y.C.; An, P.; Straka, R.J.; Tiwari, H.K.; Wood, A.C.; Kabagambe, E.K.; Hidalgo, B.; et al. Salivary AMY1 Copy Number Variation Modifies Age-Related Type 2 Diabetes Risk. Clin. Chem. 2020, 66, 718–726. [Google Scholar] [CrossRef] [PubMed]
- Dajani, R.; Li, J.; Wei, Z.; Glessner, J.T.; Chang, X.; Cardinale, C.J.; Pellegrino, R.; Wang, T.; Hakooz, N.; Khader, Y.; et al. CNV Analysis Associates AKNAD1 with Type-2 Diabetes in Jordan Subpopulations. Sci. Rep. 2015, 5, 13391. [Google Scholar] [CrossRef]
- Van, L.; Heung, T.; Malecki, S.L.; Fenn, C.; Tyrer, A.; Sanches, M.; Chow, E.W.C.; Boot, E.; Corral, M.; Dash, S.; et al. 22q11.2 microdeletion and increased risk for type 2 diabetes. EClinicalMedicine 2020, 26, 100528. [Google Scholar] [CrossRef] [PubMed]
- Glessner, J.T.; Li, J.; Desai, A.; Palmer, M.; Kim, D.; Lucas, A.M.; Chang, X.; Connolly, J.J.; Almoguera, B.; Harley, J.B.; et al. CNV Association of Diverse Clinical Phenotypes from eMERGE reveals novel disease biology underlying cardiovascular disease. Int. J. Cardiol. 2020, 298, 107–113. [Google Scholar] [CrossRef]
- Han, J.; Walters, J.T.; Kirov, G.; Pocklington, A.; Escott-Price, V.; Owen, M.J.; Holmans, P.; O’Donovan, M.C.; Rees, E. Gender differences in CNV burden do not confound schizophrenia CNV associations. Sci. Rep. 2016, 6, 25986. [Google Scholar] [CrossRef]
- American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders, 5th ed.; American Psychiatric Publishing, Inc.: Washington, DC, USA, 2013. [Google Scholar]
- Foley, C.; Heron, E.A.; Harold, D.; Walters, J.; Owen, M.; O’Donovan, M.; Sebat, J.; Kelleher, E.; Mooney, C.; Durand, A.; et al. Identifying schizophrenia patients who carry pathogenic genetic copy number variants using standard clinical assessment: Retrospective cohort study. Br. J. Psychiatry 2020, 216, 275–279. [Google Scholar] [CrossRef]
- Ahn, K.; Gotay, N.; Andersen, T.M.; Anvari, A.A.; Gochman, P.; Lee, Y.; Sanders, S.; Guha, S.; Darvasi, A.; Glessner, J.T.; et al. High rate of disease-related copy number variations in childhood onset schizophrenia. Mol. Psychiatry 2014, 19, 568–572. [Google Scholar] [CrossRef] [PubMed]
- Kahn, R.S.; Sommer, I.E.; Murray, R.M.; Meyer-Lindenberg, A.; Weinberger, D.R.; Cannon, T.D.; O’Donovan, M.; Correll, C.U.; Kane, J.M.; van Os, J.; et al. Schizophrenia. Nat. Rev. Dis. Primers 2015, 1, 15067. [Google Scholar] [CrossRef]
- Propping, P. Genetic disorders presenting as “schizophrenia”. Karl Bonhoeffer’s early view of the psychoses in the light of medical genetics. Hum. Genet. 1983, 65, 1–10. [Google Scholar] [CrossRef] [PubMed]
- deLisi, L.E.; Lovett, M. The Role of Molecular Genetics in Psychiatry: Unraveling the Etiology for Schizophrenia. In Recent Advances in Schizophrenia; Springer: New York, NY, USA, 1990; pp. 131–161. [Google Scholar]
- Gottesman, I.I.; Shields, J. Schizophrenia: The Epigenetic Puzzle. In Psychological Medicine; Roberts, D., Ed.; Cambridge University Press: Cambridge UK, 1983; pp. 690–692. [Google Scholar]
- Lowing, P.A.; Mirsky, A.F.; Pereira, R. The inheritance of schizophrenia spectrum disorders: A reanalysis of the Danish adoptee study data. Am. J. Psychiatry 1983, 140, 1167–1171. [Google Scholar]
- Kendler, K.S.; Gruenberg, A.M.; Tsuang, M.T. Psychiatric illness in first-degree relatives of schizophrenic and surgical control patients. A family study using DSM-III criteria. Arch. Gen. Psychiatry 1985, 42, 770–779. [Google Scholar] [CrossRef]
- Pulver, A.E.; Nestadt, G.; Goldberg, R.; Shprintzen, R.J.; Lamacz, M.; Wolyniec, P.S.; Morrow, B.; Karayiorgou, M.; Antonarakis, S.E.; Housman, D.; et al. Psychotic illness in patients diagnosed with velo-cardio-facial syndrome and their relatives. J. Nerv. Ment. Dis. 1994, 182, 476–478. [Google Scholar] [CrossRef] [PubMed]
- Papolos, D.F.; Faedda, G.L.; Veit, S.; Goldberg, R.; Morrow, B.; Kucherlapati, R.; Shprintzen, R.J. Bipolar spectrum disorders in patients diagnosed with velo-cardio-facial syndrome: Does a hemizygous deletion of chromosome 22q11 result in bipolar affective disorder? Am. J. Psychiatry 1996, 153, 1541–1547. [Google Scholar] [PubMed]
- Karayiorgou, M.; Morris, M.A.; Morrow, B.; Shprintzen, R.J.; Goldberg, R.; Borrow, J.; Gos, A.; Nestadt, G.; Wolyniec, P.S.; Lasseter, V.K.; et al. Schizophrenia susceptibility associated with interstitial deletions of chromosome 22q11. Proc. Natl. Acad. Sci. USA 1995, 92, 7612–7616. [Google Scholar] [CrossRef]
- Gothelf, D.; Frisch, A.; Munitz, H.; Rockah, R.; Aviram, A.; Mozes, T.; Birger, M.; Weizman, A.; Frydman, M. Velocardiofacial manifestations and microdeletions in schizophrenic inpatients. Am. J. Med. Genet. 1997, 72, 455–461. [Google Scholar] [CrossRef]
- Bassett, A.S.; Hodgkinson, K.; Chow, E.W.; Correia, S.; Scutt, L.E.; Weksberg, R. 22q11 deletion syndrome in adults with schizophrenia. Am. J. Med. Genet. 1998, 81, 328–337. [Google Scholar] [CrossRef]
- Bassett, A.S.; Chow, E.W. Schizophrenia and 22q11.2 deletion syndrome. Curr. Psychiatry Rep. 2008, 10, 148–157. [Google Scholar] [CrossRef]
- Bassett, A.S.; Scherer, S.W.; Brzustowicz, L.M. Copy number variations in schizophrenia: Critical review and new perspectives on concepts of genetics and disease. Am. J. Psychiatry 2010, 167, 899–914. [Google Scholar] [CrossRef]
- Williams, H.J.; Owen, M.J.; O’Donovan, M.C. Is COMT a susceptibility gene for schizophrenia? Schizophr. Bull. 2007, 33, 635–641. [Google Scholar] [CrossRef]
- Bani-Fatemi, A.; Adanty, C.; Dai, N.; Graff, A.; Gerretsen, P.; De Luca, V. Chromosome 22 Deletions and Suicidal Behavior in Schizophrenia. Neuropsychobiology 2021, 80, 393–400. [Google Scholar] [CrossRef]
- Murphy, K.C.; Jones, L.A.; Owen, M.J. High rates of schizophrenia in adults with velo-cardio-facial syndrome. Arch. Gen. Psychiatry 1999, 56, 940–945. [Google Scholar] [CrossRef]
- Rees, E.; Kirov, G.; Sanders, A.; Walters, J.T.; Chambert, K.D.; Shi, J.; Szatkiewicz, J.; O’Dushlaine, C.; Richards, A.L.; Green, E.K.; et al. Evidence that duplications of 22q11.2 protect against schizophrenia. Mol. Psychiatry 2014, 19, 37–40. [Google Scholar] [CrossRef]
- International Schizophrenia Consortium. Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature 2008, 455, 237–241. [Google Scholar] [CrossRef]
- Rees, E.; Walters, J.T.; Chambert, K.D.; O’Dushlaine, C.; Szatkiewicz, J.; Richards, A.L.; Georgieva, L.; Mahoney-Davies, G.; Legge, S.E.; Moran, J.L.; et al. CNV analysis in a large schizophrenia sample implicates deletions at 16p12.1 and SLC1A1 and duplications at 1p36.33 and CGNL1. Hum. Mol. Genet. 2014, 23, 1669–1676. [Google Scholar] [CrossRef]
- Merikangas, A.K.; Segurado, R.; Cormican, P.; Heron, E.A.; Anney, R.J.; Moore, S.; Kelleher, E.; Hargreaves, A.; Anderson-Schmidt, H.; Gill, M.; et al. The phenotypic manifestations of rare CNVs in schizophrenia. Schizophr. Res. 2014, 158, 255–260. [Google Scholar] [CrossRef]
- Doherty, J.L.; O’Donovan, M.C.; Owen, M.J. Recent genomic advances in schizophrenia. Clin. Genet. 2012, 81, 103–109. [Google Scholar] [CrossRef]
- Levinson, D.F.; Duan, J.; Oh, S.; Wang, K.; Sanders, A.R.; Shi, J.; Zhang, N.; Mowry, B.J.; Olincy, A.; Amin, F.; et al. Copy number variants in schizophrenia: Confirmation of five previous findings and new evidence for 3q29 microdeletions and VIPR2 duplications. Am. J. Psychiatry 2011, 168, 302–316. [Google Scholar] [CrossRef]
- Jacquemont, S.; Coe, B.P.; Hersch, M.; Duyzend, M.H.; Krumm, N.; Bergmann, S.; Beckmann, J.S.; Rosenfeld, J.A.; Eichler, E.E. A higher mutational burden in females supports a “female protective model” in neurodevelopmental disorders. Am. J. Hum. Genet. 2014, 94, 415–425. [Google Scholar] [CrossRef]
- Rees, E.; Walters, J.T.; Georgieva, L.; Isles, A.R.; Chambert, K.D.; Richards, A.L.; Mahoney-Davies, G.; Legge, S.E.; Moran, J.L.; McCarroll, S.A.; et al. Analysis of copy number variations at 15 schizophrenia-associated loci. Br. J. Psychiatry 2014, 204, 108–114. [Google Scholar] [CrossRef]
- Xu, B.; Roos, J.L.; Levy, S.; van Rensburg, E.J.; Gogos, J.A.; Karayiorgou, M. Strong association of de novo copy number mutations with sporadic schizophrenia. Nat. Genet. 2008, 40, 880–885. [Google Scholar] [CrossRef]
- Kirov, G.; Pocklington, A.J.; Holmans, P.; Ivanov, D.; Ikeda, M.; Ruderfer, D.; Moran, J.; Chambert, K.; Toncheva, D.; Georgieva, L.; et al. De novo CNV analysis implicates specific abnormalities of postsynaptic signalling complexes in the pathogenesis of schizophrenia. Mol. Psychiatry 2012, 17, 142–153. [Google Scholar] [CrossRef]
- Gulsuner, S.; McClellan, J.M. Copy number variation in schizophrenia. Neuropsychopharmacology 2015, 40, 252–254. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Malhotra, D.; Sebat, J. CNVs: Harbingers of a rare variant revolution in psychiatric genetics. Cell 2012, 148, 1223–1241. [Google Scholar] [CrossRef]
- Morris, D.W.; Pearson, R.D.; Cormican, P.; Kenny, E.M.; O’Dushlaine, C.T.; Perreault, L.P.; Giannoulatou, E.; Tropea, D.; Maher, B.S.; Wormley, B.; et al. An inherited duplication at the gene p21 Protein-Activated Kinase 7 (PAK7) is a risk factor for psychosis. Hum. Mol. Genet. 2014, 23, 3316–3326. [Google Scholar] [CrossRef] [PubMed]
- Calle Sanchez, X.; Helenius, D.; Bybjerg-Grauholm, J.; Pedersen, C.; Hougaard, D.M.; Borglum, A.D.; Nordentoft, M.; Mors, O.; Mortensen, P.B.; Geschwind, D.H.; et al. Comparing Copy Number Variations in a Danish Case Cohort of Individuals With Psychiatric Disorders. JAMA Psychiatry 2022, 79, 59–69. [Google Scholar] [CrossRef]
- Kendall, K.M.; Rees, E.; Bracher-Smith, M.; Legge, S.; Riglin, L.; Zammit, S.; O’Donovan, M.C.; Owen, M.J.; Jones, I.; Kirov, G.; et al. Association of Rare Copy Number Variants With Risk of Depression. JAMA Psychiatry 2019, 76, 818–825. [Google Scholar] [CrossRef]
- Green, E.K.; Rees, E.; Walters, J.T.; Smith, K.G.; Forty, L.; Grozeva, D.; Moran, J.L.; Sklar, P.; Ripke, S.; Chambert, K.D.; et al. Copy number variation in bipolar disorder. Mol. Psychiatry 2016, 21, 89–93. [Google Scholar] [CrossRef]
- Sonderby, I.E.; Gustafsson, O.; Doan, N.T.; Hibar, D.P.; Martin-Brevet, S.; Abdellaoui, A.; Ames, D.; Amunts, K.; Andersson, M.; Armstrong, N.J.; et al. Dose response of the 16p11.2 distal copy number variant on intracranial volume and basal ganglia. Mol. Psychiatry 2020, 25, 584–602. [Google Scholar] [CrossRef] [PubMed]
- Endres, D.; Maier, S.J.; Ziegler, C.; Nickel, K.; Riering, A.N.; Berger, B.; Lambeck, J.; Fritz, M.; Glaser, B.; Stock, F.; et al. Schizophrenia and Hereditary Polyneuropathy: PMP22 Deletion as a Common Pathophysiological Link? Front. Psychiatry 2019, 10, 270. [Google Scholar] [CrossRef]
- Moreno-De-Luca, D.; Mulle, J.G.; Simons Simplex Collection Genetics Consortium; Kaminsky, E.B.; Sanders, S.J.; Myers, S.M.; Adam, M.P.; Pakula, A.T.; Eisenhauer, N.J.; Uhas, K.; et al. Deletion 17q12 is a recurrent copy number variant that confers high risk of autism and schizophrenia. Am. J. Hum. Genet. 2010, 87, 618–630. [Google Scholar] [CrossRef]
- Hubbard, L.; Rees, E.; Morris, D.W.; Lynham, A.J.; Richards, A.L.; Pardinas, A.F.; Legge, S.E.; Harold, D.; Zammit, S.; Corvin, A.C.; et al. Rare Copy Number Variants Are Associated With Poorer Cognition in Schizophrenia. Biol. Psychiatry 2021, 90, 28–34. [Google Scholar] [CrossRef]
- Yeo, R.A.; Gangestad, S.W.; Liu, J.; Ehrlich, S.; Thoma, R.J.; Pommy, J.; Mayer, A.R.; Schulz, S.C.; Wassink, T.H.; Morrow, E.M.; et al. The impact of copy number deletions on general cognitive ability and ventricle size in patients with schizophrenia and healthy control subjects. Biol. Psychiatry 2013, 73, 540–545. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Dolcetti, A.; Silversides, C.K.; Marshall, C.R.; Lionel, A.C.; Stavropoulos, D.J.; Scherer, S.W.; Bassett, A.S. 1q21.1 Microduplication expression in adults. Genet. Med. 2013, 15, 282–289. [Google Scholar] [CrossRef]
- Isles, A.R.; Ingason, A.; Lowther, C.; Walters, J.; Gawlick, M.; Stober, G.; Rees, E.; Martin, J.; Little, R.B.; Potter, H.; et al. Parental Origin of Interstitial Duplications at 15q11.2-q13.3 in Schizophrenia and Neurodevelopmental Disorders. PLoS Genet. 2016, 12, e1005993. [Google Scholar] [CrossRef] [PubMed]
- Thygesen, J.H.; Wolfe, K.; McQuillin, A.; Vinas-Jornet, M.; Baena, N.; Brison, N.; D’Haenens, G.; Esteba-Castillo, S.; Gabau, E.; Ribas-Vidal, N.; et al. Neurodevelopmental risk copy number variants in adults with intellectual disabilities and comorbid psychiatric disorders. Br. J. Psychiatry 2018, 212, 287–294. [Google Scholar] [CrossRef]
- Lowther, C.; Merico, D.; Costain, G.; Waserman, J.; Boyd, K.; Noor, A.; Speevak, M.; Stavropoulos, D.J.; Wei, J.; Lionel, A.C.; et al. Impact of IQ on the diagnostic yield of chromosomal microarray in a community sample of adults with schizophrenia. Genome Med. 2017, 9, 105. [Google Scholar] [CrossRef]
- Derks, E.M.; Ayub, M.; Chambert, K.; Del Favero, J.; Johnstone, M.; MacGregor, S.; Maclean, A.; McKechanie, A.G.; McRae, A.F.; Moran, J.L.; et al. A genome wide survey supports the involvement of large copy number variants in schizophrenia with and without intellectual disability. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2013, 162B, 847–854. [Google Scholar] [CrossRef]
- Rees, E.; Kendall, K.; Pardinas, A.F.; Legge, S.E.; Pocklington, A.; Escott-Price, V.; MacCabe, J.H.; Collier, D.A.; Holmans, P.; O’Donovan, M.C.; et al. Analysis of Intellectual Disability Copy Number Variants for Association With Schizophrenia. JAMA Psychiatry 2016, 73, 963–969. [Google Scholar] [CrossRef]
- McKenna, K.; Gordon, C.T.; Lenane, M.; Kaysen, D.; Fahey, K.; Rapoport, J.L. Looking for childhood-onset schizophrenia: The first 71 cases screened. J. Am. Acad. Child Adolesc. Psychiatry 1994, 33, 636–644. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, A.; Ziller, M.; Spengler, D. Childhood-Onset Schizophrenia: Insights from Induced Pluripotent Stem Cells. Int. J. Mol. Sci. 2018, 19, 3829. [Google Scholar] [CrossRef] [PubMed]
- Walsh, T.; McClellan, J.M.; McCarthy, S.E.; Addington, A.M.; Pierce, S.B.; Cooper, G.M.; Nord, A.S.; Kusenda, M.; Malhotra, D.; Bhandari, A.; et al. Rare structural variants disrupt multiple genes in neurodevelopmental pathways in schizophrenia. Science 2008, 320, 539–543. [Google Scholar] [CrossRef]
- Addington, A.M.; Rapoport, J.L. The genetics of childhood-onset schizophrenia: When madness strikes the prepubescent. Curr. Psychiatry Rep. 2009, 11, 156–161. [Google Scholar] [CrossRef]
- Olsen, L.; Sparso, T.; Weinsheimer, S.M.; Dos Santos, M.B.Q.; Mazin, W.; Rosengren, A.; Sanchez, X.C.; Hoeffding, L.K.; Schmock, H.; Baekvad-Hansen, M.; et al. Prevalence of rearrangements in the 22q11.2 region and population-based risk of neuropsychiatric and developmental disorders in a Danish population: A case-cohort study. Lancet Psychiatry 2018, 5, 573–580. [Google Scholar] [CrossRef]
- Kushima, I.; Aleksic, B.; Nakatochi, M.; Shimamura, T.; Okada, T.; Uno, Y.; Morikawa, M.; Ishizuka, K.; Shiino, T.; Kimura, H.; et al. Comparative Analyses of Copy-Number Variation in Autism Spectrum Disorder and Schizophrenia Reveal Etiological Overlap and Biological Insights. Cell Rep. 2018, 24, 2838–2856. [Google Scholar] [CrossRef]
- Kushima, I.; Aleksic, B.; Nakatochi, M.; Shimamura, T.; Shiino, T.; Yoshimi, A.; Kimura, H.; Takasaki, Y.; Wang, C.; Xing, J.; et al. High-resolution copy number variation analysis of schizophrenia in Japan. Mol. Psychiatry 2017, 22, 430–440. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Q.; Li, T.; Zhao, X.; Huang, K.; Wang, T.; Li, Z.; Ji, J.; Zeng, Z.; Zhang, Z.; Li, K.; et al. Rare CNVs and tag SNPs at 15q11.2 are associated with schizophrenia in the Han Chinese population. Schizophr. Bull. 2013, 39, 712–719. [Google Scholar] [CrossRef]
- Saxena, S.; Kkani, P.; Ramasubramanian, C.; Kumar, S.G.; Monisha, R.; Prasad Rao, G.; Mohan, K.N. Analysis of 15q11.2 CNVs in an Indian population with schizophrenia. Ann. Hum. Genet. 2019, 83, 187–191. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Chen, J.; Xu, Y.; Yi, Q.; Ji, W.; Wang, P.; Shen, J.; Song, Z.; Wang, M.; Yang, P.; et al. Genome-wide Analysis of the Role of Copy Number Variation in Schizophrenia Risk in Chinese. Biol. Psychiatry 2016, 80, 331–337. [Google Scholar] [CrossRef]
- Priebe, L.; Degenhardt, F.; Strohmaier, J.; Breuer, R.; Herms, S.; Witt, S.H.; Hoffmann, P.; Kulbida, R.; Mattheisen, M.; Moebus, S.; et al. Copy number variants in German patients with schizophrenia. PLoS ONE 2013, 8, e64035. [Google Scholar] [CrossRef] [PubMed]
- Merikangas, K.R.; Jin, R.; He, J.P.; Kessler, R.C.; Lee, S.; Sampson, N.A.; Viana, M.C.; Andrade, L.H.; Hu, C.; Karam, E.G.; et al. Prevalence and correlates of bipolar spectrum disorder in the world mental health survey initiative. Arch. Gen. Psychiatry 2011, 68, 241–251. [Google Scholar] [CrossRef]
- Rowland, T.A.; Marwaha, S. Epidemiology and risk factors for bipolar disorder. Ther. Adv. Psychopharmacol. 2018, 8, 251–269. [Google Scholar] [CrossRef] [PubMed]
- Burmeister, M.; McInnis, M.G.; Zollner, S. Psychiatric genetics: Progress amid controversy. Nat. Rev. Genet. 2008, 9, 527–540. [Google Scholar] [CrossRef]
- Lachman, H.M.; Pedrosa, E.; Petruolo, O.A.; Cockerham, M.; Papolos, A.; Novak, T.; Papolos, D.F.; Stopkova, P. Increase in GSK3beta gene copy number variation in bipolar disorder. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2007, 144B, 259–265. [Google Scholar] [CrossRef]
- Zhang, D.; Cheng, L.; Qian, Y.; Alliey-Rodriguez, N.; Kelsoe, J.R.; Greenwood, T.; Nievergelt, C.; Barrett, T.B.; McKinney, R.; Schork, N.; et al. Singleton deletions throughout the genome increase risk of bipolar disorder. Mol. Psychiatry 2009, 14, 376–380. [Google Scholar] [CrossRef]
- Malhotra, D.; McCarthy, S.; Michaelson, J.J.; Vacic, V.; Burdick, K.E.; Yoon, S.; Cichon, S.; Corvin, A.; Gary, S.; Gershon, E.S.; et al. High frequencies of de novo CNVs in bipolar disorder and schizophrenia. Neuron 2011, 72, 951–963. [Google Scholar] [CrossRef]
- Noor, A.; Lionel, A.C.; Cohen-Woods, S.; Moghimi, N.; Rucker, J.; Fennell, A.; Thiruvahindrapuram, B.; Kaufman, L.; Degagne, B.; Wei, J.; et al. Copy number variant study of bipolar disorder in Canadian and UK populations implicates synaptic genes. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2014, 165B, 303–313. [Google Scholar] [CrossRef]
- Georgieva, L.; Rees, E.; Moran, J.L.; Chambert, K.D.; Milanova, V.; Craddock, N.; Purcell, S.; Sklar, P.; McCarroll, S.; Holmans, P.; et al. De novo CNVs in bipolar affective disorder and schizophrenia. Hum. Mol. Genet. 2014, 23, 6677–6683. [Google Scholar] [CrossRef]
- Chen, J.; Calhoun, V.D.; Perrone-Bizzozero, N.I.; Pearlson, G.D.; Sui, J.; Du, Y.; Liu, J. A pilot study on commonality and specificity of copy number variants in schizophrenia and bipolar disorder. Transl. Psychiatry 2016, 6, e824. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Priebe, L.; Degenhardt, F.A.; Herms, S.; Haenisch, B.; Mattheisen, M.; Nieratschker, V.; Weingarten, M.; Witt, S.; Breuer, R.; Paul, T.; et al. Genome-wide survey implicates the influence of copy number variants (CNVs) in the development of early-onset bipolar disorder. Mol. Psychiatry 2012, 17, 421–432. [Google Scholar] [CrossRef] [PubMed]
- McQuillin, A.; Bass, N.; Anjorin, A.; Lawrence, J.; Kandaswamy, R.; Lydall, G.; Moran, J.; Sklar, P.; Purcell, S.; Gurling, H. Analysis of genetic deletions and duplications in the University College London bipolar disorder case control sample. Eur. J. Hum. Genet. 2011, 19, 588–592. [Google Scholar] [CrossRef]
- Chen, Y.H.; Lu, R.B.; Hung, H.; Kuo, P.H. Identifying Potential Regions of Copy Number Variation for Bipolar Disorder. Microarrays 2014, 3, 52–71. [Google Scholar] [CrossRef]
- Kessler, R.C.; Bromet, E.J. The epidemiology of depression across cultures. Annu. Rev. Public Health 2013, 34, 119–138. [Google Scholar] [CrossRef] [PubMed]
- Glessner, J.T.; Wang, K.; Sleiman, P.M.; Zhang, H.; Kim, C.E.; Flory, J.H.; Bradfield, J.P.; Imielinski, M.; Frackelton, E.C.; Qiu, H.; et al. Duplication of the SLIT3 locus on 5q35.1 predisposes to major depressive disorder. PLoS ONE 2010, 5, e15463. [Google Scholar] [CrossRef]
- Zhang, X.; Abdellaoui, A.; Rucker, J.; de Jong, S.; Potash, J.B.; Weissman, M.M.; Shi, J.; Knowles, J.A.; Pato, C.; Pato, M.; et al. Genome-wide Burden of Rare Short Deletions Is Enriched in Major Depressive Disorder in Four Cohorts. Biol. Psychiatry 2019, 85, 1065–1073. [Google Scholar] [CrossRef]
- O’Dushlaine, C.; Ripke, S.; Ruderfer, D.M.; Hamilton, S.P.; Fava, M.; Iosifescu, D.V.; Kohane, I.S.; Churchill, S.E.; Castro, V.M.; Clements, C.C.; et al. Rare copy number variation in treatment-resistant major depressive disorder. Biol. Psychiatry 2014, 76, 536–541. [Google Scholar] [CrossRef]
- Degenhardt, F.; Priebe, L.; Herms, S.; Mattheisen, M.; Muhleisen, T.W.; Meier, S.; Moebus, S.; Strohmaier, J.; Gross, M.; Breuer, R.; et al. Association between copy number variants in 16p11.2 and major depressive disorder in a German case-control sample. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2012, 159B, 263–273. [Google Scholar] [CrossRef] [PubMed]
- Gillentine, M.A.; Schaaf, C.P. The human clinical phenotypes of altered CHRNA7 copy number. Biochem. Pharmacol. 2015, 97, 352–362. [Google Scholar] [CrossRef]
- Gillentine, M.A.; Lozoya, R.; Yin, J.; Grochowski, C.M.; White, J.J.; Schaaf, C.P.; Calarge, C.A. CHRNA7 copy number gains are enriched in adolescents with major depressive and anxiety disorders. J. Affect. Disord. 2018, 239, 247–252. [Google Scholar] [CrossRef] [PubMed]
- Rao, S.; Shi, M.; Han, X.; Lam, M.H.B.; Chien, W.T.; Zhou, K.; Liu, G.; Wing, Y.K.; So, H.C.; Waye, M.M.Y. Genome-wide copy number variation-, validation- and screening study implicates a new copy number polymorphism associated with suicide attempts in major depressive disorder. Gene 2020, 755, 144901. [Google Scholar] [CrossRef] [PubMed]
- Tansey, K.E.; Rucker, J.J.; Kavanagh, D.H.; Guipponi, M.; Perroud, N.; Bondolfi, G.; Domenici, E.; Evans, D.M.; Hauser, J.; Henigsberg, N.; et al. Copy number variants and therapeutic response to antidepressant medication in major depressive disorder. Pharmacogenomics J. 2014, 14, 395–399. [Google Scholar] [CrossRef] [PubMed]
- Hupe, P.; Stransky, N.; Thiery, J.P.; Radvanyi, F.; Barillot, E. Analysis of array CGH data: From signal ratio to gain and loss of DNA regions. Bioinformatics 2004, 20, 3413–3422. [Google Scholar] [CrossRef]
- Wang, P.; Kim, Y.; Pollack, J.; Narasimhan, B.; Tibshirani, R. A method for calling gains and losses in array CGH data. Biostatistics 2005, 6, 45–58. [Google Scholar] [CrossRef]
- Picard, F.; Robin, S.; Lavielle, M.; Vaisse, C.; Daudin, J.J. A statistical approach for array CGH data analysis. BMC Bioinform. 2005, 6, 27. [Google Scholar] [CrossRef]
- Zhang, X.; Du, R.; Li, S.; Zhang, F.; Jin, L.; Wang, H. Evaluation of copy number variation detection for a SNP array platform. BMC Bioinform. 2014, 15, 50. [Google Scholar] [CrossRef]
- Wang, K.; Li, M.; Hadley, D.; Liu, R.; Glessner, J.; Grant, S.; Hakonarson, H.; Bucan, M. PennCNV: An integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Res. 2007, 17, 1665–1674. [Google Scholar] [CrossRef]
- Zhang, Z.; Cheng, H.; Hong, X.; Di Narzo, A.F.; Franzen, O.; Peng, S.; Ruusalepp, A.; Kovacic, J.C.; Bjorkegren, J.; Wang, X.; et al. EnsembleCNV: An ensemble machine learning algorithm to identify and genotype copy number variation using SNP array data. Nucleic Acids Res. 2019, 47, e39. [Google Scholar] [CrossRef]
- Forsingdal, A.; Jorgensen, T.N.; Olsen, L.; Werge, T.; Didriksen, M.; Nielsen, J. Can Animal Models of Copy Number Variants That Predispose to Schizophrenia Elucidate Underlying Biology? Biol. Psychiatry 2019, 85, 13–24. [Google Scholar] [CrossRef]
- Nakatani, J.; Tamada, K.; Hatanaka, F.; Ise, S.; Ohta, H.; Inoue, K.; Tomonaga, S.; Watanabe, Y.; Chung, Y.J.; Banerjee, R.; et al. Abnormal behavior in a chromosome-engineered mouse model for human 15q11-13 duplication seen in autism. Cell 2009, 137, 1235–1246. [Google Scholar] [CrossRef]
- Fujitani, M.; Zhang, S.; Fujiki, R.; Fujihara, Y.; Yamashita, T. A chromosome 16p13.11 microduplication causes hyperactivity through dysregulation of miR-484/protocadherin-19 signaling. Mol. Psychiatry 2017, 22, 364–374. [Google Scholar] [CrossRef]
- Mulle, J.G.; Sullivan, P.F.; Hjerling-Leffler, J. Editorial overview: Rare CNV disorders and neuropsychiatric phenotypes: Opportunities, challenges, solutions. Curr. Opin. Genet. Dev. 2021, 68, iii–ix. [Google Scholar] [CrossRef]
- Zhao, D.; Lin, M.; Chen, J.; Pedrosa, E.; Hrabovsky, A.; Fourcade, H.M.; Zheng, D.; Lachman, H.M. MicroRNA Profiling of Neurons Generated Using Induced Pluripotent Stem Cells Derived from Patients with Schizophrenia and Schizoaffective Disorder, and 22q11.2 Del. PLoS ONE 2015, 10, e0132387. [Google Scholar] [CrossRef]
- Toyoshima, M.; Akamatsu, W.; Okada, Y.; Ohnishi, T.; Balan, S.; Hisano, Y.; Iwayama, Y.; Toyota, T.; Matsumoto, T.; Itasaka, N.; et al. Analysis of induced pluripotent stem cells carrying 22q11.2 deletion. Transl. Psychiatry 2016, 6, e934. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, S. Integrated pathophysiology of schizophrenia, major depression, and bipolar disorder as monoamine axon disorder. Front. Biosci. 2022, 14, 4. [Google Scholar] [CrossRef] [PubMed]
- Wyatt, R.J.; Murphy, D.L.; Belmaker, R.; Cohen, S.; Donnelly, C.H.; Pollin, W. Reduced monoamine oxidase activity in platelets: A possible genetic marker for vulnerability to schizophrenia. Science 1973, 179, 916–918. [Google Scholar] [CrossRef]
- Nielsen, J.; Fejgin, K.; Sotty, F.; Nielsen, V.; Mork, A.; Christoffersen, C.T.; Yavich, L.; Lauridsen, J.B.; Clausen, D.; Larsen, P.H.; et al. A mouse model of the schizophrenia-associated 1q21.1 microdeletion syndrome exhibits altered mesolimbic dopamine transmission. Transl. Psychiatry 2017, 7, 1261. [Google Scholar] [CrossRef] [PubMed]
- Di Giovanni, G.; Svob Strac, D.; Sole, M.; Unzeta, M.; Tipton, K.F.; Muck-Seler, D.; Bolea, I.; Della Corte, L.; Nikolac Perkovic, M.; Pivac, N.; et al. Monoaminergic and Histaminergic Strategies and Treatments in Brain Diseases. Front. Neurosci. 2016, 10, 541. [Google Scholar] [CrossRef]
- Gonzalez, M.M.; Aston-Jones, G. Light deprivation damages monoamine neurons and produces a depressive behavioral phenotype in rats. Proc. Natl. Acad. Sci. USA 2008, 105, 4898–4903. [Google Scholar] [CrossRef] [PubMed]
- Maillet, A.; Krack, P.; Lhommee, E.; Metereau, E.; Klinger, H.; Favre, E.; Le Bars, D.; Schmitt, E.; Bichon, A.; Pelissier, P.; et al. The prominent role of serotonergic degeneration in apathy, anxiety and depression in de novo Parkinson’s disease. Brain 2016, 139, 2486–2502. [Google Scholar] [CrossRef] [PubMed]
- Hughes, R.B.; Whittingham-Dowd, J.; Clapcote, S.J.; Broughton, S.J.; Dawson, N. Altered medial prefrontal cortex and dorsal raphe activity predict genotype and correlate with abnormal learning behavior in a mouse model of autism-associated 2p16.3 deletion. Autism Res. 2022, 15, 614–627. [Google Scholar] [CrossRef] [PubMed]
- Euston, D.R.; Gruber, A.J.; McNaughton, B.L. The role of medial prefrontal cortex in memory and decision making. Neuron 2012, 76, 1057–1070. [Google Scholar] [CrossRef]
- Ostlund, S.B.; Balleine, B.W. Lesions of medial prefrontal cortex disrupt the acquisition but not the expression of goal-directed learning. J. Neurosci. 2005, 25, 7763–7770. [Google Scholar] [CrossRef]
- Mehta, D.; Iwamoto, K.; Ueda, J.; Bundo, M.; Adati, N.; Kojima, T.; Kato, T. Comprehensive survey of CNVs influencing gene expression in the human brain and its implications for pathophysiology. Neurosci. Res. 2014, 79, 22–33. [Google Scholar] [CrossRef] [PubMed]
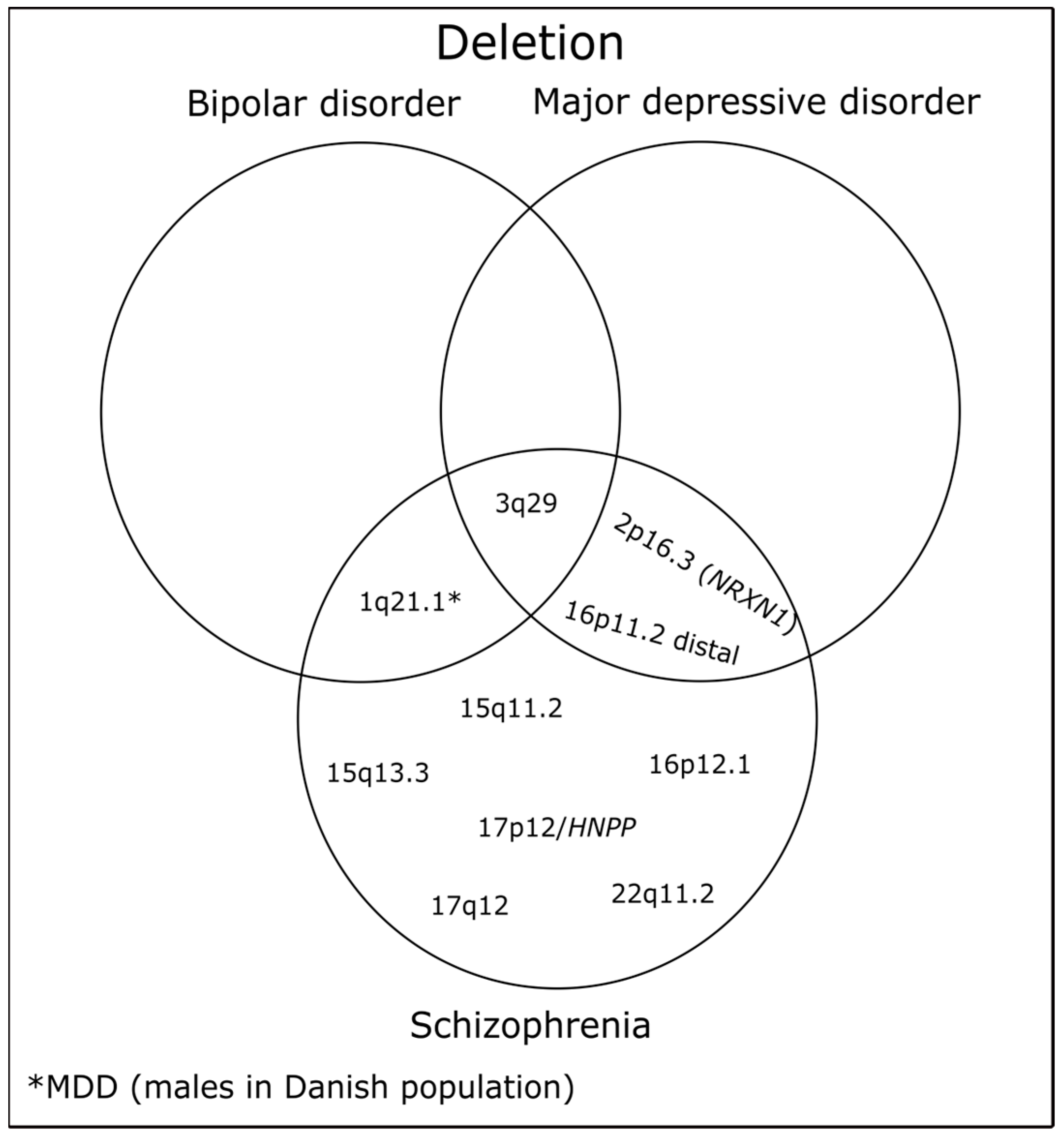
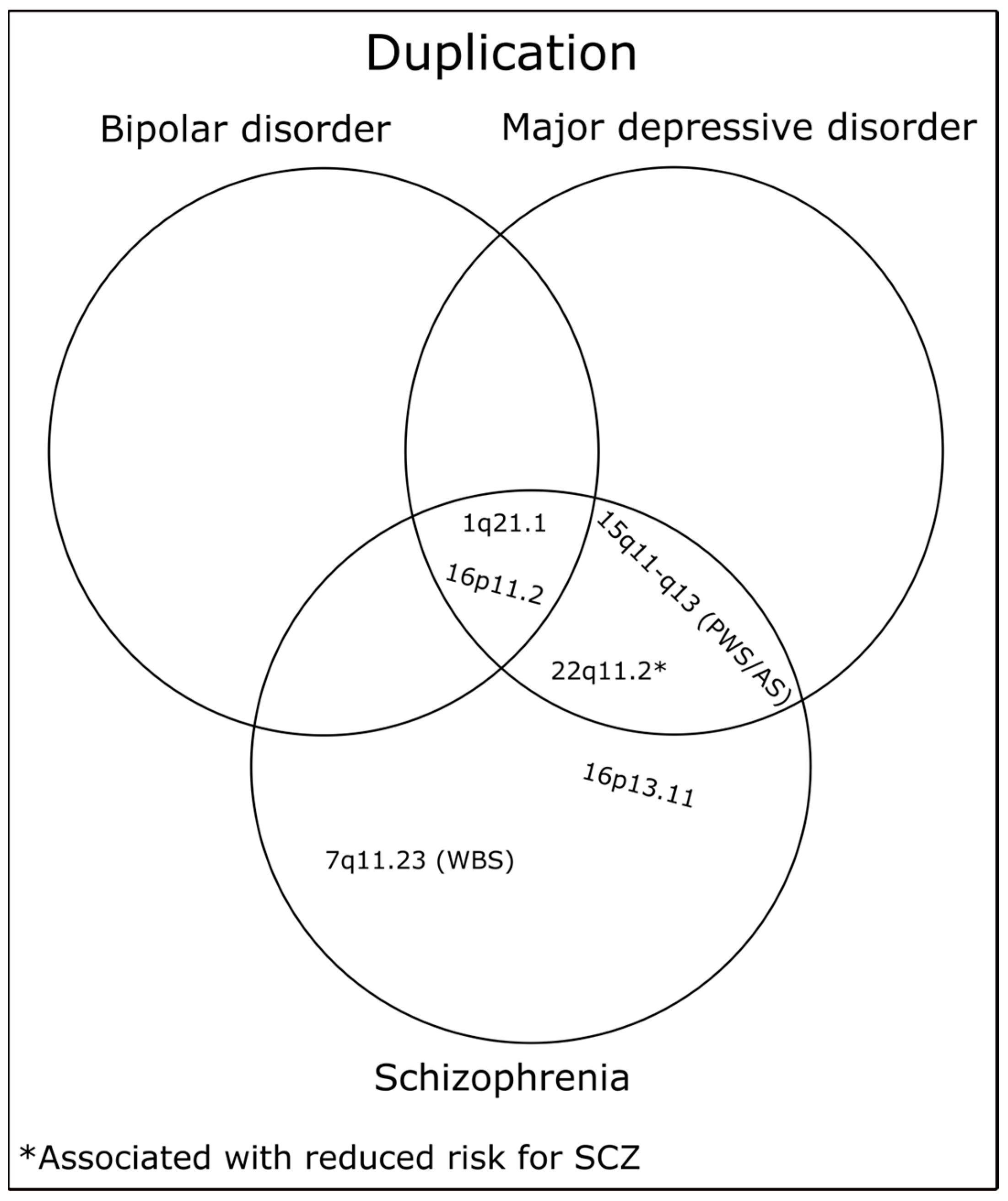
| Localization | Neurodevelopmental Associations | Position in Mb * | Protein Coding Genes | No. of Genes | Refs. |
|---|---|---|---|---|---|
| 1q21.1 | SCZ, BD, MDD (males in Danish population) | chr1:146.53–147.39 | PRKAB2, FMO5, CHD1L, BCL9, ACP6, GJA5, GJA8 | 7 | [56,62] |
| 2p16.3 (NRXN1) | SCZ, MDD | chr2:50.15–51.26 | NRXN1 | 1 | [56,63] |
| 3q29 | SCZ, BD, MDD | chr3:195.72–197.35 | TFRC, ZDHH19, SLC51A, PCYT1A, TCTEX1D2, TM4SF19, UBXN7, RNF168, SMCO1, WDR53, FBXO45, NRROS, CEP19, PIGX, PAK2, SENP5, NCBP2, NCBP2AS2, PIGZ, MELTF, DLG1, BDH1 | 22 | [56,63,64] |
| 15q11.2 | SCZ | chr15:22.81–23.09 | TUBGCP5, CYFIP1, NIPA1, NIPA2 | 4 | [56] |
| 15q13.3 | SCZ | chr15:31.08–32.46 | FAN1, MTMR10, TRPM1, KLF13, OTUD7A, CHRNA7 | 6 | [56] |
| 16p12.1 | SCZ | chr16:21.95–22.43 | UQCRC2, PDZD9, MOSMO, VWA3A, EEF2K, POLR3E, CDR2 | 7 | [56] |
| 16p11.2 distal | SCZ, MDD | chr16:28.82–29.04 | ATXN2L, TUFM, SH2B1, ATP2A1, RABEP2, CD19, NFATC2IP, SPNS1, LAT | 9 | [56,65] |
| 17p12/HNPP | SCZ | chr17:14.16–15.43 | HS3ST3B1, PMP22, TEKT3, CDRT4, TVP23C | 5 | [66] |
| 17q12 deletion | SCZ | chr17:34.81–36.20 | ZNHIT3, MYO19, PIGW, GGNBP2, DHRS11, MRM1, LHX1, AATF, ACACA, C17orf78, TADA2A, DUSP14, SYNRG, DDX52, HNF1B | 15 | [67] |
| 22q11.2 | SCZ | chr22:19.04–21.47 | DGCR2, ESS2, TSSK2, GSC2, SLC25A1, CLTCL1, HIRA, MRPL40, C22orf39, UFD1, CDC45, CLDN5, SEPTIN5, GP1BB, TBX1, GNB1L, RTL10, TXNRD2, COMT, ARVCF, TANGO2, DGCR8, TRMT2A, RANBP1, ZDHHC8, CCDC188, RTN4R, DGCR6L, GGTLC3, TMEM191B, RIMP3, ZNF74, SCARF2, KLHL22, MED15, PI4KA, SERPIND1, SNAP29, CRKL, AIFM3, LZTR1, THAP7, P2RX6, SLC7A4, LRRC74B | 45 | [11] |
| Localization | Neurodevelopmental Associations | Position in Mb * | Protein Coding Genes | No. of Genes | Refs. |
|---|---|---|---|---|---|
| 1q21.1 | SCZ, BD, MDD | chr1:146.53–147.39 | PRKAB2, FMO5, CHD1L, BCL9, ACP6, GJA5, GJA8 | 7 | [56,63,64] |
| 7q11.23 (WBS) | SCZ | chr7:72.74–74.14 | TRIM50, FKBP6, FZD9, BAZ1B, BCL7B, TBL2, MLXIPL, VPS37D, DNAJC30, BUD23, STX1A, ABHD11, CLDN3, CLDN4, METTL27, TMEM270, ELN, LIMK1, EIF4H, LAT2, RFC2, CLIP2, GTF2IRD1, GTF2I | 24 | [56] |
| 15q11-q13 (PWS/AS) | SCZ, MDD | chr15:22.81–28.39 | TUBGCP5, CYFIP1, NIPA2, NIPA1, GOLGA8S, GOLGA6L2, MKRN3, MAGEL2, NDN, NPAP1, SNRPN, SNURF, UBE3A, ATP10A, GABRB3, GABRA5, GABRG3, OCA2, HERC2 | 19 | [56,63] |
| 16p13.11 | SCZ | chr16:15.51–16.29 | BMERB1, MARF1, NDE1, MYH11, CEP20, ABCC1, ABCC6 | 7 | [56] |
| 16p11.2 | SCZ, BD, MDD | chr16:29.65–30.20 | SPN, QPRT, C16orf54, ZG16, KIF22, MAZ, PRRT2, PAGR1, MVP, CDIPT, SEZ6L2, ASPHD1, KCTD13, TMEM219, TAOK2, HIRIP3, INO80E, DOC2A, C16orf92, TLCD3B, ALDOA, PPP4C, TBX6, YPEL3, GDPD3, MAPK3, CORO1A | 27 | [56,63,64] |
| 22q11.2 | # SCZ, MDD | chr22:19.04–21.47 | DGCR2, ESS2, TSSK2, GSC2, SLC25A1, CLTCL1, HIRA, MRPL40, C22orf39, UFD1, CDC45, CLDN5, SEPTIN5, GP1BB, TBX1, GNB1L, RTL10, TXNRD2, COMT, ARVCF, TANGO2, DGCR8, TRMT2A, RANBP1, ZDHHC8, CCDC188, RTN4R, DGCR6L, GGTLC3, TMEM191B, RIMBP3, ZNF74, SCARF2, KLHL22, MED15, PI4KA, SERPIND1, SNAP29, CRKL, AIFM3, LZTR1, THAP7, P2RX6, SLC7A4, LRRC74B | 45 | [56,63] |
| Location | Disease | CNV Frequency (%) | Odds Ratio (95% CI) | p | References | ||
|---|---|---|---|---|---|---|---|
| Cases | Controls | ||||||
| Unique CNVs | 15q11.2 | SCZ | 0.642 | 0.368 | 1.8 (1.35, 2.38) | 0.000023 | [56] |
| 15q13.3 | SCZ | 0.098 | 0.019 | 4.6 (1.64, 16.22) | 0.0015 | [56] | |
| 16p12.1 | SCZ | 0.162 | 0.045 | 3.3 (1.61, 7.05) | 0.00034 | [56] | |
| 17p12/HNPP | SCZ | N/A | N/A | N/A | N/A | [66] | |
| 17q12 deletion | SCZ | N/A | N/A | N/A | N/A | [67] | |
| 22q11.2 | SCZ | 0.303 | 0.0049 | 67.7 (9.3–492.8) | 5.70 × 10−18 | [11] | |
| Shared CNVs | 1q21.1 | SCZ | 0.172 | 0.026 | 6.8 (2.9, 18.51) | 2.10 × 10−7 | [56] |
| BD | 0.033 | 0.021 | 1.61 (0.47, 5.5) | 0.44 | [64] | ||
| 2p16.3 | SCZ | 0.152 | 0.034 | 4.5 (2.03, 10.94) | 0.000028 | [56] | |
| MDD | 0.075 | 0.037 | 2.01 (1.18, 3.19) | 5.70 × 10−3 | [63] | ||
| 3q29 | SCZ | 0.069 | 0.004 | 18 (2.66, 763.34) | 0.0002 | [56] | |
| BD | 0.025 | 0.001 | 17.31 (1.57, 190.97) | 0.03 | [64] | ||
| MDD | 0.013 | 0.001 | 11.22 (2.27, 46.52) | 1.00 × 10−3 | [63] | ||
| 16p11.2 distal | SCZ | 0.025 | 0.019 | 1.7 (0.37, 7.6) | 0.51 | [56] | |
| MDD | 0.029 | 0.013 | 2.23 (0.92, 4.63) | 5.00 × 10−2 | [63] | ||
| Location | Disease | CNV Frequency (%) | Odds Ratio (95% CI) | p | References | ||
|---|---|---|---|---|---|---|---|
| Cases | Controls | ||||||
| Unique CNVs | 7q11.23 (WBS) | SCZ | 0.083 | 0.000 | NA | 4.00 × 10−7 | [56] |
| 16p13.11 | SCZ | 0.377 | 0.222 | 1.7 (1.2, 2.52) | 0.0022 | [56] | |
| 22q11.2 | MDD | 0.011 | 0.063 | 1.72 (1.12, 2.53) | 9.00 × 10−3 | [63] | |
| Shared CNVs | 1q21.1 | SCZ | 0.108 | 0.049 | 2.3 (1.08, 5.09) | 0.022 | [56] |
| BD | 0.099 | 0.037 | 2.64 (1.19, 5.88) | 0.022 | [64] | ||
| MDD | 0.088 | 0.040 | 2.17 (1.34, 3.36) | 9.10 × 10−4 | [63] | ||
| 15q11-q13 (PWS/AS) | SCZ | 0.083 | 0.000 | NA | 4.00 × 10−7 | [56] | |
| MDD | 0.025 | 0.003 | 8.14 (2.77, 21.69) | 4.61 × 10−5 | [63] | ||
| 16p11.2 | SCZ | 0.304 | 0.030 | 11 (5.08, 26.43) | 3.70 × 10−15 | [56] | |
| BD | 0.130 | 0.030 | 4.37 (2.12, 9) | 0.00023 | [64] | ||
| MDD | 0.071 | 0.028 | 2.65 (1.53, 4.31) | 2.04 × 10−4 | [63] | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Büki, G.; Hadzsiev, K.; Bene, J. Copy Number Variations in Neuropsychiatric Disorders. Int. J. Mol. Sci. 2023, 24, 13671. https://doi.org/10.3390/ijms241813671
Büki G, Hadzsiev K, Bene J. Copy Number Variations in Neuropsychiatric Disorders. International Journal of Molecular Sciences. 2023; 24(18):13671. https://doi.org/10.3390/ijms241813671
Chicago/Turabian StyleBüki, Gergely, Kinga Hadzsiev, and Judit Bene. 2023. "Copy Number Variations in Neuropsychiatric Disorders" International Journal of Molecular Sciences 24, no. 18: 13671. https://doi.org/10.3390/ijms241813671
APA StyleBüki, G., Hadzsiev, K., & Bene, J. (2023). Copy Number Variations in Neuropsychiatric Disorders. International Journal of Molecular Sciences, 24(18), 13671. https://doi.org/10.3390/ijms241813671








