Proteomic Analysis Reveals Salt-Tolerant Mechanism in Soybean Applied with Plant-Derived Smoke Solution
Abstract
:1. Introduction
2. Results
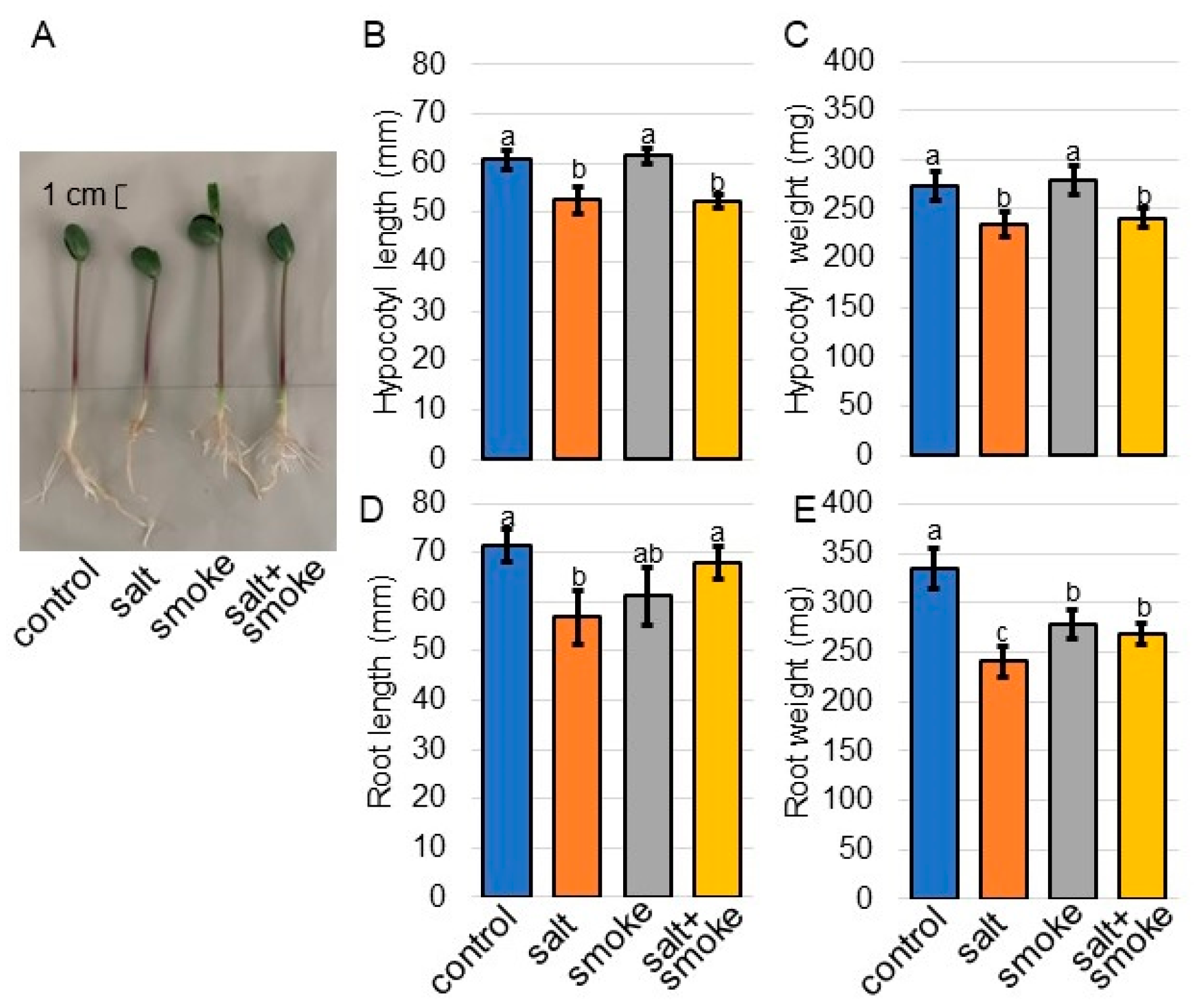
2.1. Morphological Analysis of Soybean Treated with Plant-Derived Smoke Solution under Salt Stress
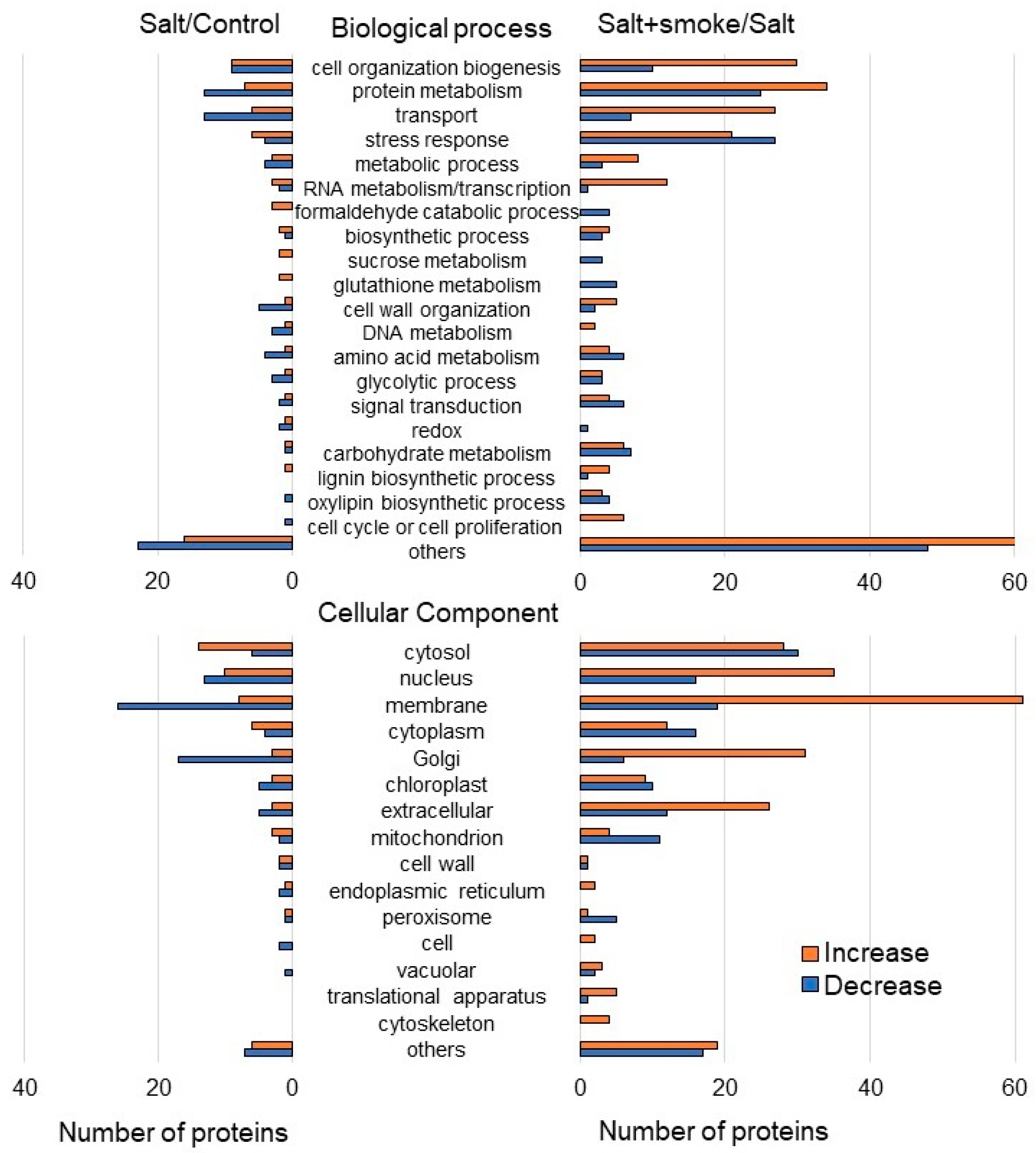
2.2. Identification and Functional Investigation of Proteins in Soybean Treated with Plant-Derived Smoke Solution under Salt Stress
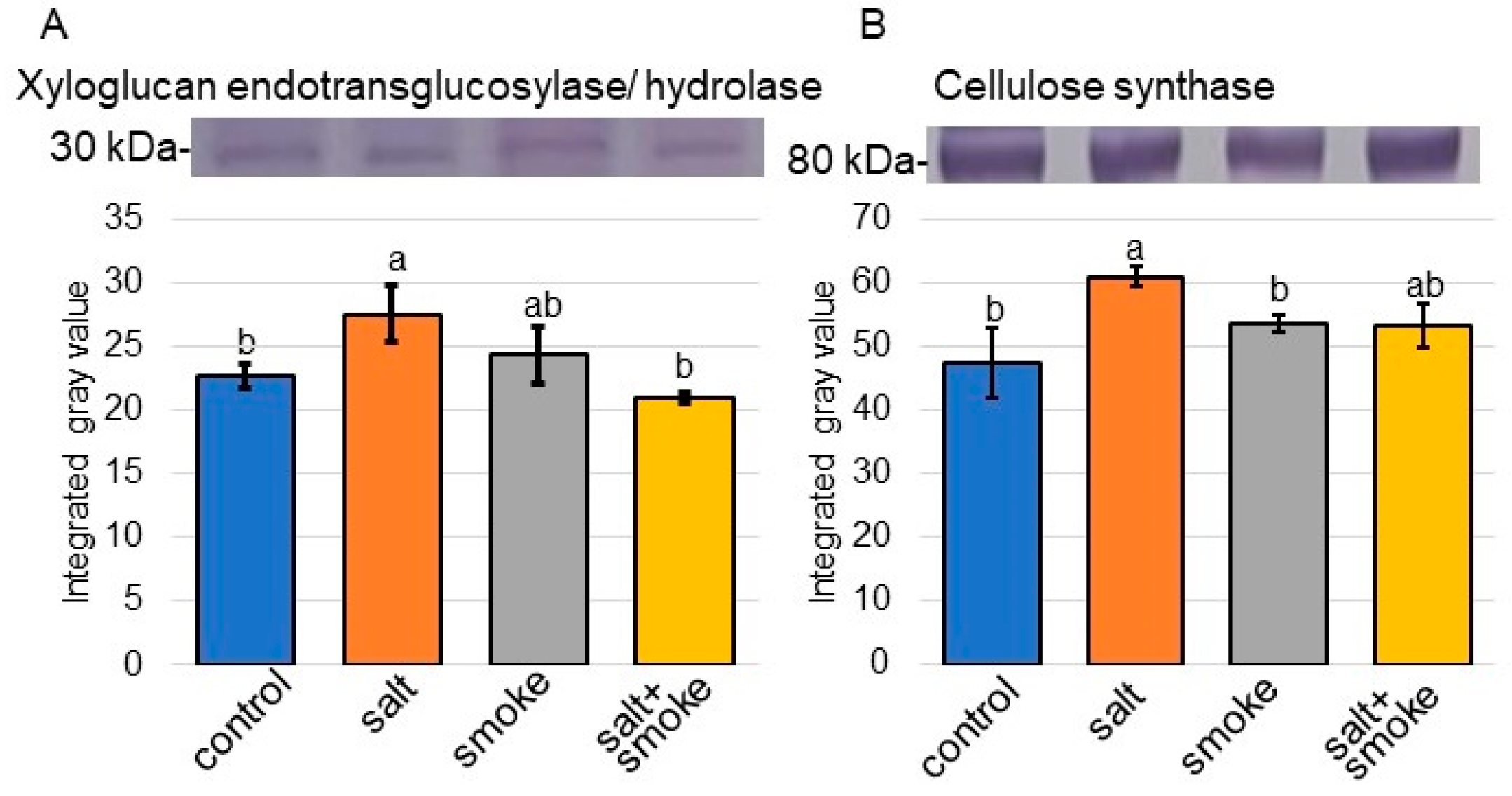
2.3. Immuno-Blot Analysis of Proteins Involved in Transport, Stress Response, Sucrose Metabolism, and Cell Wall Organization in Soybean with Application of Plant-Derived Smoke Solution under Salt Stress
2.4. Lectin Blot Analysis of Proteins Involved in Soybean with the Application of Plant-Derived Smoke Solution under Salt Stress
2.5. The Contents of ATP and Gamma-Aminobutyric Acid in Soybean with Application of Plant-Derived Smoke Solution under Salt Stress
3. Discussion
3.1. Cell Wall Organization Is Related to Salt-Tolerant Mechanism in Soybean with Plant-Derived Smoke Solution
3.2. Glycoprotein Folding Is Related to Salt-Tolerance Mechanism in Soybean Treated with Plant-Derived Smoke Solution
3.3. Energy Metabolism Iis Related to Salt-Tolerant Mechanism in Soybean Treated with Plant-Derived Smoke Solution
4. Materials and Methods
4.1. Plant Material and Treatment
4.2. Protein Extraction
4.3. Protein Enrichment, Reduction, Alkylation, and Digestion
4.4. Protein Identification Using nano-Liquid Chromatography Mass Spectrometry
4.5. Mass-Spectrometry Data Analysis
4.6. Differential Analysis of Proteins using Mass-Spectrometry Data
4.7. Immuno-Blot Analysis
4.8. Measurement of ATP Contents
4.9. Measurement of gamma-Aminobutyric Acid Contents
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Bhoite, R.; Han, Y.; Chaitanya, A.K.; Varshney, R.K.; Sharma, D.L. Genomic approaches to enhance adaptive plasticity to cope with soil constraints amidst climate change in wheat. Plant Genome 2023, e20358. [Google Scholar] [CrossRef]
- Li, X.; Wang, A.; Wan, W.; Luo, X.; Zheng, L.; He, G.; Huang, D.; Chen, W.; Huang, Q. High salinity inhibits soil bacterial community mediating nitrogen cycling. Appl. Environ. Microbiol. 2021, 87, e0136621. [Google Scholar] [CrossRef]
- Sarri, E.; Termentzi, A.; Abraham, E.M.; Papadopoulos, G.K.; Baira, K.; Machera, K.; Loukas, V.; Komaitis, F.; Tani, E. Salinity stress alters the secondary metabolic profile of M. sativa, M. arborea and their hybrid (Alborea). Int. J. Mol. Sci. 2021, 22, 4882. [Google Scholar] [CrossRef]
- Singh, A. Soil salinization management for sustainable development: A review. J. Environ. Manag. 2021, 277, 111383. [Google Scholar] [CrossRef] [PubMed]
- Yuvaraj, M.; Bose, K.S.C.; Elavarasi, P.; Tawfik, E. Soil salinity and its management. In Soil Moisture Importance; Meena, R.S., Datta, R., Eds.; IntechOpen: London, UK, 2020. [Google Scholar]
- McDonald, G.K.; Tavakkoli, E.; Rengasamy, P. Commentary: Bread wheat with high salinity and sodicity tolerance. Front. Plant Sci. 2020, 11, 1194. [Google Scholar] [CrossRef] [PubMed]
- Phang, T.-H.; Shao, G.; Lam, H.-M. Salt tolerance in soybean. J. Integr. Plant Biol. 2008, 50, 1196–1212. [Google Scholar] [CrossRef]
- Chen, R.; Li, M.; Zhang, H.; Duan, L.; Sun, X.; Jiang, Q.; Zhang, H.; Hu, Z. Continuous salt stress-induced long non-coding RNAs and DNA methylation patterns in soybean roots. BMC Genomics 2019, 20, 730. [Google Scholar] [CrossRef] [PubMed]
- Liu, A.; Xiao, Z.; Wang, Z.; Lam, H.M.; Chye, M.L. Galactolipid and phospholipid profile and proteome alterations in soybean leaves at the onset of salt stress. Front. Plant Sci. 2021, 12, 644408. [Google Scholar] [CrossRef] [PubMed]
- Yuan, M.; Zhang, D.; Wang, Z.; Zhu, Z.; Sun, H.; Wang, W.; Han, D.; Qu, Z.; Ma, B.; Wang, J.; et al. Salt altered rhizosphere fungal community and induced soybean recruit specific species to ameliorate salt stress. Front. Microbiol. 2023, 14, 1142780. [Google Scholar] [CrossRef]
- Hanin, M.; Ebel, C.; Ngom, M.; Laplaze, L.; Masmoudi, K. New insights on plant salt tolerance mechanisms and their potential use for breeding. Front. Plant Sci. 2016, 7, 1787. [Google Scholar] [CrossRef] [PubMed]
- Sheteiwy, M.S.; Shao, H.; Qi, W.; Daly, P.; Sharma, A.; Shaghaleh, H.; Hamoud, Y.A.; El-Esawi, M.A.; Pan, R.; Wan, Q.; et al. Seed priming and foliar application with jasmonic acid enhance salinity stress tolerance of soybean (Glycine max L.) seedlings. J. Sci. Food Agric. 2021, 101, 2027–2041. [Google Scholar] [CrossRef] [PubMed]
- Kelly, L.T.; Giljohann, K.M.; Duane, A.; Aquilué, N.; Archibald, S.; Batllori, E.; Bennett, A.F.; Buckland, S.T.; Canelles, Q.; Clarke, M.F.; et al. Fire and biodiversity in the Anthropocene. Science 2020, 370, 355. [Google Scholar] [CrossRef] [PubMed]
- Dixon, K.W.; Merritt, D.J.; Flematti, G.R.; Ghisalberti, E.L. Karrikinolide—A phytoreactive compound derived from smoke with applications in horticulture, ecological restoration and agriculture. Acta Hortic. 2009, 813, 155–170. [Google Scholar] [CrossRef]
- Khatoon, A.; Rehman, S.U.; Aslam, M.M.; Jamil, M.; Komatsu, S. Plant-derived smoke affects biochemical mechanism on plant growth and seed germination. Int. J. Mol. Sci. 2020, 21, 7760. [Google Scholar] [CrossRef] [PubMed]
- Bose, U.; Juhász, A.; Broadbent, J.A.; Komatsu, S.; Colgrave, M.L. Multi-omics strategies for decoding smoke-assisted germina- tion pathways and seed vigour. Int. J. Mol. Sci. 2020, 21, 7512. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.M.; To, T.K.; Matsui, A.; Tanoi, K.; Kobayashi, N.I.; Matsuda, F.; Habu, Y.; Ogawa, D.; Sakamoto, T.; Matsunaga, S.; et al. Acetate-mediated novel survival strategy against drought in plants. Nat. Plants 2017, 3, 17119. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, A.; Tripathi, D.K.; Roychoudhury, A. The karrikin “calisthenics”: Can compounds derived from smoke help in stress tolerance? Physiol. Plant. 2019, 165, 290–302. [Google Scholar] [CrossRef] [PubMed]
- Jia, K.P.; Baz, L.; Al-Babili, S. From carotenoids to strigolactones. J. Exp. Bot. 2018, 69, 2189–2204. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.; Lian, Y.; Wang, C. Comparing and Contrasting the Multiple Roles of Butenolide Plant Growth Regulators: Strigolactones and Karrikins in Plant Development and Adaptation to Abiotic Stresses. Int. J. Mol. Sci. 2019, 20, 6270. [Google Scholar] [CrossRef]
- Khan, M.H.U.; Khattak, J.Z.K.; Jamil, M.; Malook, I.; Khan, S.U.; Jan, M.; Din, I.; Saud, S.; Kamran, M.; Alharby, H.; et al. Bacillus safensis with plant-derived smoke stimulates rice growth under saline conditions. Environ. Sci. Pollut. Res. Int. 2017, 24, 23850–23863. [Google Scholar] [CrossRef] [PubMed]
- Otori, M.; Murashita, Y.; Rehman, S.; Komatsu, S. Proteomic study to understand promotive effects of plant-derived smoke on soybean root growth under flooding stress. Plant Mol. Biol. Rep. 2021, 39, 24–33. [Google Scholar] [CrossRef]
- Murashita, Y.; Nishiuchi, T.; Rehman, S.U.; Komatsu, S. Subcellular proteomics to understand promotive effect of plant-derived smoke solution on soybean root. Proteomes 2021, 9, 39. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Z.; Kobayashi, T.; Zhu, W.; Imai, H.; Zhao, R.; Ohno, T.; Rehman, S.U.; Uemura, M.; Tian, J.; Komatsu, S. Plant-derived smoke enhances plant growth through ornithine-synthesis pathway and ubiquitin-proteasome pathway in soybean. J. Proteomics 2020, 221, 103781. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Rehman, S.U.; Yamaguchi, H.; Hitachi, K.; Tsuchida, K.; Yamaguchi, T.; Sunohara, Y.; Matsumoto, H.; Komatsu, S. Proteomic analysis of the effect of plant-derived smoke on soybean during recovery from flooding stress. J. Proteomics 2018, 181, 238–248. [Google Scholar] [CrossRef]
- Aslam, M.M.; Rehman, S.; Khatoon, A.; Jamil, M.; Yamaguchi, H.; Hitachi, K.; Tsuchida, K.; Li, X.; Sunohara, Y.; Matsumoto, H.; et al. Molecular responses of maize shoot to a plant derived smoke solution. Int. J. Mol. Sci. 2019, 20, 1319. [Google Scholar] [CrossRef] [PubMed]
- Rehman, A.; Rehman, S.U.; Khatoon, A.; Qasim, M.; Itoh, T.; Iwasaki, Y.; Wang, X.; Sunohara, Y.; Matsumoto, H.; Komatsu, S. Proteomic analysis of the promotive effect of plant-derived smoke on plant growth of chickpea. J. Proteom. 2018, 176, 56–70. [Google Scholar] [CrossRef] [PubMed]
- Cosgrove, D.J. Growth of the plant cell wall. Nat. Rev. Mol. Cell Biol. 2005, 6, 850–861. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Li, Y.; Lu, C.; Tang, Y.; Jiang, X.; Gai, Y. Isolation and characterization of Populus xyloglucan endotransglycosylase/hydrolase (XTH) involved in osmotic stress responses. Int. J. Biol. Macromol. 2020, 155, 1277–1287. [Google Scholar] [CrossRef] [PubMed]
- Van Sandt, V.S.T.; Suslov, D.; Verbelen, J.-P.; Vissenberg, K. Xyloglucan endotransglucosylase activity loosens a plant cell wall. Ann. Bot. 2007, 100, 1467–1473. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.F.; Shi, Y.Z.; Lei, G.J.; Fry, S.C.; Zhang, B.C.; Zhou, Y.H.; Braam, J.; Jiang, T.; Xu, X.Y.; Mao, C.Z.; et al. XTH31, Encoding an in vitro XEH/XET-active enzyme, regulates aluminum sensitivity by modulating in vivo XET action, cell wall xyloglucan content, and aluminum binding capacity in Arabidopsis. Plant Cell. 2012, 24, 4731–4747. [Google Scholar] [CrossRef]
- Xuan, Y.; Zhou, Z.S.; Li, H.B.; Yang, Z.M. Identification of a group of XTHs genes responding to heavy metal mercury, salinity and drought stresses in Medicago truncatula. Ecotoxicol. Environ. Saf. 2016, 132, 153–163. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Ma, Y.; Horst, W.J.; Yang, Z.-B. Spatial–temporal analysis of polyethylene glycol-reduced aluminium accumulation and xyloglucan endotransglucosylase action in root tips of common bean (Phaseolus vulgaris). Ann. Bot. 2016, 118, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.Y.; Seo, Y.S.; Kim, S.J.; Kim, W.T.; Shin, J.S. Constitutive expression of CaXTH3, a hot pepper xyloglucan endotransglucosylase/hydrolase, enhanced tolerance to salt and drought stresses without phenotypic defects in tomato plants (Solanum lycopersicum cv. Dotaerang). Plant Cell Rep. 2011, 30, 867–877. [Google Scholar] [CrossRef] [PubMed]
- Song, L.; Valliyodan, B.; Prince, S.; Wan, J.; Nguyen, H.T. Characterization of the XTH gene family: New insight to the roles in soybean flooding tolerance. Int. J. Mol. Sci. 2018, 19, 2705. [Google Scholar] [CrossRef]
- Paredez, A.R.; Somerville, C.R.; Ehrhardt, D.W. Visualization of cellulose synthase demonstrates functional association with microtubules. Science 2006, 312, 1491–1495. [Google Scholar] [CrossRef] [PubMed]
- Pear, J.R.; Kawagoe, Y.; Schreckengost, W.E.; Delmer, D.P.; Stalker, D.M. Higher plants contain homologs of the bacterial celA genes encoding the catalytic subunit of cellulose synthase. Proc. Natl. Acad. Sci. USA 1996, 93, 12637–12642. [Google Scholar] [CrossRef] [PubMed]
- Vain, T.; Crowell, E.F.; Timpano, H.; Biot, E.; Desprez, T.; Mansoori, N.; Trindade, L.M.; Pagant, S.; Robert, S.; Höfte, H.; et al. The cellulase KORRIGAN is part of the cellulose synthase complex. Plant Physiol. 2014, 165, 1521–1532. [Google Scholar] [CrossRef] [PubMed]
- Gu, Y.; Kaplinsky, N.; Bringmann, M.; Cobb, A.; Carroll, A.; Sampathkumar, A.; Baskin, T.I.; Persson, S.; Somerville, C.R. Identification of a cellulose synthase-associated protein required for cellulose biosynthesis. Proc. Natl. Acad. Sci. USA 2010, 107, 12866–12871. [Google Scholar] [CrossRef] [PubMed]
- Endler, A.; Kesten, C.; Schneider, R.; Zhang, Y.; Ivakov, A.; Froehlich, A.; Funke, N.; Persson, S. A mechanism for sustained cellulose synthesis during salt stress. Cell 2015, 162, 1353–1364. [Google Scholar] [CrossRef] [PubMed]
- Kesten, C.; Wallmann, A.; Schneider, R.; McFarlane, H.E.; Diehl, A.; Khan, G.A.; van Rossum, B.-J.; Lampugnani, E.R.; Szymanski, W.G.; Cremer, N.; et al. The companion of cellulose synthase 1 confers salt tolerance through a tau-like mechanism in plants. Nat. Comm. 2019, 10, 857. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Zhang, L.; Wang, Y.; Xu, F.; Liu, M.; Lin, P.; Ren, S.; Ma, R.; Guo, Y.D. Knockdown of a cellulose synthase gene BoiCesA affects the leaf anatomy, cellulose content and salt tolerance in broccoli. Sci. Rep. 2017, 7, 41397. [Google Scholar] [CrossRef] [PubMed]
- Moremen, K.W.; Tiemeyer, M.; Nairn, A.V. Vertebrate protein glycosylation: Diversity, synthesis and function. Nat. Rev. Mol. Cell Biol. 2012, 13, 448–462. [Google Scholar] [CrossRef] [PubMed]
- Yan, Q.; Lennarz, W.J. Studies on the function of oligosaccharyl transferase subunits. Stt3p is directly involved in the glycosylation process. J. Biol. Chem. 2002, 277, 47692–47700. [Google Scholar] [CrossRef] [PubMed]
- Kelleher, D.J.; Gilmore, R. An evolving view of the eukaryotic oligosaccharyltransferase. Glycobiology 2006, 16, 47R–62R. [Google Scholar] [CrossRef] [PubMed]
- Nagashima, Y.; von Schaewen, A.; Koiwa, H. Function of N-glycosylation in plants. Plant Sci. 2018, 274, 70–79. [Google Scholar] [CrossRef]
- Kajiura, H.; Koiwa, H.; Nakazawa, Y.; Okazawa, A.; Kobayashi, A.; Seki, T.; Fujiyama, K. Two Arabidopsis thaliana Golgi alpha-mannosidase I enzymes are responsible for plant N-glycan maturation. Glycobiology 2010, 20, 235–247. [Google Scholar] [CrossRef] [PubMed]
- Strasser, R.; Steinkellner, H.; Borén, M.; Altmann, F.; Mach, L.; Glössl, J.; Mucha, J. Molecular cloning of cDNA encoding N-acetylglucosaminyltransferase II from Arabidopsis thaliana. Glycoconj. J. 1999, 16, 787–791. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Niu, G.; Zhang, H.; Sun, Y.; Sun, S.; Yu, F.; Lu, S.; Yang, Y.; Li, J.; Hong, Z. Trimming of N-glycans by the Golgi-Localized α-1,2-Mannosidases, MNS1 and MNS2, is crucial for maintaining RSW2 protein abundance during salt stress in Arabidopsis. Mol. Plant. 2018, 11, 678–690. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Niu, G.; Li, X.; Zhang, H.; Chen, H.; Hou, D.; Lan, P.; Hong, Z. Comparative label-free quantitative proteomics analysis reveals the essential roles of N-glycans in salt tolerance by modulating protein abundance in Arabidopsis. Front. Plant Sci. 2021, 12, 646425. [Google Scholar] [CrossRef]
- Çatav, Ş.S.; Elgin, E.S.; Dağ, Ç.; Stark, J.L.; Küçükakyüz, K. NMR-based metabolomics reveals that plant-derived smoke stimulates root growth via affecting carbohydrate and energy metabolism in maize. Metabolomics 2018, 14, 143. [Google Scholar] [CrossRef]
- Munns, R.; Tester, M. Mechanisms of salinity tolerance. Ann. Rev. Plant Biol. 2008, 64, 651–681. [Google Scholar] [CrossRef] [PubMed]
- Jacoby, R.P.; Millar, A.H.; Taylor, N.L. Investigating the role of respiration in plant salinity tolerance by analyzing mitochondrial proteomes from wheat and a salinity-tolerant Amphiploid (wheat × Lophopyrum elongatum). J Proteome Res. 2013, 12, 4807–4829. [Google Scholar] [CrossRef] [PubMed]
- Carillo, P. GABA shunt in durum wheat. Front. Plant Sci. 2018, 9, 100. [Google Scholar] [CrossRef] [PubMed]
- Che-Othman, M.H.; Jacoby, R.P.; Millar, A.H.; Taylor, N.L. Wheat mitochondrial respiration shifts from the tricarboxylic acid cycle to the GABA shunt under salt stress. New Phytol. 2020, 225, 1166–1180. [Google Scholar] [CrossRef] [PubMed]
- Stein, O.; Granot, D. An overview of sucrose synthases in plants. Front. Plant Sci. 2019, 10, 95. [Google Scholar] [CrossRef] [PubMed]
- Yin, Y.; Qi, F.; Gao, L.; Rao, S.; Yang, Z.; Fang, W. iTRAQ-based quantitative proteomic analysis of dark-germinated soybeans in response to salt stress. RSC Adv. 2018, 8, 17905–17913. [Google Scholar] [CrossRef]
- Al-Quraan, N.A.; Samarah, N.H.; Tanash, A.A. Effect of drought stress on wheat (Triticum durum) growth and metabolism: Insight from GABA shunt, reactive oxygen species and dehydrin genes expression. Funct. Plant Biol. 2022. [Google Scholar] [CrossRef]
- Samarah, N.H.; Al-Quraan, N.A.; Al-Wraikat, B.S. Ultrasonic treatment to enhance seed germination and vigour of wheat (Triticum durum) in association with γ-aminobutyric acid (GABA) shunt pathway. Funct. Plant Biol. 2023, 50, 277–293. [Google Scholar] [CrossRef]
- Tieu, A.; Plummer, J.A.; Dixon, K.A.; Sivasithamparam, K.; Sieler, I.M. Germination of four species of native Western Australian plants using plant-derived smoke. Aust. J. Bot. 1999, 47, 207. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Komatsu, S.; Han, C.; Nanjo, Y.; Altaf-Un-Nahar, M.; Wang, K.; He, D.; Yang, P. Label-free quantitative proteomic analysis of abscisic acid effect in early-stage soybean under flooding. J. Proteome Res. 2013, 12, 4769–4784. [Google Scholar] [CrossRef] [PubMed]
- Tyanova, S.; Temu, T.; Sinitcyn, P.; Carlson, A.; Hein, M.Y.; Geiger, T.; Mann, M.; Cox, J. The Perseus computational platform for comprehensive analysis of (prote) omics data. Nat. Methods 2016, 13, 731–740. [Google Scholar] [CrossRef] [PubMed]
- Konishi, H.; Ishiguro, K.; Komatsu, S. A proteomics approach towards understanding blast fungus infection of rice grown under different levels of nitrogen fertilization. Proteomics 2001, 1, 1162–1171. [Google Scholar] [CrossRef] [PubMed]
- Komatsu, S.; Deschamps, T.; Hiraga, S.; Kato, M.; Chiba, M.; Hashiguchi, A.; Tougou, M.; Shimamura, S.; Yasue, H. Characterization of a novel flooding stress-responsive alcohol dehydrogenase expressed in soybean roots. Plant Mol. Biol. 2011, 77, 309–322. [Google Scholar] [CrossRef]
- Nanjo, Y.; Skultety, L.; Ashraf, Y.; Komatsu, S. Comparative proteomic analysis of early-stage soybean seedlings responses to flooding by using gel and gel-free techniques. J. Proteome Res. 2010, 9, 3989–4002. [Google Scholar] [CrossRef]
- Nishiyama, T.; Sulistyaningdyah, W.T.; Ueda, K.; Kusakabe, H. GABA enzymatic assay kit. Biosci. Biotech. Biochem. 2020, 84, 118–125. [Google Scholar] [CrossRef]
- Vizcaíno, J.A.; Côté, R.G.; Csordas, A.; Dianes, J.A.; Fabregat, A.; Foster, J.M.; Griss, J.; Alpi, E.; Birim, M.; Contell, J.; et al. PRoteomics IDEntifications (PRIDE) database and associated tools: Status in 2013. Nucleic Acids Res. 2013, 41, D1063–D1069. [Google Scholar] [CrossRef]
- Okuda, S.; Watanabe, Y.; Moriya, Y.; Kawano, S.; Yamamoto, T.; Matsumoto, M.; Takami, T.; Kobayashi, D.; Araki, N.; Yoshizawa, A.C.; et al. jPOSTTrepo: An international standard data repository for proteomes. Nucleic Acids Res. 2017, 45, D1107–D1111. [Google Scholar] [CrossRef]




| Salt/Control | |||||||
| Difference | Accession | Description | Cov | MP | AAs | MW | pI |
| 7.2714 | K7MB33 | Uncharacterized protein | 13 | 7 | 838 | 92.4 | 4.69 |
| 4.0604 | I1KF11 | Dihydroorotase | 24 | 6 | 346 | 38.4 | 6.98 |
| 4.0325 | A0A0R0HMK4 | Phosphatidylinositol-specific phospholipase C | 8 | 2 | 421 | 46.1 | 7.50 |
| 3.6190 | A0A0R0IZE4 | 40S ribosomal protein SA | 48 | 15 | 310 | 33.9 | 5.26 |
| 3.1335 | I1JY29 | Alcohol dehydrogenase | 27 | 6 | 381 | 41.2 | 6.00 |
| 3.0291 | I1KC24 | CBFD_NFYB_HMF | 7 | 2 | 229 | 25.0 | 5.38 |
| 2.9526 | K7MJ40 | GOLD domain-containing protein | 22 | 6 | 433 | 49.5 | 5.87 |
| 2.6089 | I1L2Y7 | Glycosyltransferase | 8 | 3 | 473 | 52.7 | 6.02 |
| 2.6088 | A0A0R0I6S7 | Uncharacterized protein | 11 | 3 | 461 | 52.9 | 7.66 |
| 2.4850 | I1KN97 | DUF3700 domain-containing protein | 32 | 5 | 235 | 25.4 | 5.83 |
| −3.3697 | I1L0J1 | CRAL-TRIO domain-containing protein | 16 | 5 | 467 | 52.9 | 5.16 |
| −3.4240 | K7LMI3 | K Homology domain-containing protein | 11 | 6 | 794 | 84.2 | 5.06 |
| −3.4587 | I1M1F3 | alpha-1,2-Mannosidase | 5 | 2 | 610 | 67.8 | 7.09 |
| −3.6234 | A0A0R0GMV1 | Cupin type-1 domain-containing protein | 17 | 4 | 489 | 55.1 | 5.60 |
| −3.7702 | I1N036 | Proliferating cell nuclear antigen | 55 | 9 | 266 | 29.5 | 4.79 |
| −3.8266 | I1KWV5 | Ubiquitin receptor RAD23 | 24 | 7 | 401 | 42.0 | 4.84 |
| −3.8568 | C6TBW8 | Dihydrodipicolinate reductase | 11 | 3 | 344 | 37.4 | 6.95 |
| −3.9165 | A0A0R0IL99 | Glutamate--cysteine ligase | 26 | 10 | 510 | 57.6 | 8.07 |
| −3.9998 | A0A0R0KVB4 | Peptidase A1 domain-containing protein | 29 | 7 | 427 | 46.4 | 8.32 |
| −6.1145 | A0A0R0KRW0 | Cupin type-1 domain-containing protein | 56 | 14 | 387 | 43.4 | 5.22 |
| Salt + Smoke/Salt | |||||||
| Difference | Accession | Description | Cov | MP | AAs | MW | pI |
| 4.9048 | C6TI83 | BURP domain-containing protein | 21 | 5 | 276 | 30.9 | 6.21 |
| 4.8781 | I1MDT8 | Serine decarboxylase | 16 | 5 | 485 | 54.7 | 6.14 |
| 4.8530 | I1JQB4 | FAS1 domain-containing protein | 15 | 3 | 453 | 50.1 | 6.46 |
| 4.7338 | I1KWV5 | Ubiquitin receptor RAD23 | 24 | 7 | 401 | 42.0 | 4.84 |
| 4.1979 | A0A0R0EAL6 | PKS_ER domain-containing protein | 50 | 13 | 357 | 39.0 | 5.80 |
| 4.0866 | K7KZF3 | Fe2OG dioxygenase | 12 | 3 | 314 | 35.9 | 6.09 |
| 3.8789 | C6TBW8 | Dihydrodipicolinate reductase | 11 | 3 | 344 | 37.4 | 6.95 |
| 3.8521 | I1K0I8 | AT-hook motif nuclear-localized protein | 14 | 2 | 327 | 33.4 | 8.95 |
| 3.8462 | I1MDR1 | Transmembrane protein 87B | 6 | 2 | 516 | 58.2 | 6.46 |
| 3.8164 | C6TJ36 | Xyloglucan endotransglucosylase/hydrolase | 7 | 2 | 302 | 34.2 | 5.76 |
| −3.4548 | C6T2R8 | Glutathione S-transferase | 31 | 8 | 216 | 25.0 | 5.57 |
| −3.7336 | A0A0R0KV96 | Cytochrome P450 | 11 | 3 | 510 | 58.4 | 8.60 |
| −3.8375 | I1LYU9 | Arginine biosynthesis bifunctional protein | 14 | 3 | 464 | 48.5 | 6.37 |
| −4.2521 | Q42785 | Nonsymbiotic hemoglobin | 12 | 2 | 161 | 18.0 | 8.92 |
| −4.3069 | I1JE14 | Glycosyltransferase | 11 | 3 | 475 | 53.2 | 6.01 |
| −4.3759 | I1M4F8 | Uncharacterized protein | 12 | 2 | 302 | 32.9 | 5.29 |
| −4.4426 | A0A0R0F139 | PKS_ER domain-containing protein | 69 | 14 | 361 | 39.3 | 7.28 |
| −5.4892 | I1JY29 | Alcohol dehydrogenase | 27 | 6 | 381 | 41.2 | 6.00 |
| −5.6909 | Q9FQE8 | Glutathione S-transferase | 35 | 7 | 219 | 25.6 | 5.97 |
| −6.1355 | C6TMH1 | ZnMc domain-containing protein | 12 | 2 | 357 | 40.1 | 5.63 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Komatsu, S.; Kimura, T.; Rehman, S.U.; Yamaguchi, H.; Hitachi, K.; Tsuchida, K. Proteomic Analysis Reveals Salt-Tolerant Mechanism in Soybean Applied with Plant-Derived Smoke Solution. Int. J. Mol. Sci. 2023, 24, 13734. https://doi.org/10.3390/ijms241813734
Komatsu S, Kimura T, Rehman SU, Yamaguchi H, Hitachi K, Tsuchida K. Proteomic Analysis Reveals Salt-Tolerant Mechanism in Soybean Applied with Plant-Derived Smoke Solution. International Journal of Molecular Sciences. 2023; 24(18):13734. https://doi.org/10.3390/ijms241813734
Chicago/Turabian StyleKomatsu, Setsuko, Taiki Kimura, Shafiq Ur Rehman, Hisateru Yamaguchi, Keisuke Hitachi, and Kunihiro Tsuchida. 2023. "Proteomic Analysis Reveals Salt-Tolerant Mechanism in Soybean Applied with Plant-Derived Smoke Solution" International Journal of Molecular Sciences 24, no. 18: 13734. https://doi.org/10.3390/ijms241813734
APA StyleKomatsu, S., Kimura, T., Rehman, S. U., Yamaguchi, H., Hitachi, K., & Tsuchida, K. (2023). Proteomic Analysis Reveals Salt-Tolerant Mechanism in Soybean Applied with Plant-Derived Smoke Solution. International Journal of Molecular Sciences, 24(18), 13734. https://doi.org/10.3390/ijms241813734








