Cerebral Organoid Arrays for Batch Phenotypic Analysis in Sections and Three Dimensions
Abstract
:1. Introduction
2. Results
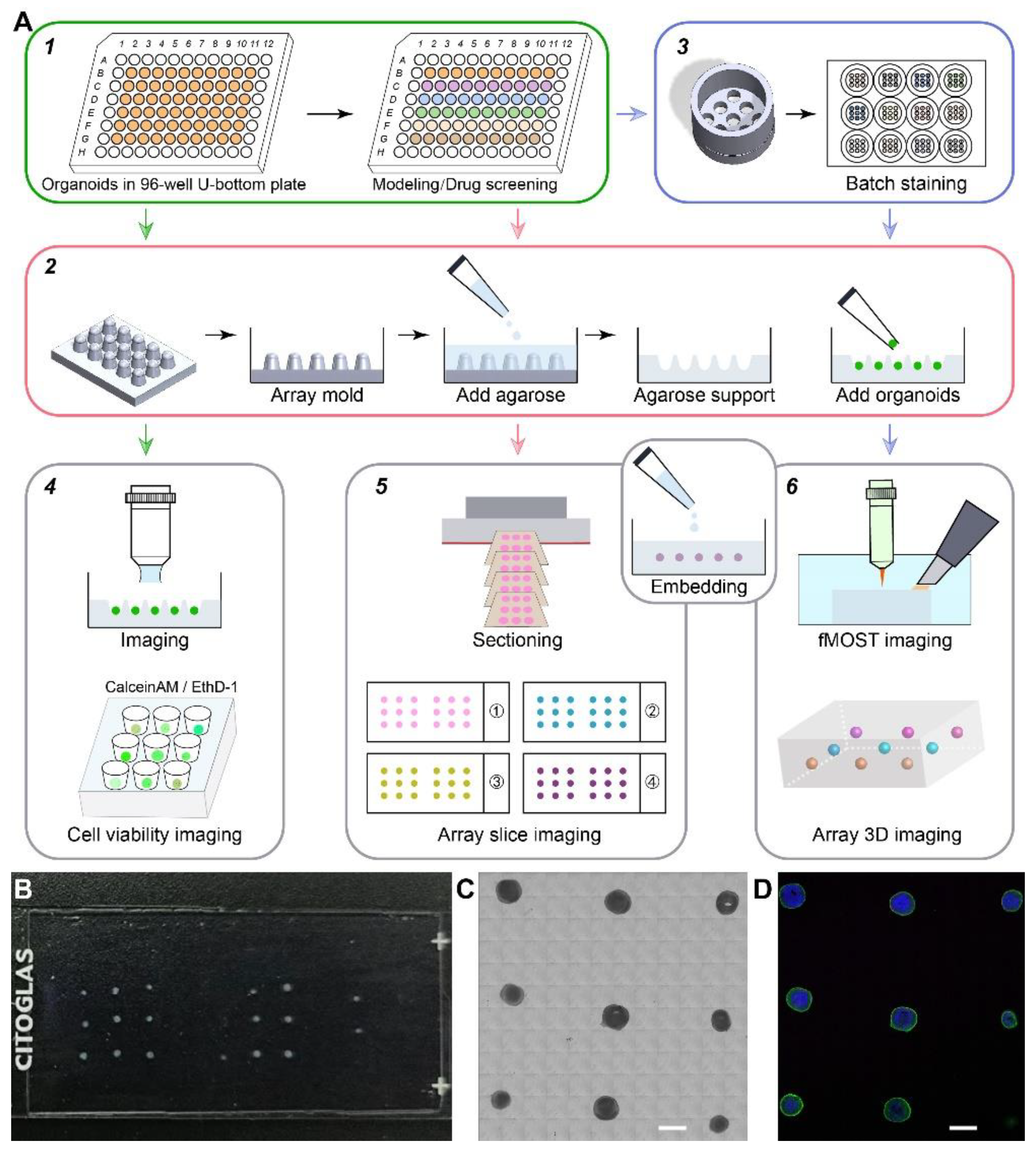
2.1. Integrated Strategy for Organoids Phenotypic Analysis in Batch
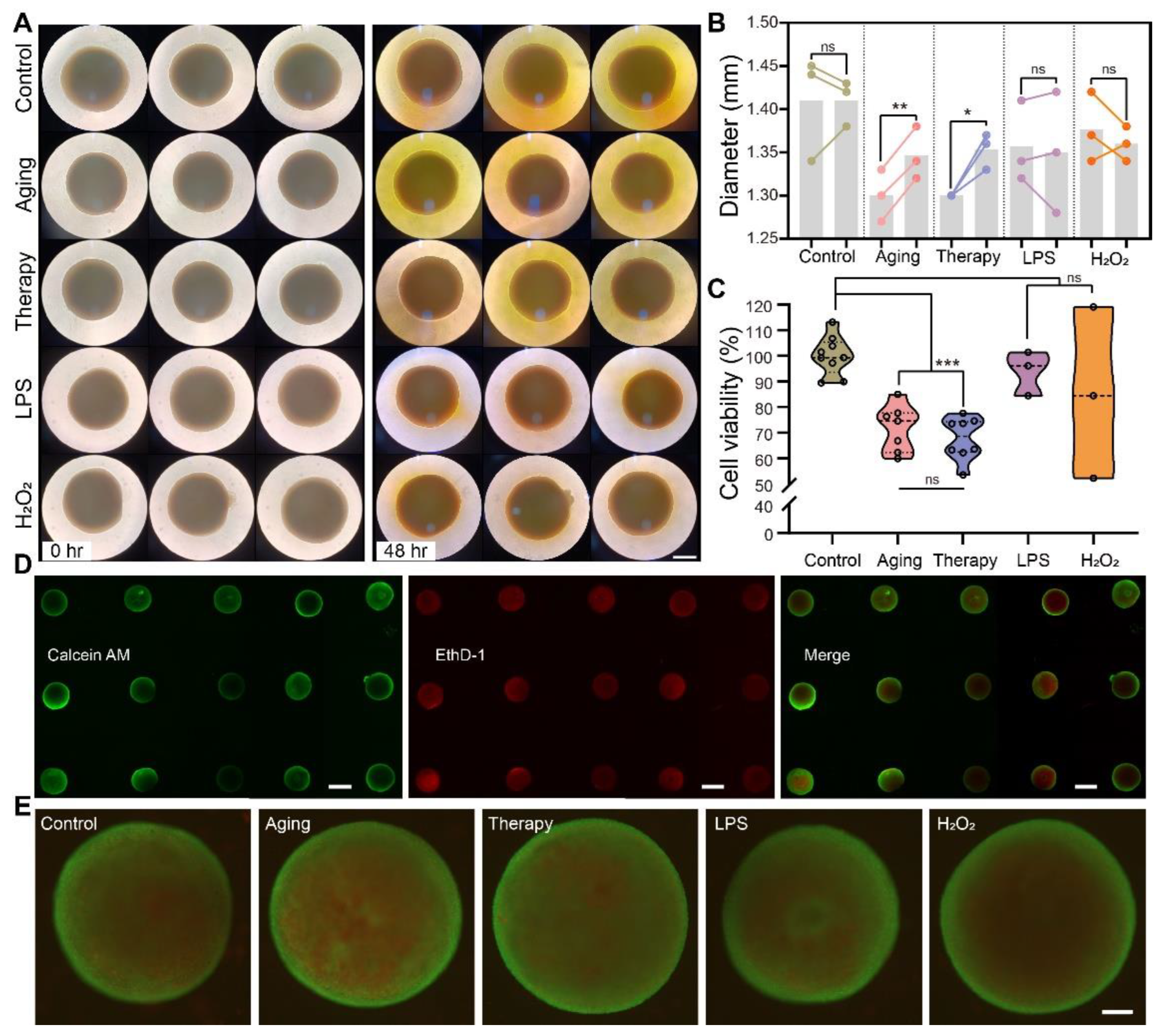
2.2. Cell Viability Assay and Live/Dead Imaging of Cerebral Organoids in Disease Modeling
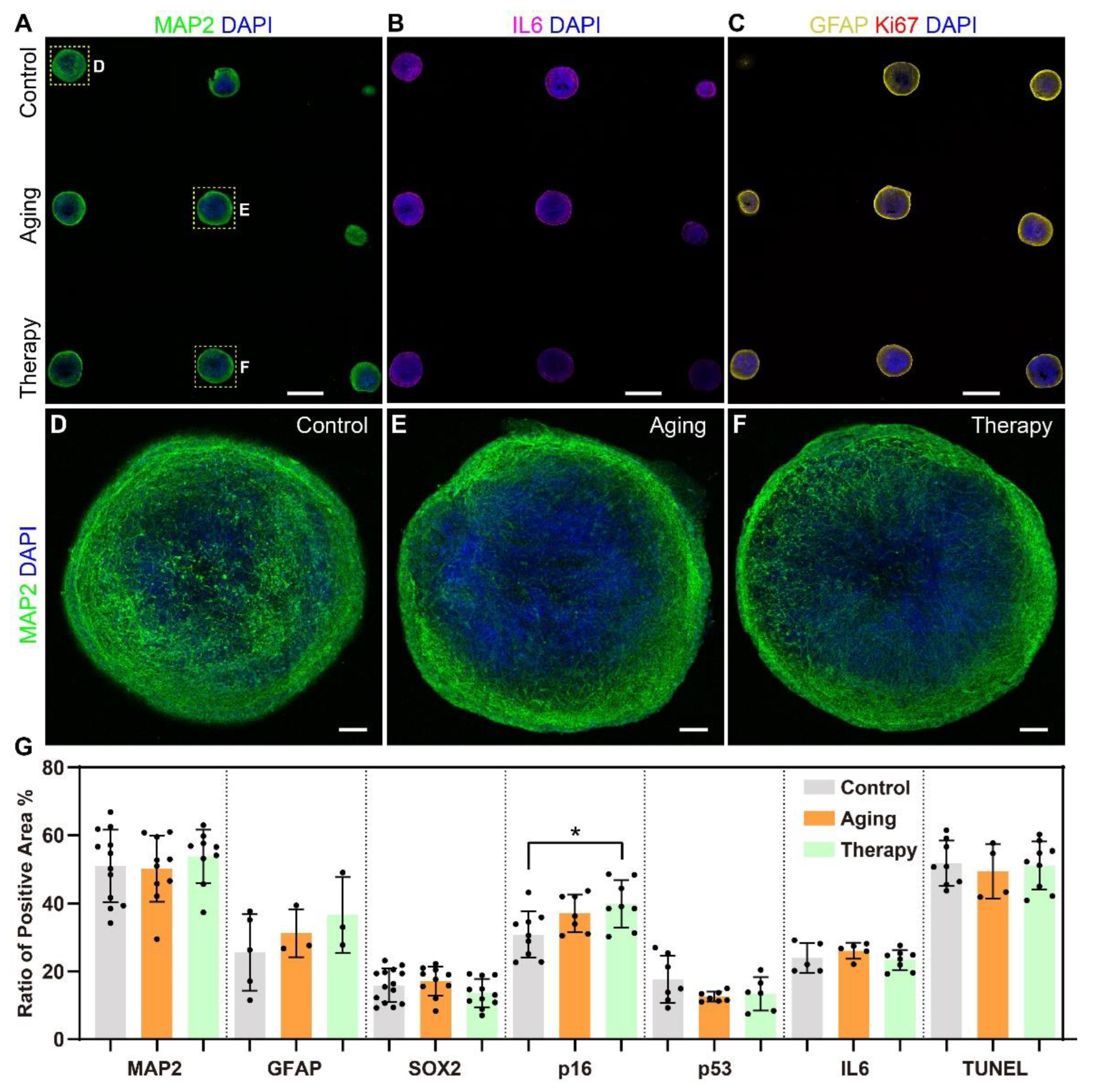
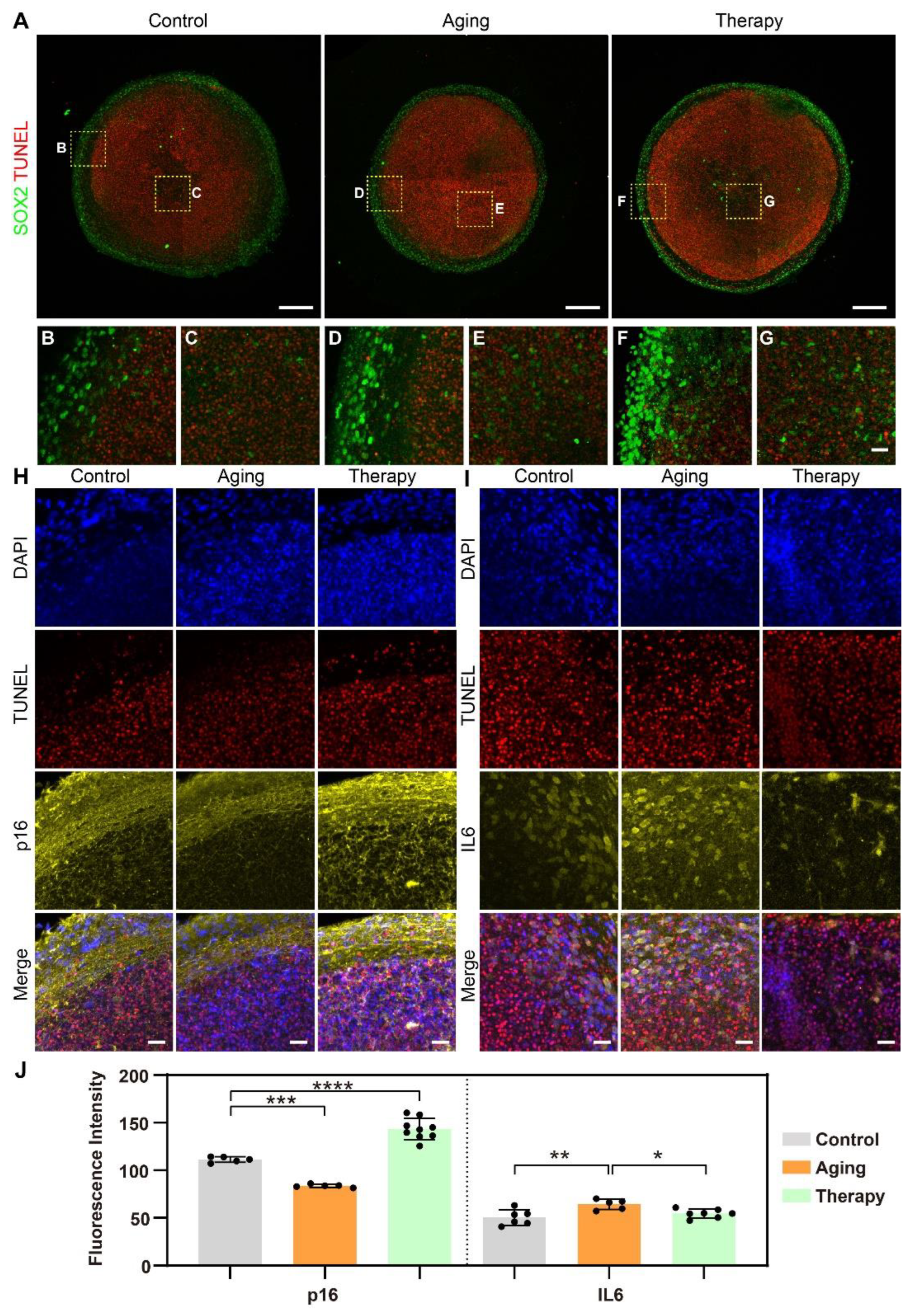
2.3. In Situ Phenotypic Analysis in Sectioned Agarose Arrays of Cerebral Organoids in Aging Models
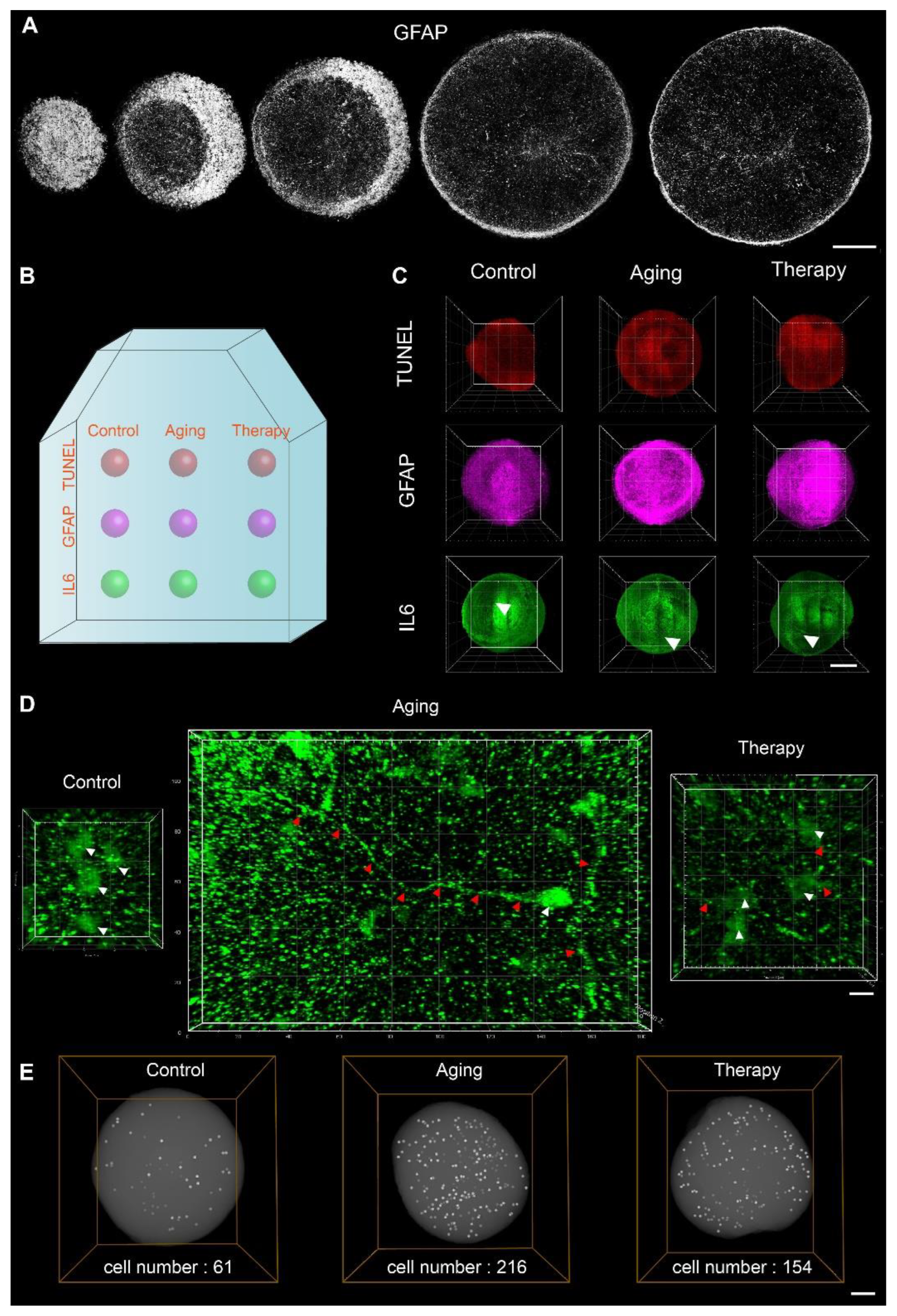
2.4. Three-Dimensional In Situ Phenotypic Analysis Showed Changes at Single-Cell Levels and Subcellular Levels
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Drug Treatment
5.2. Cell Viability Assay and Live Imaging
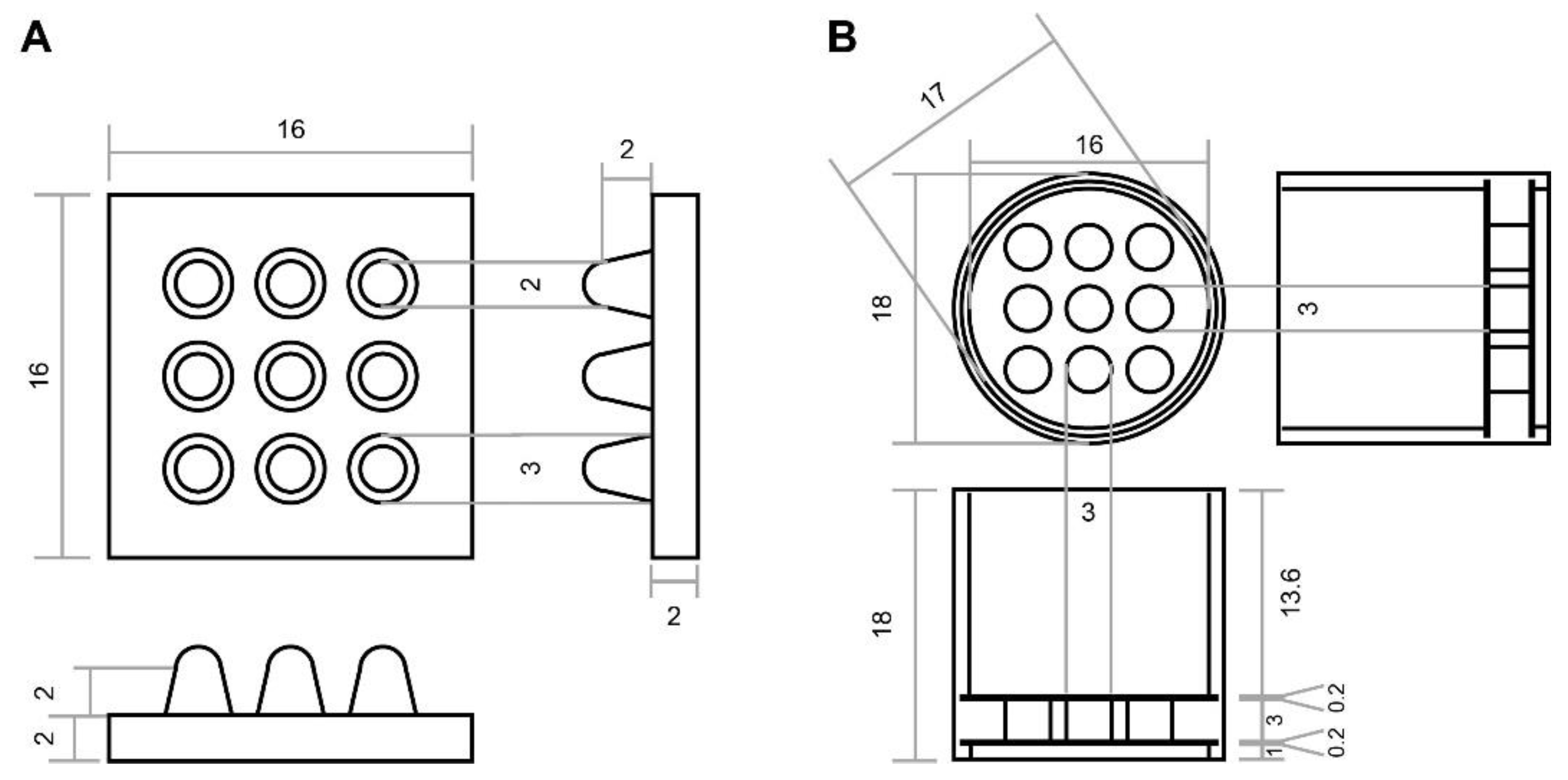
5.3. Designing and Rapid Prototyping of the Array Mold
5.4. Array Agarose Embedding
5.5. Cell Apoptosis Detection Using the TUNEL Assay
5.6. Immunostaining
5.7. Three-Dimensional Immunostaining
5.8. Microscopic Imaging
5.9. Visualization and Reconstruction
5.10. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
References
- Papaspyropoulos, A.; Tsolaki, M.; Foroglou, N.; Pantazaki, A. Modeling and targeting Alzheimer’s disease with organoids. Front. Pharmacol. 2020, 11, 396. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, C.; Armijo, E.; Bravo-Alegria, J.; Becerra-Calixto, A.; Mays, C.; Soto, C. Modeling amyloid beta and tau pathology in human cerebral organoids. Mol. Psychiatry 2018, 23, 2363–2374. [Google Scholar] [CrossRef] [PubMed]
- Raja, W.K.; Mungenast, A.E.; Lin, Y.T.; Ko, T.; Abdurrob, F.; Seo, J.; Tsai, L.H. Self-organizing 3D human neural tissue derived from induced pluripotent stem cells recapitulate Alzheimer’s disease phenotypes. PLoS ONE 2016, 11, e0161969. [Google Scholar] [CrossRef]
- Wulansari, N.; Darsono, W.H.W.; Woo, H.J.; Chang, M.Y.; Kim, J.; Bae, E.J.; Sun, W.; Lee, J.H.; Cho, I.J.; Shin, H.; et al. Neurodevelopmental defects and neurodegenerative phenotypes in human brain organoids carrying Parkinson’s disease-linked DNAJC6 mutations. Sci. Adv. 2021, 7, abb1540. [Google Scholar] [CrossRef]
- Kim, S.W.; Woo, H.J.; Kim, E.H.; Kim, H.S.; Suh, H.N.; Kim, S.H.; Song, J.J.; Wulansari, N.; Kang, M.; Choi, S.Y.; et al. Neural stem cells derived from human midbrain organoids as a stable source for treating Parkinson’s disease: Midbrain organoid-NSCs (Og-NSC) as a stable source for PD treatment. Prog. Neurobiol. 2021, 204, 102086. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wei, T.; Wang, Q.; Zhang, C.; Li, K.; Deng, J. Resveratrol’s neural protective effects for the injured embryoid body and cerebral organoid. BMC Pharmacol. Toxicol. 2022, 23, 47. [Google Scholar] [CrossRef]
- Seo, W.M.; Yoon, J.; Lee, J.H.; Lee, Y.; Lee, H.; Geum, D.; Sun, W.; Song, M.R. Modeling axonal regeneration by changing cytoskeletal dynamics in stem cell-derived motor nerve organoids. Sci. Rep. 2022, 12, 2082. [Google Scholar] [CrossRef]
- Abreu, C.M.; Gama, L.; Krasemann, S.; Chesnut, M.; Odwin-Dacosta, S.; Hogberg, H.T.; Hartung, T.; Pamies, D. Microglia increase inflammatory responses in iPSC-derived human brainspheres. Front. Microbiol. 2018, 9, 2766. [Google Scholar] [CrossRef]
- Arzua, T.; Yan, Y.; Jiang, C.; Logan, S.; Allison, R.L.; Wells, C.; Kumar, S.N.; Schäfer, R.; Bai, X. Modeling alcohol-induced neurotoxicity using human induced pluripotent stem cell-derived three-dimensional cerebral organoids. Transl. Psychiatry 2020, 10, 347. [Google Scholar] [CrossRef]
- Notaras, M.; Lodhi, A.; Barrio-Alonso, E.; Foord, C.; Rodrick, T.; Jones, D.; Fang, H.; Greening, D.; Colak, D. Neurodevelopmental signatures of narcotic and neuropsychiatric risk factors in 3D human-derived forebrain organoids. Mol. Psychiatry 2021, 26, 7760–7783. [Google Scholar] [CrossRef]
- Yang, X.; Jiang, T.; Liu, L.; Zhao, X.; Yu, X.; Yang, M.; Liu, G.; Luo, Q. Observing single cells in whole organs with optical imaging. J. Innov. Opt. Health Sci. 2023, 16, 2330002. [Google Scholar] [CrossRef]
- Lancaster, M.A.; Corsini, N.S.; Wolfinger, S.; Gustafson, E.H.; Phillips, A.W.; Burkard, T.R.; Otani, T.; Livesey, F.J.; Knoblich, J.A. Guided self-organization and cortical plate formation in human brain organoids. Nat. Biotechnol. 2017, 35, 659–666. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Muffat, J.; Omer, A.; Bosch, I.; Lancaster, M.A.; Sur, M.; Gehrke, L.; Knoblich, J.A.; Jaenisch, R. Induction of expansion and folding in human cerebral organoids. Cell Stem Cell 2017, 20, 385–396. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.; Chen, J.; Deng, Z.; Sun, T.; Luo, Q.; Gong, H.; Li, X.; Long, B. Multiscale analysis of cellular composition and morphology in intact cerebral organoids. Biology 2022, 11, 1270. [Google Scholar] [CrossRef] [PubMed]
- Mora-Bermudez, F.; Badsha, F.; Kanton, S.; Camp, J.G.; Vernot, B.; Köhler, K.; Voigt, B.; Okita, K.; Maricic, T.; He, Z.; et al. Differences and similarities between human and chimpanzee neural progenitors during cerebral cortex development. eLife 2016, 5, 18683. [Google Scholar] [CrossRef]
- Knight, G.T.; Lundin, B.F.; Iyer, N.; Ashton, L.M.; Sethares, W.; Willett, R.M.; Ashton, R.S. Engineering induction of singular neural rosette emergence within hPSC-derived tissues. eLife 2018, 7, 37549. [Google Scholar] [CrossRef] [PubMed]
- Kallioniemi, O.P.; Wagner, U.; Kononen, J.; Sauter, G. Tissue microarray technology for high-throughput molecular profiling of cancer. Hum. Mol. Genet. 2001, 10, 657–662. [Google Scholar] [CrossRef]
- Ivanov, D.P.; Grabowska, A.M. In vitro tissue microarrays for quick and efficient spheroid characterization. SLAS Discov. 2018, 23, 211–217. [Google Scholar] [CrossRef]
- Ivanov, D.P.; Grabowska, A.M. Spheroid arrays for high-throughput single-cell analysis of spatial patterns and biomarker expression in 3D. Sci. Rep. 2017, 7, 41160. [Google Scholar] [CrossRef] [PubMed]
- Gabriel, J.; Brennan, D.; Elisseeff, J.H.; Beachley, V. Microarray embedding/sectioning for parallel analysis of 3D cell spheroids. Sci. Rep. 2019, 9, 16287. [Google Scholar] [CrossRef]
- Moraes, G.S.; Wink, M.R.; Klamt, F.; Silva, A.O.; Fernandes, M.C. Simplified low-cost methodology to establish, histologically process and analyze three-dimensional cancer cell spheroid arrays. Eur. J. Cell Biol. 2020, 99, 151095. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhu, L.; Li, H.; Xie, W.; Liu, J.; Zhang, Y.; Li, Y.; Wang, C. In vivo and in vitro neuroprotective effects of maca polysaccharide. Front. Biosci. 2022, 27, 8. [Google Scholar] [CrossRef]
- Goshi, N.; Morgan, R.K.; Lein, P.J.; Seker, E. A primary neural cell culture model to study neuron, astrocyte, and microglia interactions in neuroinflammation. J. Neuroinflammation 2020, 17, 155. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Chen, Z.; Liang, W.; Yang, Z.; Du, Z.; Gong, S. D-galactose-induced accelerated aging model on auditory cortical neurons by regulating oxidative stress and apoptosis in vitro. J. Nutr. Health Aging 2022, 26, 13–22. [Google Scholar] [CrossRef]
- Colardo, M.; Petraroia, M.; Lerza, L.; Pensabeme, D.; Martella, N.; Pallottini, V.; Segatto, M. NGF modulates cholesterol metabolism and stimulates APOE secretion in glial cells conferring neuroprotection against oxidative stress. Int. J. Mol. Sci. 2022, 23, 4842. [Google Scholar] [CrossRef]
- Czerniecki, S.M.; Cruz, N.M.; Harder, J.L.; Menon, R.; Annis, J.; Otto, E.A.; Gulieva, R.E.; Islas, L.V.; Kim, Y.K.; Tran, L.M.; et al. High-throughput screening enhances kidney organoid differentiation from human pluripotent stem cells and enables automated multidimensional phenotyping. Cell Stem Cell 2018, 22, 929–940. [Google Scholar] [CrossRef] [PubMed]
- Beghin, A.; Grenci, G.; Sahni, G.; Guo, S.; Rajendiran, H.; Delaire, T.; Raffi, S.B.M.; Blanc, D.; Mets, R.; Ong, H.T.; et al. Automated high-speed 3D imaging of organoid cultures with multi-scale phenotypic quantification. Nat. Methods 2022, 19, 881–892. [Google Scholar] [CrossRef]
- Ivanov, D.P.; Grabowska, A.M.; Garnett, M.C. High-throughput spheroid screens using volume, resazurin reduction, and acid phosphatase activity. Methods Mol. Biol. 2017, 1601, 43–59. [Google Scholar] [PubMed]
- Choi, S.J.; Choi, Y.I.; Kim, L.; Park, I.S.; Han, J.Y.; Kim, J.M.; Chu, Y.C. Preparation of compact agarose cell blocks from the residues of liquid-based cytology samples. Korean J. Pathol. 2014, 48, 351–360. [Google Scholar] [CrossRef]
- Yan, P.; Seelentag, W.; Bachmann, A.; Bosman, F.T. An agarose matrix facilitates sectioning of tissue microarray blocks. J. Histochem. Cytochem. 2007, 55, 21–24. [Google Scholar] [CrossRef]
- Xu, J.C.; Fan, J.; Wang, X.; Eacker, S.M.; Kam, T.I.; Chen, L.; Yin, X.; Zhu, J.; Chi, Z.; Jiang, H. Cultured networks of excitatory projection neurons and inhibitory interneurons for studying human cortical neurotoxicity. Sci. Transl. Med. 2016, 8, 333ra348. [Google Scholar] [CrossRef] [PubMed]
- Renier, N.; Wu, Z.; Simon, D.J.; Yang, J.; Ariel, P.; Lavigne, M.T. iDISCO: A simple, rapid method to immunolabel large tissue samples for volume imaging. Cell 2014, 159, 896–910. [Google Scholar] [CrossRef]
- Gong, H.; Xu, D.L.; Yuan, J.; Li, X.; Guo, C.; Peng, J.; Li, Y.; Schwarz, L.; Li, A.; Hu, B.; et al. High-throughput dual-colour precision imaging for brain-wide connectome with cytoarchitectonic landmarks at the cellular level. Nat. Commun. 2016, 7, 12142. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Q.; Li, A.; Jin, R.; Zhang, D.; Li, X.; Jia, X.; Ding, Z.; Luo, P.; Zhou, C.; Jiang, C.; et al. High-definition imaging using line-illumination modulation microscopy. Nat. Methods 2021, 18, 309–315. [Google Scholar] [CrossRef] [PubMed]
| Antibody | Dilution for Sections | Dilution for 3D Immunostaining | Cat. No. | Company |
|---|---|---|---|---|
| Anti-p16, mouse | 1:100 | - | Sc-1661 | Santa Cruz Bio, Dallas, Texas, USA |
| Anti-p53, mouse | 1:100 | - | Sc-126 | Santa Cruz Bio, Dallas, Texas, USA |
| Anti-IL6, mouse | 1:100 | 1:10 | Sc-57315 | Santa Cruz Bio, Dallas, Texas, USA |
| Anti-Ki67, mouse | 1:20 | 1:10 | 550609 | BD Pharmingen, San Diego, CA, USA |
| Anti-SOX2, rabbit | 1:500 | 1:50 | ab97959 | Abcam, Cambridge, UK |
| Anti-MAP2, rabbit | 1:500 | 1:50 | AB5622 | Millipore, Darmstadt, Germany |
| Anti-GFAP, chicken | 1:500 | 1:50 | C9205 | Millipore, Darmstadt, Germany |
| Alexa Fluor 488 donkey anti-chicken lgG (H + L) | 1:1000 | 1:100 | A78948 | Invitrogen, Waltham, MA, USA |
| Alexa Fluor 488 donkey anti-rabbit lgG (H + L) | 1:1000 | 1:100 | A21206 | Invitrogen, Waltham, MA, USA |
| Alexa Fluor 594 donkey anti-mouse lgG (H + L) | 1:1000 | 1:100 | A21203 | Invitrogen, Waltham, MA, USA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, J.; Ma, H.; Deng, Z.; Luo, Q.; Gong, H.; Long, B.; Li, X. Cerebral Organoid Arrays for Batch Phenotypic Analysis in Sections and Three Dimensions. Int. J. Mol. Sci. 2023, 24, 13903. https://doi.org/10.3390/ijms241813903
Chen J, Ma H, Deng Z, Luo Q, Gong H, Long B, Li X. Cerebral Organoid Arrays for Batch Phenotypic Analysis in Sections and Three Dimensions. International Journal of Molecular Sciences. 2023; 24(18):13903. https://doi.org/10.3390/ijms241813903
Chicago/Turabian StyleChen, Juan, Haihua Ma, Zhiyu Deng, Qingming Luo, Hui Gong, Ben Long, and Xiangning Li. 2023. "Cerebral Organoid Arrays for Batch Phenotypic Analysis in Sections and Three Dimensions" International Journal of Molecular Sciences 24, no. 18: 13903. https://doi.org/10.3390/ijms241813903
APA StyleChen, J., Ma, H., Deng, Z., Luo, Q., Gong, H., Long, B., & Li, X. (2023). Cerebral Organoid Arrays for Batch Phenotypic Analysis in Sections and Three Dimensions. International Journal of Molecular Sciences, 24(18), 13903. https://doi.org/10.3390/ijms241813903






