Revisiting Assessment of Computational Methods for Hi-C Data Analysis
Abstract
:1. Introduction
2. Results
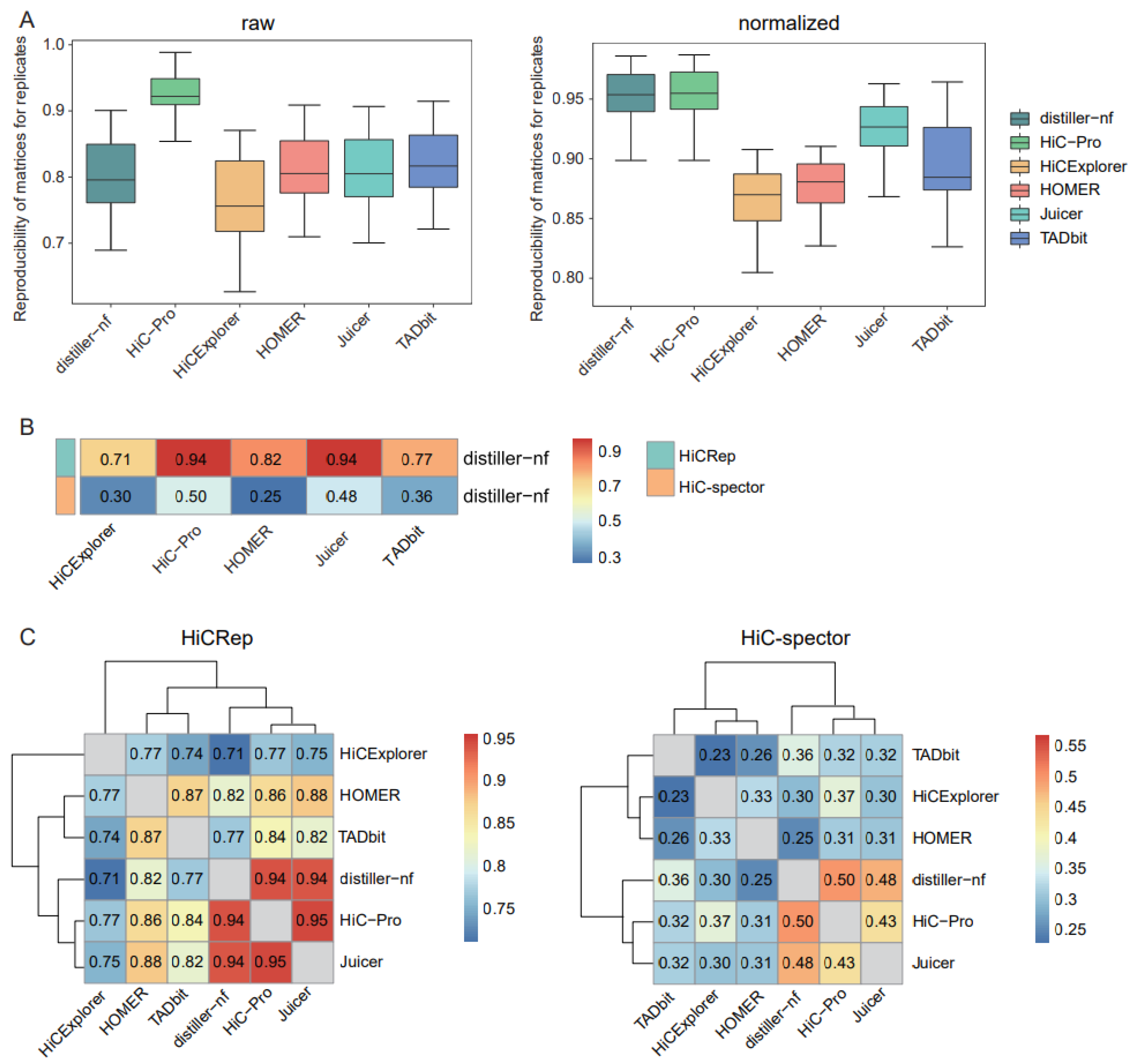
2.1. Data Processing
2.2. Comparison of TAD Identification Tools
2.3. Identification of PEIs
2.4. Identification of CIs
3. Discussion
4. Materials and Methods
4.1. Input Data
4.2. Methods for Data Preprocessing
4.2.1. Tool Usage
4.2.2. The Reproducibility of Hi-C Interaction Matrix
4.3. Methods for the Analysis of TADs
4.3.1. Tool Usage
4.3.2. The Concordance of TAD Intervals
4.3.3. The Concordance of TAD Boundaries
4.3.4. Manually Curated TADs
4.4. Methods for the Analysis of CIs and PEIs
4.4.1. Tool Usage
4.4.2. PEIs and Random PEIs
4.4.3. CRISPR Dataset for PEI Validation
4.4.4. Validated CIs Interactions
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lieberman-Aiden, E.; van Berkum, N.L.; Williams, L.; Imakaev, M.; Ragoczy, T.; Telling, A.; Amit, I.; Lajoie, B.R.; Sabo, P.J.; Dorschner, M.O.; et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 2009, 326, 289–293. [Google Scholar] [CrossRef] [PubMed]
- Dixon, J.R.; Selvaraj, S.; Yue, F.; Kim, A.; Li, Y.; Shen, Y.; Hu, M.; Liu, J.S.; Ren, B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 2012, 485, 376–380. [Google Scholar] [CrossRef]
- Cavalli, G.; Misteli, T. Functional implications of genome topology. Nat. Struct. Mol. Biol. 2013, 20, 290–299. [Google Scholar] [CrossRef] [PubMed]
- Jin, F.; Li, Y.; Dixon, J.R.; Selvaraj, S.; Ye, Z.; Lee, A.Y.; Yen, C.A.; Schmitt, A.D.; Espinoza, C.A.; Ren, B. A high-resolution map of the three-dimensional chromatin interactome in human cells. Nature 2013, 503, 290–294. [Google Scholar] [CrossRef]
- Rao, S.S.; Huntley, M.H.; Durand, N.C.; Stamenova, E.K.; Bochkov, I.D.; Robinson, J.T.; Sanborn, A.L.; Machol, I.; Omer, A.D.; Lander, E.S.; et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 2014, 159, 1665–1680. [Google Scholar] [CrossRef] [PubMed]
- Schmitt, A.D.; Hu, M.; Ren, B. Genome-wide mapping and analysis of chromosome architecture. Nat. Rev. Mol. Cell Biol. 2016, 17, 743–755. [Google Scholar] [CrossRef]
- Ay, F.; Noble, W.S. Analysis methods for studying the 3D architecture of the genome. Genome Biol. 2015, 16, 183. [Google Scholar] [CrossRef]
- Dali, R.; Blanchette, M. A critical assessment of topologically associating domain prediction tools. Nucleic Acids Res. 2017, 45, 2994–3005. [Google Scholar] [CrossRef]
- Forcato, M.; Nicoletti, C.; Pal, K.; Livi, C.M.; Ferrari, F.; Bicciato, S. Comparison of computational methods for Hi-C data analysis. Nat. Methods 2017, 14, 679–685. [Google Scholar] [CrossRef]
- Zufferey, M.; Tavernari, D.; Oricchio, E.; Ciriello, G. Comparison of computational methods for the identification of topologically associating domains. Genome Biol. 2018, 19, 217. [Google Scholar] [CrossRef]
- Aljogol, D.; Thompson, I.R.; Osborne, C.S.; Mifsud, B. Comparison of Capture Hi-C Analytical Pipelines. Front. Genet. 2022, 13, 786501. [Google Scholar] [CrossRef] [PubMed]
- Durand, N.C.; Shamim, M.S.; Machol, I.; Rao, S.S.; Huntley, M.H.; Lander, E.S.; Aiden, E.L. Juicer Provides a One-Click System for Analyzing Loop-Resolution Hi-C Experiments. Cell Syst. 2016, 3, 95–98. [Google Scholar] [CrossRef]
- Servant, N.; Varoquaux, N.; Lajoie, B.R.; Viara, E.; Chen, C.J.; Vert, J.P.; Heard, E.; Dekker, J.; Barillot, E. HiC-Pro: An optimized and flexible pipeline for Hi-C data processing. Genome Biol. 2015, 16, 259. [Google Scholar] [CrossRef]
- Ramírez, F.; Bhardwaj, V.; Arrigoni, L.; Lam, K.C.; Grüning, B.A.; Villaveces, J.; Habermann, B.; Akhtar, A.; Manke, T. High-resolution TADs reveal DNA sequences underlying genome organization in flies. Nat. Commun. 2018, 9, 189. [Google Scholar] [CrossRef]
- Heinz, S.; Benner, C.; Spann, N.; Bertolino, E.; Lin, Y.C.; Laslo, P.; Cheng, J.X.; Murre, C.; Singh, H.; Glass, C.K. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 2010, 38, 576–589. [Google Scholar] [CrossRef]
- Serra, F.; Baù, D.; Goodstadt, M.; Castillo, D.; Filion, G.J.; Marti-Renom, M.A. Automatic analysis and 3D-modelling of Hi-C data using TADbit reveals structural features of the fly chromatin colors. PLoS Comput. Biol. 2017, 13, e1005665. [Google Scholar] [CrossRef]
- Yang, T.; Zhang, F.; Yardımcı, G.G.; Song, F.; Hardison, R.C.; Noble, W.S.; Yue, F.; Li, Q. HiCRep: Assessing the reproducibility of Hi-C data using a stratum-adjusted correlation coefficient. Genome Res. 2017, 27, 1939–1949. [Google Scholar] [CrossRef] [PubMed]
- Filippova, D.; Patro, R.; Duggal, G.; Kingsford, C. Identification of alternative topological domains in chromatin. Algorithms Mol. Biol. AMB 2014, 9, 14. [Google Scholar] [CrossRef] [PubMed]
- Lévy-Leduc, C.; Delattre, M.; Mary-Huard, T.; Robin, S. Two-dimensional segmentation for analyzing Hi-C data. Bioinformatics 2014, 30, i386–i392. [Google Scholar] [CrossRef]
- Weinreb, C.; Raphael, B.J. Identification of hierarchical chromatin domains. Bioinformatics 2016, 32, 1601–1609. [Google Scholar] [CrossRef]
- An, L.; Yang, T.; Yang, J.; Nuebler, J.; Xiang, G.; Hardison, R.C.; Li, Q.; Zhang, Y. OnTAD: Hierarchical domain structure reveals the divergence of activity among TADs and boundaries. Genome Biol. 2019, 20, 282. [Google Scholar] [CrossRef]
- Crane, E.; Bian, Q.; McCord, R.P.; Lajoie, B.R.; Wheeler, B.S.; Ralston, E.J.; Uzawa, S.; Dekker, J.; Meyer, B.J. Condensin-driven remodelling of X chromosome topology during dosage compensation. Nature 2015, 523, 240–244. [Google Scholar] [CrossRef]
- Shin, H.; Shi, Y.; Dai, C.; Tjong, H.; Gong, K.; Alber, F.; Zhou, X.J. TopDom: An efficient and deterministic method for identifying topological domains in genomes. Nucleic Acids Res. 2016, 44, e70. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Li, G.; Zhang, M.Q.; Chen, Y. HiCDB: A sensitive and robust method for detecting contact domain boundaries. Nucleic Acids Res. 2018, 46, 11239–11250. [Google Scholar] [CrossRef]
- Despang, A.; Schöpflin, R.; Franke, M.; Ali, S.; Jerković, I.; Paliou, C.; Chan, W.L.; Timmermann, B.; Wittler, L.; Vingron, M.; et al. Functional dissection of the Sox9-Kcnj2 locus identifies nonessential and instructive roles of TAD architecture. Nat. Genet. 2019, 51, 1263–1271. [Google Scholar] [CrossRef]
- Ron, G.; Globerson, Y.; Moran, D.; Kaplan, T. Promoter-enhancer interactions identified from Hi-C data using probabilistic models and hierarchical topological domains. Nat. Commun. 2017, 8, 2237. [Google Scholar] [CrossRef]
- Sahin, M.; Wong, W.; Zhan, Y.; Van Deynze, K.; Koche, R.; Leslie, C.S. HiC-DC+ enables systematic 3D interaction calls and differential analysis for Hi-C and HiChIP. Nat. Commun. 2021, 12, 3366. [Google Scholar] [CrossRef]
- Cao, Y.; Chen, Z.; Chen, X.; Ai, D.; Chen, G.; McDermott, J.; Huang, Y.; Guo, X.; Han, J.J. Accurate loop calling for 3D genomic data with cLoops. Bioinformatics 2020, 36, 666–675. [Google Scholar] [CrossRef] [PubMed]
- Rowley, M.J.; Poulet, A.; Nichols, M.H.; Bixler, B.J.; Sanborn, A.L.; Brouhard, E.A.; Hermetz, K.; Linsenbaum, H.; Csankovszki, G.; Lieberman Aiden, E.; et al. Analysis of Hi-C data using SIP effectively identifies loops in organisms from C. elegans to mammals. Genome Res. 2020, 30, 447–458. [Google Scholar] [CrossRef]
- Mifsud, B.; Tavares-Cadete, F.; Young, A.N.; Sugar, R.; Schoenfelder, S.; Ferreira, L.; Wingett, S.W.; Andrews, S.; Grey, W.; Ewels, P.A.; et al. Mapping long-range promoter contacts in human cells with high-resolution capture Hi-C. Nat. Genet. 2015, 47, 598–606. [Google Scholar] [CrossRef] [PubMed]
- Kaul, A.; Bhattacharyya, S.; Ay, F. Identifying statistically significant chromatin contacts from Hi-C data with FitHiC2. Nat. Protoc. 2020, 15, 991–1012. [Google Scholar] [CrossRef]
- Fulco, C.P.; Nasser, J.; Jones, T.R.; Munson, G.; Bergman, D.T.; Subramanian, V.; Grossman, S.R.; Anyoha, R.; Doughty, B.R.; Patwardhan, T.A.; et al. Activity-by-contact model of enhancer-promoter regulation from thousands of CRISPR perturbations. Nat. Genet. 2019, 51, 1664–1669. [Google Scholar] [CrossRef] [PubMed]
- Marco-Sola, S.; Sammeth, M.; Guigó, R.; Ribeca, P. The GEM mapper: Fast, accurate and versatile alignment by filtration. Nat. Methods 2012, 9, 1185–1188. [Google Scholar] [CrossRef] [PubMed]
- Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv 2013, arXiv:1303.3997. [Google Scholar]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Abdennur, N.; Mirny, L.A. Cooler: Scalable storage for Hi-C data and other genomically labeled arrays. Bioinformatics 2019, 36, 311–316. [Google Scholar] [CrossRef] [PubMed]
- Imakaev, M.; Fudenberg, G.; McCord, R.P.; Naumova, N.; Goloborodko, A.; Lajoie, B.R.; Dekker, J.; Mirny, L.A. Iterative correction of Hi-C data reveals hallmarks of chromosome organization. Nat. Methods 2012, 9, 999–1003. [Google Scholar] [CrossRef]
- Yan, K.K.; Yardimci, G.G.; Yan, C.; Noble, W.S.; Gerstein, M. HiC-spector: A matrix library for spectral and reproducibility analysis of Hi-C contact maps. Bioinformatics 2017, 33, 2199–2201. [Google Scholar] [CrossRef] [PubMed]
- Yardımcı, G.G.; Ozadam, H.; Sauria, M.E.G.; Ursu, O.; Yan, K.K.; Yang, T.; Chakraborty, A.; Kaul, A.; Lajoie, B.R.; Song, F.; et al. Measuring the reproducibility and quality of Hi-C data. Genome Biol. 2019, 20, 57. [Google Scholar] [CrossRef] [PubMed]
- Quinlan, A.R.; Hall, I.M. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 2010, 26, 841–842. [Google Scholar] [CrossRef] [PubMed]
- Sanyal, A.; Lajoie, B.R.; Jain, G.; Dekker, J. The long-range interaction landscape of gene promoters. Nature 2012, 489, 109–113. [Google Scholar] [CrossRef] [PubMed]






| Method | Percentage of Aligned Read Pairs (%) | Percentage of Valid Mapped Reads (%) | |
|---|---|---|---|
| bwa | distiller-nf | 87.21 | 72.37 |
| Juicer | 94.15 | 71.58 | |
| Bowtie2 | HiC-Pro | 89.18 | 58.38 |
| HiCExplorer | 62.57 | 40.34 | |
| HOMER | 73.29 | 60.96 | |
| GEM | TADbit | 80.67 | 71.35 |
| Cell Type | Restriction Enzyme | SRA Accession Number |
|---|---|---|
| GM12878 | MboI | SRR1658572 |
| GM12878 | MboI | SRR1658592 |
| GM12878 | MboI | SRR1658593 |
| GM12879 | MboI | SRR1658594, SRR1658595 |
| GM12880 | MboI | SRR1658596, SRR1658597 |
| GM12878 | MboI | SRR1658598 |
| GM12878 | MboI | SRR1658599 |
| GM12878 | MboI | SRR1658600 |
| GM12878 | MboI | SRR1658601 |
| GM12878 | MboI | SRR1658602 |
| GM12878 | MboI | SRR1658603 |
| K562 (CCL-243) | MboI | SRR1658693 |
| K562 (CCL-243) | MboI | SRR1658694 |
| K562 (CCL-243) | MboI | SRR1658695 |
| K562 (CCL-243) | MboI | SRR1658696 |
| K562 (CCL-243) | MboI | SRR1658697 |
| K562 (CCL-243) | MboI | SRR1658698 |
| K562 (CCL-243) | MboI | SRR1658699 |
| K562 (CCL-243) | MboI | SRR1658700 |
| K562 (CCL-243) | MboI | SRR1658701 |
| K562 (CCL-243) | MboI | SRR1658702 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, J.; Zhu, X.; Wang, R.; Li, M.; Tang, Q. Revisiting Assessment of Computational Methods for Hi-C Data Analysis. Int. J. Mol. Sci. 2023, 24, 13814. https://doi.org/10.3390/ijms241813814
Yang J, Zhu X, Wang R, Li M, Tang Q. Revisiting Assessment of Computational Methods for Hi-C Data Analysis. International Journal of Molecular Sciences. 2023; 24(18):13814. https://doi.org/10.3390/ijms241813814
Chicago/Turabian StyleYang, Jing, Xingxing Zhu, Rui Wang, Mingzhou Li, and Qianzi Tang. 2023. "Revisiting Assessment of Computational Methods for Hi-C Data Analysis" International Journal of Molecular Sciences 24, no. 18: 13814. https://doi.org/10.3390/ijms241813814






