Investigation into Molecular Brain Aging in Senescence-Accelerated Mouse (SAM) Model Employing Whole Transcriptomic Analysis in Search of Potential Molecular Targets for Therapeutic Interventions
Abstract
:1. Introduction
2. Results
2.1. Characterization of Gene Expression Profiles in SAMR1 and SAMP8 Mice
2.2. Overview of Biological Events in SAMR1 and SAMP8 Mice
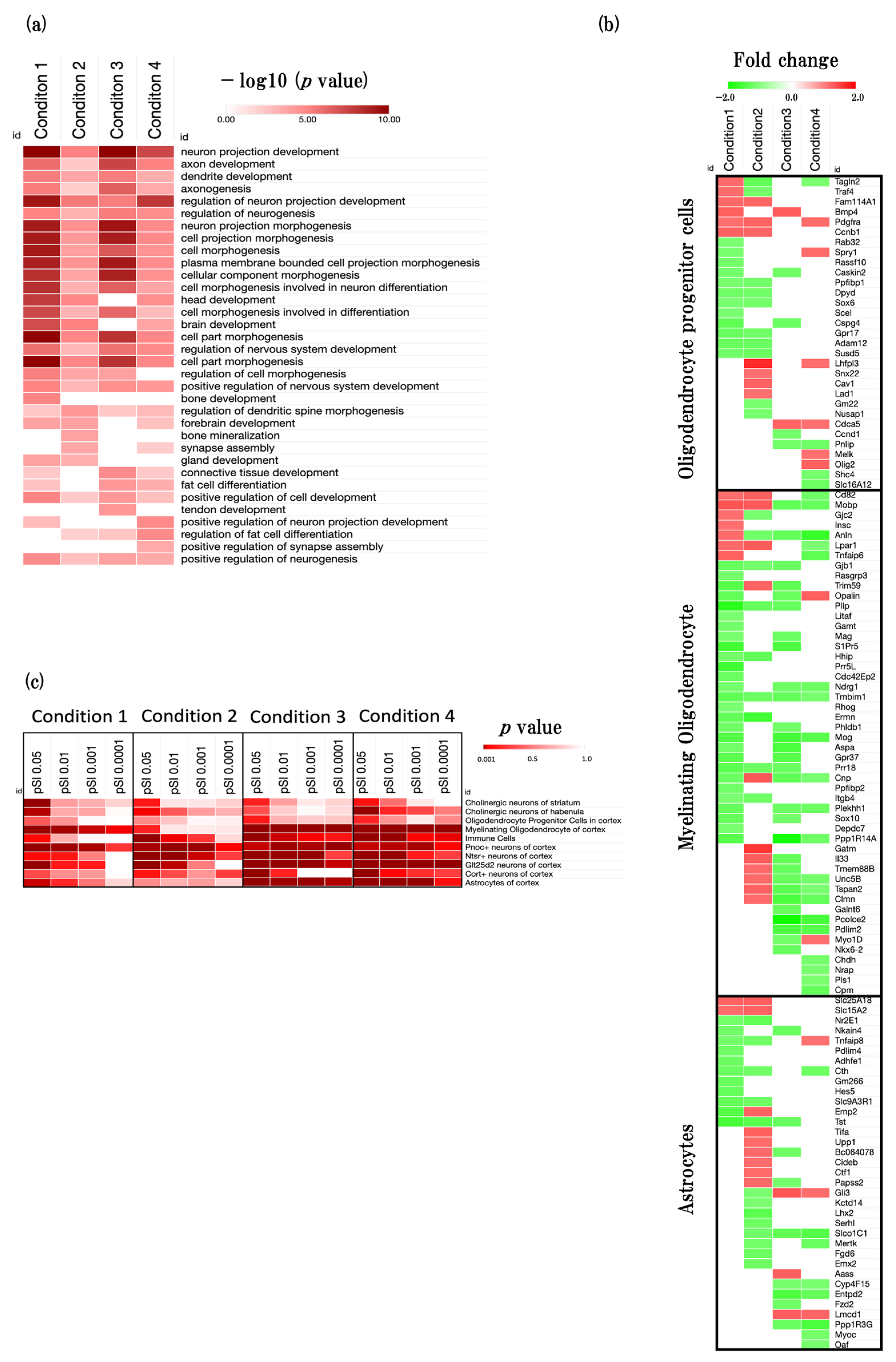
2.3. Effects of Aging on Neurogenesis
2.3.1. Overview of the Neurodevelopment-Related GOBPs
2.3.2. Cell-Type Changes during the Aging Process
2.4. Effects of Aging on Synapse
2.4.1. Overview of the Synapse-Related GOBPs
2.4.2. Functional Changes during Aging Process of Synapse and Related Predicted Genes
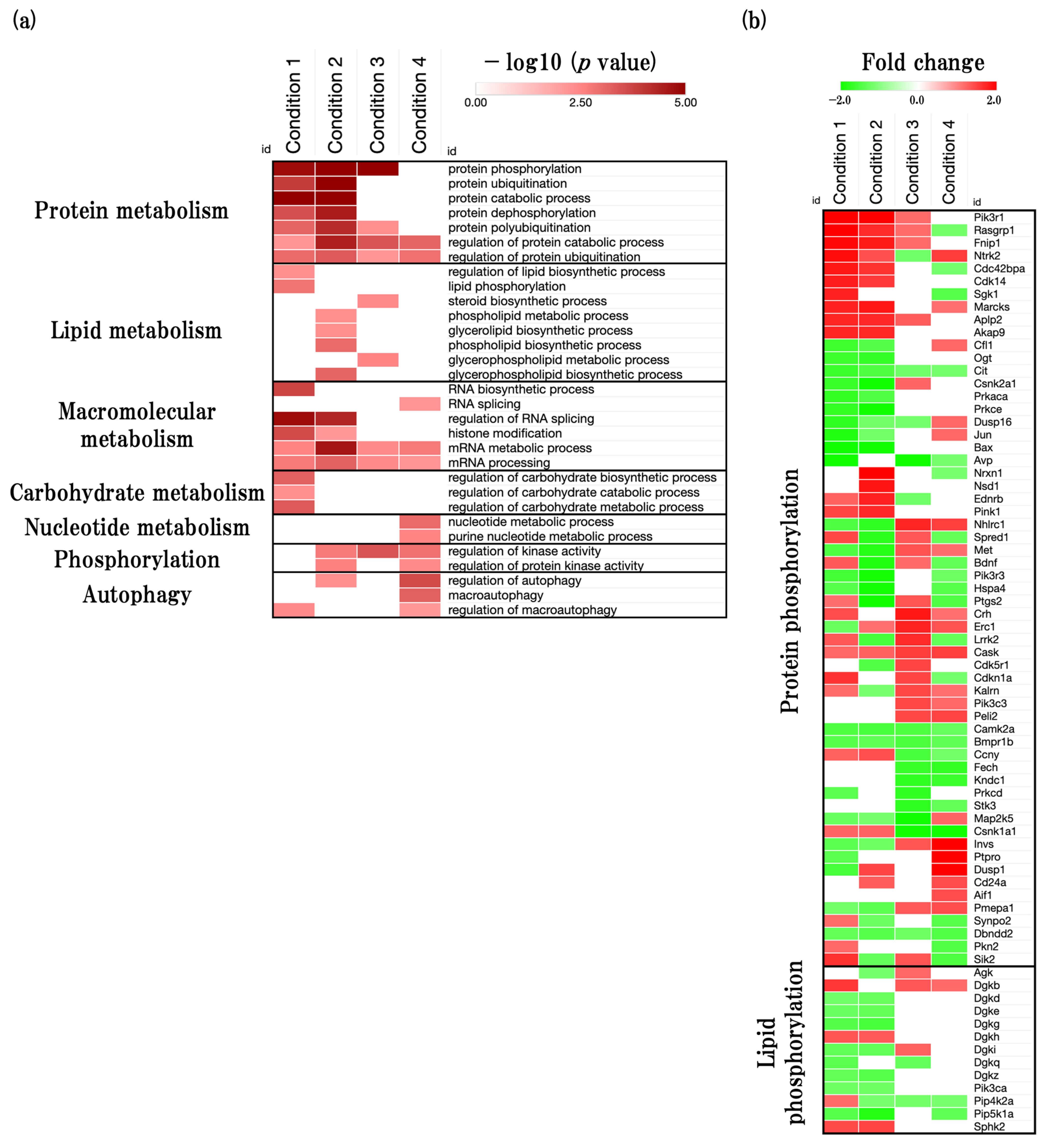
2.5. Effects of Aging on Neuro-Metabolism
2.5.1. Overview of the Neurometabolism Related GOBPs
2.5.2. Functional Changes during Aging Process of Neurometabolism and Related Predicted Gene
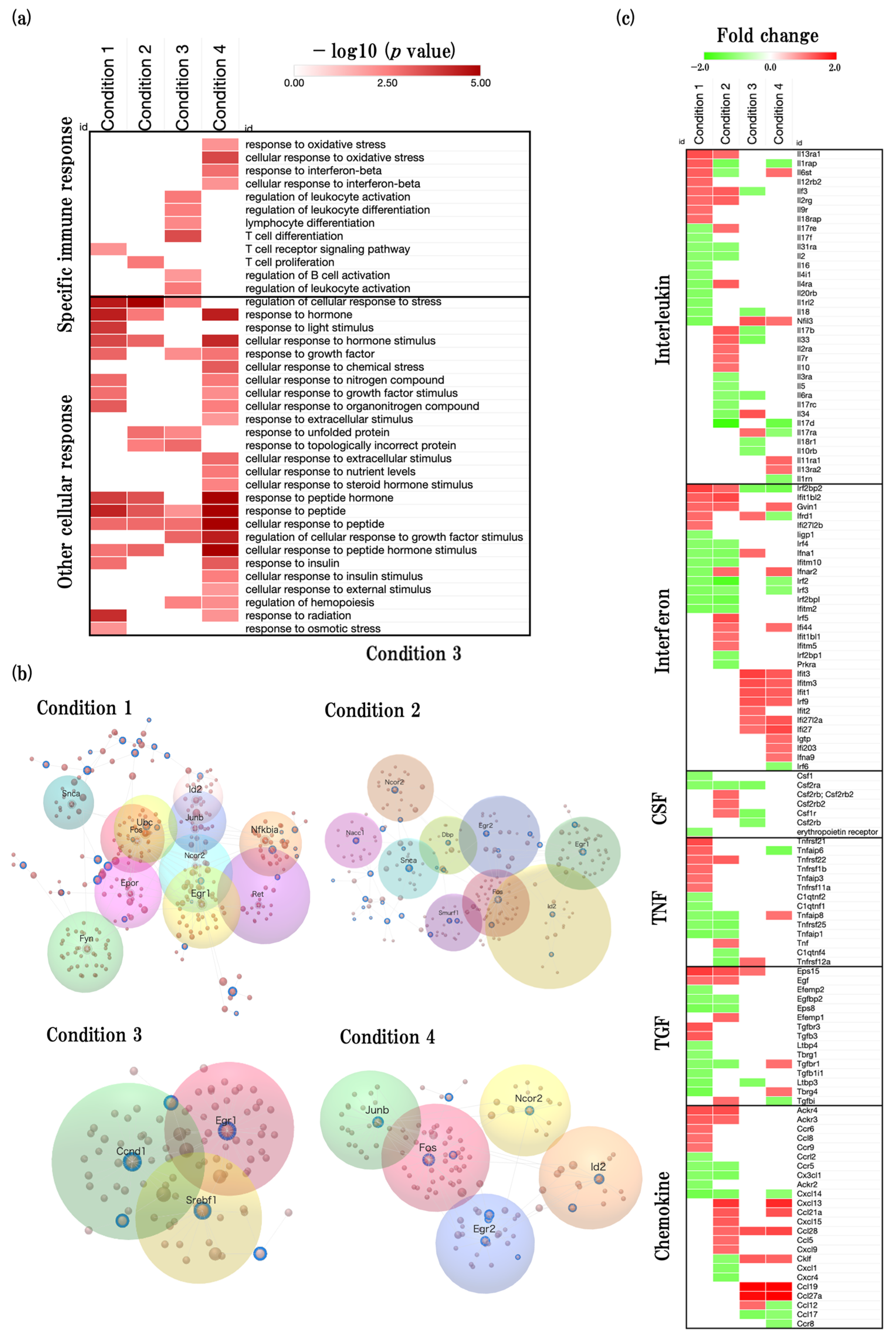
2.6. Effects of Aging on Neuroinflammation
2.6.1. Overview of the Neuroinflammation-Related GOBPs
2.6.2. Functional Changes during Aging Process of Neuroinflammation and Related Predicted Genes
2.7. Protein–Protein Interaction Network Analysis
2.8. Gene-Set Perturbation Analysis
3. Discussion
4. Materials and Methods
4.1. Animals
4.2. RNA Extraction and Quantification
4.3. Microarray Experiment
4.4. Data Processing
4.5. Date Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Vaupel, J.W. Biodemography of human ageing. Nature 2010, 464, 536–542. [Google Scholar] [CrossRef]
- Yankner, B.A.; Lu, T.; Loerch, P. The aging brain. Annu. Rev. Pathol. Mech. Dis. 2008, 3, 41–66. [Google Scholar] [CrossRef]
- Grimm, A.; Eckert, A. Brain aging and neurodegeneration: From a mitochondrial point of view. J. Neurochem. 2017, 143, 418–431. [Google Scholar] [CrossRef] [PubMed]
- Mattson, M.P.; Arumugam, T.V. Hallmarks of brain aging: Adaptive and pathological modification by metabolic states. Cell Metab. 2018, 27, 1176–1199. [Google Scholar] [CrossRef]
- Barrientos, R.; Kitt, M.; Watkins, L.; Maier, S. Neuroinflammation in the normal aging hippocampus. Neuroscience 2015, 309, 84–99. [Google Scholar] [CrossRef] [PubMed]
- Harada, C.N.; Love, M.C.N.; Triebel, K.L. Normal cognitive aging. Clin. Geriatr. Med. 2013, 29, 737–752. [Google Scholar] [CrossRef]
- Blazer, D.G.; Yaffe, K.; Karlawish, J. Cognitive aging: A report from the Institute of Medicine. JAMA 2015, 313, 2121–2122. [Google Scholar] [CrossRef] [PubMed]
- Gray, J.A.; Roth, B.L. The pipeline and future of drug development in schizophrenia. Mol. Psychiatry 2007, 12, 904–922. [Google Scholar] [CrossRef]
- Van Dam, D.; De Deyn, P.P. Animal models in the drug discovery pipeline for Alzheimer’s disease. Br. J. Pharmacol. 2011, 164, 1285–1300. [Google Scholar] [CrossRef]
- Takeda, T.; Hosokawa, M.; Higuchi, K. Senescence-accelerated mouse (SAM): A novel murine model of senescence. Exp. Gerontol. 1997, 32, 105–109. [Google Scholar] [CrossRef] [PubMed]
- Flood, J.F.; Morley, J.E. Learning and memory in the SAMP8 mouse. Neurosci. Biobehav. Rev. 1997, 22, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Takeda, T. Senescence-accelerated mouse (SAM): A biogerontological resource in aging research. Neurobiol. Aging 1999, 20, 105–110. [Google Scholar] [CrossRef]
- Canudas, A.M.; Gutierrez-Cuesta, J.; Rodríguez, M.I.; Acuña-Castroviejo, D.; Sureda, F.X.; Camins, A.; Pallàs, M. Hyperphosphorylation of microtubule-associated protein tau in senescence-accelerated mouse (SAM). Mech. Ageing Dev. 2005, 126, 1300–1304. [Google Scholar] [CrossRef] [PubMed]
- Manich, G.; Mercader, C.; Del Valle, J.; Duran-Vilaregut, J.; Camins, A.; Pallàs, M.; Vilaplana, J.; Pelegri, C. Characterization of amyloid-β granules in the hippocampus of SAMP8 mice. J. Alzheimers Dis. 2011, 25, 535–546. [Google Scholar] [CrossRef]
- Zhang, S.; Li, H.; Zheng, L.; Li, H.; Feng, C.; Zhang, W. Identification of functional tRNA-derived fragments in senescence-accelerated mouse prone 8 brain. Aging 2019, 11, 10485. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Zhu, D.; Li, H.; Li, H.; Feng, C.; Zhang, W. Characterization of circRNA-associated-ceRNA networks in a senescence-accelerated mouse prone 8 brain. Mol. Ther. 2017, 25, 2053–2061. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.-C.; Lu, G.; Chan, C.-Y.; Chen, Y.; Wang, H.; Yew, D.T.-W.; Feng, Z.-T.; Kung, H.-F. Microarray profile of brain aging-related genes in the frontal cortex of SAMP8. J. Mol. Neurosci. 2010, 41, 12–16. [Google Scholar] [CrossRef]
- Piñero, J.; Bravo, À.; Queralt-Rosinach, N.; Gutiérrez-Sacristán, A.; Deu-Pons, J.; Centeno, E.; García-García, J.; Sanz, F.; Furlong, L.I. DisGeNET: A comprehensive platform integrating information on human disease-associated genes and variants. Nucleic Acids Res. 2016, 45, D833–D839. [Google Scholar] [CrossRef]
- Han, H.; Shim, H.; Shin, D.; Shim, J.E.; Ko, Y.; Shin, J.; Kim, H.; Cho, A.; Kim, E.; Lee, T. TRRUST: A reference database of human transcriptional regulatory interactions. Sci. Rep. 2015, 5, 11432. [Google Scholar] [CrossRef]
- Pan, J.; Ma, N.; Yu, B.; Zhang, W.; Wan, J. Transcriptomic profiling of microglia and astrocytes throughout aging. J. Neuroinflamm. 2020, 17, 97. [Google Scholar] [CrossRef]
- Miyamoto, M.; Kiyota, Y.; Yamazaki, N.; Nagaoka, A.; Matsuo, T.; Nagawa, Y.; Takeda, T. Age-related changes in learning and memory in the senescence-accelerated mouse (SAM). Physiol. Behav. 1986, 38, 399–406. [Google Scholar] [CrossRef]
- Akiguchi, I.; Pallàs, M.; Budka, H.; Akiyama, H.; Ueno, M.; Han, J.; Yagi, H.; Nishikawa, T.; Chiba, Y.; Sugiyama, H. SAMP8 mice as a neuropathological model of accelerated brain aging and dementia: Toshio Takeda’s legacy and future directions. Neuropathology 2017, 37, 293–305. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Chen, H.; Sun, X. Age-related spatial cognitive impairment is correlated with a decrease in ChAT in the cerebral cortex, hippocampus and forebrain of SAMP8 mice. Neurosci. Lett. 2009, 454, 212–217. [Google Scholar] [CrossRef]
- Yanai, S.; Endo, S. Early onset of behavioral alterations in senescence-accelerated mouse prone 8 (SAMP8). Behav. Brain Res. 2016, 308, 187–195. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.D.; Bordey, A. The astrocyte odyssey. Prog. Neurobiol. 2008, 86, 342–367. [Google Scholar] [CrossRef] [PubMed]
- Saab, A.S.; Nave, K.-A. Myelin dynamics: Protecting and shaping neuronal functions. Curr. Opin. Neurobiol. 2017, 47, 104–112. [Google Scholar] [CrossRef]
- Fancy, S.P.; Zhao, C.; Franklin, R.J. Increased expression of Nkx2.2 and Olig2 identifies reactive oligodendrocyte progenitor cells responding to demyelination in the adult CNS. Mol. Cell. Neurosci. 2004, 27, 247–254. [Google Scholar] [CrossRef] [PubMed]
- Buée, L.; Bussière, T.; Buée-Scherrer, V.; Delacourte, A.; Hof, P.R. Tau protein isoforms, phosphorylation and role in neurodegenerative disorders. Brain Res. Rev. 2000, 33, 95–130. [Google Scholar] [CrossRef]
- Grundke-Iqbal, I.; Iqbal, K.; Tung, Y.-C.; Quinlan, M.; Wisniewski, H.M.; Binder, L.I. Abnormal phosphorylation of the microtubule-associated protein tau (tau) in Alzheimer cytoskeletal pathology. Proc. Natl. Acad. Sci. USA 1986, 83, 4913–4917. [Google Scholar] [CrossRef] [PubMed]
- Braak, H.; Braak, E. Staging of Alzheimer’s disease-related neurofibrillary changes. Neurobiol. Aging 1995, 16, 271–278. [Google Scholar] [CrossRef]
- Wegmann, S.; Biernat, J.; Mandelkow, E. A current view on Tau protein phosphorylation in Alzheimer’s disease. Curr. Opin. Neurobiol. 2021, 69, 131–138. [Google Scholar] [CrossRef] [PubMed]
- Trimbuch, T.; Beed, P.; Vogt, J.; Schuchmann, S.; Maier, N.; Kintscher, M.; Breustedt, J.; Schuelke, M.; Streu, N.; Kieselmann, O. Synaptic PRG-1 modulates excitatory transmission via lipid phosphate-mediated signaling. Cell 2009, 138, 1222–1235. [Google Scholar] [CrossRef]
- Vogt, J.; Yang, J.W.; Mobascher, A.; Cheng, J.; Li, Y.; Liu, X.; Baumgart, J.; Thalman, C.; Kirischuk, S.; Unichenko, P. Molecular cause and functional impact of altered synaptic lipid signaling due to a prg—1 gene SNP. EMBO Mol. Med. 2016, 8, 25–38. [Google Scholar] [CrossRef] [PubMed]
- Giusto, N.; Salvador, G.; Castagnet, P.; Pasquare, S.; Ilincheta de Boschero, M. Age-associated changes in central nervous system glycerolipid composition and metabolism. Neurochem. Res. 2002, 27, 1513–1523. [Google Scholar] [CrossRef] [PubMed]
- Zheng, P. Neuroactive steroid regulation of neurotransmitter release in the CNS: Action, mechanism and possible significance. Prog. Neurobiol. 2009, 89, 134–152. [Google Scholar] [CrossRef]
- Aman, Y.; Schmauck-Medina, T.; Hansen, M.; Morimoto, R.I.; Simon, A.K.; Bjedov, I.; Palikaras, K.; Simonsen, A.; Johansen, T.; Tavernarakis, N. Autophagy in healthy aging and disease. Nat. Aging 2021, 1, 634–650. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q.; Qiang, J.; Gu, P.; Wang, Y.; Geng, Y.; Wang, M. Age-related autophagy alterations in the brain of senescence accelerated mouse prone 8 (SAMP8) mice. Exp. Gerontol. 2011, 46, 533–541. [Google Scholar] [CrossRef] [PubMed]
- Thiery, J.P. Epithelial–mesenchymal transitions in development and pathologies. Curr. Opin. Cell Biol. 2003, 15, 740–746. [Google Scholar] [CrossRef]
- Krabbe, C.; Zimmer, J.; Meyer, M. Neural transdifferentiation of mesenchymal stem cells—A critical review. APMIS 2005, 113, 831–844. [Google Scholar] [CrossRef]
- Hetz, C.; Zhang, K.; Kaufman, R.J. Mechanisms, regulation and functions of the unfolded protein response. Nat. Rev. Mol. Cell Biol. 2020, 21, 421–438. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Kaufman, R.J. Protein misfolding in the endoplasmic reticulum as a conduit to human disease. Nature 2016, 529, 326–335. [Google Scholar] [CrossRef] [PubMed]
- Wallace, D.C. Mitochondrial genetics: A paradigm for aging and degenerative diseases? Science 1992, 256, 628–632. [Google Scholar] [CrossRef] [PubMed]
- Boland, M.L.; Chourasia, A.H.; Macleod, K.F. Mitochondrial dysfunction in cancer. Front. Oncol. 2013, 3, 292. [Google Scholar] [CrossRef] [PubMed]
- Karsenti, E.; Vernos, I. The mitotic spindle: A self-made machine. Science 2001, 294, 543–547. [Google Scholar] [CrossRef] [PubMed]
- Yasutis, K.; Kozminski, K. Cell cycle checkpoint regulators reach a zillion. Cell Cycle 2013, 12, 1501–1509. [Google Scholar] [CrossRef]
- Ali, A.T.; Hochfeld, W.E.; Myburgh, R.; Pepper, M.S. Adipocyte and adipogenesis. Eur. J. Cell Biol. 2013, 92, 229–236. [Google Scholar] [CrossRef]
- Laplante, M.; Sabatini, D.M. mTOR signaling at a glance. J. Cell Sci. 2009, 122, 3589–3594. [Google Scholar] [CrossRef] [PubMed]
- Saxton, R.A.; Sabatini, D.M. mTOR signaling in growth, metabolism, and disease. Cell 2017, 168, 960–976. [Google Scholar] [CrossRef] [PubMed]
- Alers, S.; Löffler, A.S.; Wesselborg, S.; Stork, B. Role of AMPK-mTOR-Ulk1/2 in the regulation of autophagy: Cross talk, shortcuts, and feedbacks. Mol. Cell. Biol. 2012, 32, 2–11. [Google Scholar] [CrossRef] [PubMed]
- Eltzschig, H.K.; Carmeliet, P. Hypoxia and inflammation. N. Engl. J. Med. 2011, 364, 656–665. [Google Scholar] [CrossRef]
- Bottazzi, B.; Riboli, E.; Mantovani, A. Aging, inflammation and cancer. In Seminars in Immunology; Elsevier: Amsterdam, The Netherlands, 2018; pp. 74–82. [Google Scholar]
- Goedhart, J.; Luijsterburg, M.S. VolcaNoseR is a web app for creating, exploring, labeling and sharing volcano plots. Sci. Rep. 2020, 10, 20560. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef] [PubMed]
- Dai, Y.; Hu, R.; Liu, A.; Cho, K.S.; Manuel, A.M.; Li, X.; Dong, X.; Jia, P.; Zhao, Z. WebCSEA: Web-based cell-type-specific enrichment analysis of genes. Nucleic Acids Res. 2022, 50, W782–W790. [Google Scholar] [CrossRef]
- Koopmans, F.; van Nierop, P.; Andres-Alonso, M.; Byrnes, A.; Cijsouw, T.; Coba, M.P.; Cornelisse, L.N.; Farrell, R.J.; Goldschmidt, H.L.; Howrigan, D.P. SynGO: An evidence-based, expert-curated knowledge base for the synapse. Neuron 2019, 103, 217–234.e4. [Google Scholar] [CrossRef]
- Zhou, G.; Pang, Z.; Lu, Y.; Ewald, J.; Xia, J. OmicsNet 2.0: A web-based platform for multi-omics integration and network visual analytics. Nucleic Acids Res. 2022, 50, W527–W533. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.; Benner, M.J.; Hancock, R.E. NetworkAnalyst-integrative approaches for protein–protein interaction network analysis and visual exploration. Nucleic Acids Res. 2014, 42, W167–W174. [Google Scholar] [CrossRef]
- Guo, S.; Xu, Z.; Dong, X.; Hu, D.; Jiang, Y.; Wang, Q.; Zhang, J.; Zhou, Q.; Liu, S.; Song, W. GPSAdb: A comprehensive web resource for interactive exploration of genetic perturbation RNA-seq datasets. Nucleic Acids Res. 2023, 51, D964–D968. [Google Scholar] [CrossRef] [PubMed]





| Condition | Comparison | Explanation |
|---|---|---|
| Condition 1 | SAMR1: 1 year old vs. 16 weeks old | physiological aging |
| Condition 2 | SAMP8: 1 year old vs. 16 weeks old | accelerated aging |
| Condition 3 | 16 weeks old: SAMP8 vs. SAMR1 | early events in an accelerated aging |
| Condition 4 | 1 year old: SAMP8 vs. SAMR1 | late events in an accelerated aging |
| Gene Symbol | Description | Fold Change | p Value | Biological Functions |
|---|---|---|---|---|
| Condition 1 | ||||
| Hnrnpab | heterogeneous nuclear ribonucleoprotein A/B | 3.98 | 2.0 × 10−4 | DNA binding, RNA binding, protein binding, nucleus, epithelial to mesenchymal transition, and dendrite |
| Pik3r1 | phosphatidylinositol 3-kinase, regulatory subunit, polypeptide 1 (p85 alpha) | 3.28 | 1.74 × 10−5 | kinase activator activity, protein binding, ATPase binding, insulin receptor signaling pathway, and B cell differentiation |
| Lrrtm2 | leucine rich repeat transmembrane neuronal 2 | 2.81 | 2.24 × 10−5 | synapse organization, excitatory synapse, glutamatergic synapse, GABA-ergic synapse |
| Ano3 | anoctamin 3 | 2.78 | 7.93 × 10−5 | chloride channel activity, lipid transport, and calcium-activated phosphatidylcholine scrambling |
| Homer1 | homer homolog 1 (Drosophila) | 2.77 | 9.0 × 10−4 | protein binding, molecular adaptor activity, dendrite, structural constituent of postsynapse, postsynaptic density, and excitatory synapse |
| Zcchc24 | zinc finger, CCHC domain containing 24 | 2.61 | 2.64 × 10−6 | nucleic acid binding |
| Rasgrp1 | RAS guanyl releasing protein 1 | 2.55 | 3.0 × 10−4 | calcium ion binding, inflammatory response to antigenic stimulus, natural killer cell activation, cell differentiation, T cell activation, and B cell activation |
| Klhl24 | kelch-like 24 | 2.55 | 3.5 × 10−3 | protein ubiquitination, regulation of kainate selective glutamate receptor activity, cytoplasm, andcell projection |
| Etnk1 | ethanolamine kinase 1 | 2.48 | 3.54 × 10−6 | nucleotide binding, ethanolamine kinase activity, ATP binding, kinase activity, lipid metabolic process, biosynthetic process, and phosphorylation |
| Map2 | microtubule-associated protein 2 | 2.44 | 1.69 × 10−2 | Protein binding, microtubule binding, axon genesis, dendrite development, and postsynapse |
| Condition 2 | ||||
| Ttr | transthyretin | 10.94 | 3.5 × 10−3 | protein binding, purine nucleobase metabolic process, extracellular region, and extracellular space |
| Apold1 | apolipoprotein L domain containing 1 | 1.82 | 5.72 × 10−5 | lipid binding, angiogenesis, and regulation of endothelial cell differentiation |
| Cyp2c29 | cytochrome P450, family 2, subfamily c, polypeptide 29 | 1.76 | 3.36 × 10−5 | steroid hydroxylase activity, long-chain fatty acid omega-1 hydroxylase activity, lipid metabolic process, cytoplasm, and endoplasmic reticulum |
| Lncpint | long non-protein coding RNA, Trp53 induced transcript | 1.74 | 2.0 × 10−4 | skeletal system development, tissue development, and adipose tissue development |
| Rbm12b1 | RNA binding motif protein 12 B1 | 1.66 | 2.66 × 10−5 | nucleic acid binding, RNA binding, regulation of RNA splicing, and nucleoplasm |
| Itih3 | inter-alpha trypsin inhibitor, heavy chain 3 | 1.62 | 1.4 × 10−3 | peptidase inhibitor activity, extracellular region, and collagen-containing extracellular matrix |
| Gatm | glycine amidinotransferase (L-arginine:glycine amidinotransferase) | 1.6 | 5.0 × 10−4 | amidinotransferase activity, creatine biosynthetic process, learning or memory, and mitochondrion |
| Btg2 | B cell translocation gene 2, anti-proliferative | 1.59 | 1.06 × 10−2 | protein binding, DNA damage response, neuron differentiation, and apoptotic process |
| Defa15 | Defensin, alpha,15 | 1.53 | 2.2 × 10−3 | defense response and extracellular region |
| Fos | FBJ osteosarcoma oncogene | 1.52 | 4.4 × 10−3 | DNA binding and cellular response to reactive oxygen species |
| Condition 3 | ||||
| Prss22 | protease, serine 22 | 4.69 | 9.43 × 10−6 | peptidase activity and extracellular space |
| Dctn3 | dynactin 3 | 2.91 | 7.96 × 10−5 | cell cycle, cell division, kinetochore, nucleolus, and cytosol |
| Adat2 | adenosine deaminase, tRNA-specific 2 | 2.84 | 2.39 × 10−5 | catalytic activity and tRNA processing |
| Vps52 | vacuolar protein sorting 52 (yeast) | 2.82 | 8.21 × 10−5 | protein targeting, Golgi to vacuole transport, protein transport, endosome |
| Rpp25l | ribonuclease P/MRP 25 subunit-like | 1.91 | 1.7 × 10−3 | nucleic acid binding, RNA binding, and tRNA 5′-leader removal |
| Hspa1b | heat shock protein 1B | 1.91 | 2.4 × 10−3 | protease binding, ATP binding, protein folding, nucleus, mitochondrion, and cell body |
| Abcb1a | ATP-binding cassette, sub-family B (MDR/TAP), member 1A | 1.82 | 2.0 × 10−4 | nucleotide binding, ATP binding, and G2/M transition of mitotic cell cycle |
| Klf10 | Kruppel-like factor 10 | 1.81 | 2.9 × 10−3 | DNA binding, protein binding, and rhythmic process |
| Crh | corticotropin releasing hormone | 1.81 | 5.5 × 10−3 | positive regulation of protein phosphorylation, inflammatory response, and learning and memory |
| Vip | vasoactive intestinal polypeptide | 1.76 | 2.0 × 10−4 | signaling receptor binding and learning and memory |
| Condition 4 | ||||
| Prss22 | protease, serine 22 | 6.1 | 2.59 × 10−7 | peptidase activity |
| Galc | galactosylceramidase | 5.06 | 4.3 × 10−6 | galactosylceramidase activity, lipid metabolic process, myelination, and mitochondrion |
| Vps41 | vacuolar protein sorting 41 (yeast) | 4.91 | 2.08 × 10−8 | protein binding, microtubule binding, autophagy, protein transport, cytoplasm, and endosome |
| Abcb1a | ATP-binding cassette, sub-family B (MDR/TAP), member 1A | 4.72 | 9.37 × 10−7 | nucleotide binding, ATP binding, transmembrane transporter activity, G2/M transition of mitotic cell cycle, phospholipid translocation, and cytoplasm |
| Pigo | phosphatidylinositol glycan anchor biosynthesis, class O | 4.38 | 4.08 × 10−7 | protein binding, transferase activity, endoplasmic reticulum, and membrane |
| Soga3 | SOGA family member 3 | 4.28 | 5.69 × 10−8 | regulation of autophagy, extracellular space, and membrane |
| Cdhr1 | cadherin-related family member 1 | 2.92 | 3.37 × 10−2 | calcium ion binding, protein binding, cell adhesion, cell-cell adhesion, and membrane |
| Npas4 | neuronal PAS domain protein 4 | 2.86 | 1.63 × 10−2 | DNA binding, protein binding, learning, cell differentiation, and inhibitory synapse assembly |
| Btg2 | B cell translocation gene 2, anti-proliferative | 2.81 | 3.2 × 10−3 | protein binding, neuron differentiation, and apoptotic process |
| Dctn3 | dynactin 3 | 2.75 | 2.24 × 10−6 | cell cycle, cell division, kinetochore, and cytosol |
| Gene Symbol | Description | Fold Change | p Value | Biological Functions |
|---|---|---|---|---|
| Condition 1 | ||||
| Pmch | pro-melanin-concentrating hormone | −4.88 | 1.29 × 10−2 | signaling receptor binding, positive regulation of cytosolic calcium ion concentration, chemical synaptic transmission, and dopaminergic |
| Avp | arginine vasopressin | −4.23 | 9.3 × 10−3 | protein kinase activity, positive regulation of cytosolic calcium ion concentration, locomotory behavior, negative regulation of apoptotic process, ERK1 and ERK2 cascade, and dendrite |
| Atp1a3 | ATPase, Na+/K+ transporting, alpha 3 polypeptide | −2.79 | 8.0 × 10−4 | nucleotide binding, amyloid-beta binding, transporter activity, potassium ion transport, memory, intracellular potassium ion homeostasis, myelin sheath, and synapse |
| Npas4 | neuronal PAS domain protein 4 | −2.4 | 2.58 × 10−5 | DNA binding, protein binding, cell differentiation, regulation of synaptic transmission, GABAergic, excitatory postsynaptic potential, inhibitory postsynaptic potential, and nucleus |
| Fjx1 | four jointed box 1 (Drosophila) | −2.28 | 2.0 × 10−4 | cell–cell signaling, extracellular region, and extracellular space |
| Slc17a6 | solute carrier family 17 (sodium-dependent inorganic phosphate cotransporter), member 6 | −2.16 | 1.93 × 10−2 | chloride channel activity, neurotransmitter transmembrane transporter activity, neurotransmitter transport, synaptic transmission, glutamatergic, and synaptic vesicle |
| Bax | BCL2-associated X protein | −2.15 | 8.65 × 10−5 | protein binding, channel activity, leukocyte homeostasis, T cell homeostatic proliferation, B cell homeostasis, myeloid cell homeostasis, apoptotic process, and mitochondrial fusion |
| Nell2 | NEL-like 2 | −2.12 | 1.15 × 10−5 | protein kinase C binding, neuron cellular homeostasis, extracellular region, and dendrite |
| Junb | jun B proto-oncogene | −2.11 | 7.0 × 10−3 | DNA binding, transcription factor binding, osteoblast differentiation, cell differentiation, regulation of cell cycle, nucleus, and nucleoplasm |
| Cabp7 | calcium binding protein 7 | −2.1 | 3.2 × 10−2 | calcium ion binding and Golgi apparatus |
| Condition 2 | ||||
| Homer1 | homer homolog 1 (Drosophila) | −3.52 | 7.0 × 10−4 | signaling receptor binding, protein binding, molecular adaptor activity, structural constituent of postsynapse, regulation of postsynaptic neurotransmitter receptor activity, regulation of dendritic spine maintenance, axon, dendrite, neuron projection, excitatory synapse, and glutamatergic synapse |
| Fjx1 | four jointed box 1 (Drosophila) | −2.31 | 8.46 × 10−6 | cell–cell signaling, extracellular region, and extracellular space |
| Bax | BCL2-associated X protein | −2.31 | 2.0 × 10−4 | protein binding, lipid binding, neuron migration, leukocyte homeostasis, T cell homeostatic proliferation, B cell homeostasis, myeloid cell homeostasis, and apoptotic process |
| Slc23a2 | solute carrier family 23 (nucleobase transporters), member 2 | −2.2 | 9.38 × 10−5 | transporter activity, transmembrane transporter activity, brain development, and cytoplasm |
| Atp1a3 | ATPase, Na+/K+ transporting, alpha 3 polypeptide | −2.09 | 2.0 × 10−4 | nucleotide binding, amyloid-beta binding, transporter activity, ATP binding, protein-folding chaperone binding, potassium ion transport, memory, cellular response to amyloid-beta, nucleus, cytoplasm, Golgi apparatus, axon, myelin sheath, synapse, and postsynapse |
| Hspa4 | heat shock protein 4 | −2.08 | 9.06 × 10−6 | nucleotide binding, protein binding, ATP binding, negative regulation of protein phosphorylation, protein folding, neuron apoptotic process, regulation of microglial cell activation, nucleus, cytoplasm, lipid droplet, cytosol, and extracellular exosome |
| Csnk2a1 | casein kinase 2, alpha 1 polypeptide | −2.07 | 2.0 × 10−4 | nucleotide binding, protein kinase activity, protein binding, ATP binding, kinase activity, protein phosphorylation, apoptotic process, cell cycle, Wnt signaling pathway, phosphorylation, rhythmic process, regulation of cell cycle, chromatin, nucleus, and cytosol |
| Rab4a | RAB4A, member RAS oncogene family | −2.01 | 3.0 × 10−4 | nucleotide binding, ATPase activator activity, GTPase activity, cytoplasm, endosome, membrane, synaptic vesicle membrane, recycling endosome, and extracellular exosome |
| Nedd4l | neural precursor cell expressed, developmentally down-regulated gene 4-like | −2.01 | 3.43 × 10−6 | ubiquitin-protein transferase activity, protein binding, regulation of membrane depolarization, protein ubiquitination, cell differentiation, cytoplasm, endosome, Golgi apparatus, and plasma membrane |
| Swi5 | SWI5 recombination repair homolog (yeast) | −2 | 2.53 × 10−5 | Protein binding, DNA repair, and DNA damage response |
| Condition 3 | ||||
| Peg3 | paternally expressed 3 | −6.63 | 9.43 × 10−7 | DNA binding, regulation of transcription by RNA polymerase II, and apoptotic process, |
| Pmch | pro-melanin-concentrating hormone | −5.26 | 1.61 × 10−2 | signaling receptor binding, positive regulation of cytosolic calcium ion concentration, chemical synaptic transmission, regulation of neuronal synaptic plasticity, extracellular region, and extracellular space |
| Avp | arginine vasopressin | −4.59 | 5.3 × 10−3 | protein kinase activity, neuropeptide hormone activity, signal transduction, locomotory behavior, positive regulation of cell growth, apoptotic process, ERK1 and ERK2 cascade, dendrite, and neuronal dense core vesicle |
| Kif5b | kinesin family member 5B | −4.05 | 1.34 × 10−6 | nucleotide binding, protein binding, ATP binding, microtubule binding, mitochondrial transport, axon guidance, synaptic vesicle transport, axonal growth cone, and endocytic vesicle |
| Csnk1a1 | casein kinase 1, alpha 1 | −3.94 | 9.54 × 10−6 | nucleotide binding, protein kinase activity, ATP binding, cell morphogenesis, Golgi organization, cell cycle, signal transduction, cell division, chromosome, centromeric region, spindle, and nucleus |
| Vcp | valosin containing protein | −3.85 | 1.0 × 10−4 | protein binding, ATP binding, lipid binding, DNA repair, autophagy, apoptotic process, DNA damage response, nucleus, cytosol, myelin sheath, synapse, and glutamatergic synapse |
| Ppp1r1a | protein phosphatase 1, regulatory (inhibitor) subunit 1A | −3.8 | 5.02 × 10−6 | Protein phosphatase inhibitor activity, protein binding, signal transduction, extracellular space, and cytoplasm |
| Klf12 | Kruppel-like factor 12 | −3.38 | 6.94 × 10−7 | DNA binding, regulation of transcription by RNA polymerase II, apoptotic process, protein binding, DNA binding, regulation of transcription by RNA polymerase II, nucleus, and cytosol |
| Papola | poly (A) polymerase alpha | −3.31 | 3.74 × 10−5 | nucleotide binding, RNA binding, protein binding, ATP binding, mRNA polyadenylation, and nucleus |
| Gng12 | guanine nucleotide binding protein (G protein), gamma 12 | −2.98 | 6.72 × 10−5 | G-protein beta-subunit binding, signal transduction, actin filament, and membrane |
| Condition 4 | ||||
| Peg3 | paternally expressed 3 | −4.79 | 3.88 × 10−7 | DNA-binding transcription repressor activity, RNA polymerase II-specific, apoptotic process, nucleus, cytoplasm, and autophagosome |
| Vcp | valosin containing protein | −3.69 | 1.86 × 10−7 | protein binding, ATP binding, lipid binding, DNA repair, autophagy, apoptotic process, DNA damage response, nucleus, cytosol, myelin sheath, synapse, and glutamatergic synapse |
| Papola | poly (A) polymerase alpha | −3.49 | 4.62 × 10−6 | nucleotide binding, RNA binding, protein binding, ATP binding, mRNA processing, and nucleus |
| Rassf3 | Ras association (RalGDS/AF-6) domain family member 3 | −3.48 | 9.25 × 10−6 | signal transduction, cytoplasm, cytosol, and cytoskeleton |
| Erdr1 | erythroid differentiation regulator 1 | −3.31 | 4.32 × 10−5 | negative regulation of cell population proliferation and negative regulation of cell migration |
| Ppp1r1a | protein phosphatase 1, regulatory (inhibitor) subunit 1A | −3.11 | 1.29 × 10−5 | protein phosphatase inhibitor activity, protein binding, carbohydrate metabolic process, signal transduction, extracellular space, and cytoplasm |
| Klf12 | Kruppel-like factor 12 | −2.98 | 2.85 × 10−6 | DNA binding, protein binding, nucleus, and cytosol |
| Gpr158 | G protein-coupled receptor 158 | −2.97 | 1.0 × 10−6 | transmembrane signaling receptor activity, positive regulation of neurotransmitter secretion, signal transduction, brain development, regulation of synapse organization, cognition, and postsynaptic membrane |
| Gng12 | guanine nucleotide binding protein (G protein), gamma 12 | −2.9 | 3.44 × 10−5 | G-protein beta-subunit binding, signal transduction, actin filament, and membrane |
| Nedd4l | neural precursor cell expressed, developmentally down-regulated gene 4-like | −2.84 | 5.62 × 10−6 | ubiquitin-protein transferase activity, protein binding, transferase activity, ubiquitin-dependent protein catabolic process, protein ubiquitination, cell differentiation, cytoplasm, and Golgi apparatus |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fujiwara, M.; Ferdousi, F.; Isoda, H. Investigation into Molecular Brain Aging in Senescence-Accelerated Mouse (SAM) Model Employing Whole Transcriptomic Analysis in Search of Potential Molecular Targets for Therapeutic Interventions. Int. J. Mol. Sci. 2023, 24, 13867. https://doi.org/10.3390/ijms241813867
Fujiwara M, Ferdousi F, Isoda H. Investigation into Molecular Brain Aging in Senescence-Accelerated Mouse (SAM) Model Employing Whole Transcriptomic Analysis in Search of Potential Molecular Targets for Therapeutic Interventions. International Journal of Molecular Sciences. 2023; 24(18):13867. https://doi.org/10.3390/ijms241813867
Chicago/Turabian StyleFujiwara, Michitaka, Farhana Ferdousi, and Hiroko Isoda. 2023. "Investigation into Molecular Brain Aging in Senescence-Accelerated Mouse (SAM) Model Employing Whole Transcriptomic Analysis in Search of Potential Molecular Targets for Therapeutic Interventions" International Journal of Molecular Sciences 24, no. 18: 13867. https://doi.org/10.3390/ijms241813867
APA StyleFujiwara, M., Ferdousi, F., & Isoda, H. (2023). Investigation into Molecular Brain Aging in Senescence-Accelerated Mouse (SAM) Model Employing Whole Transcriptomic Analysis in Search of Potential Molecular Targets for Therapeutic Interventions. International Journal of Molecular Sciences, 24(18), 13867. https://doi.org/10.3390/ijms241813867







