Deciphering the Enigmatic Influence: Non-Coding RNAs Orchestrating Wnt/β-Catenin Signaling Pathway in Tumor Progression
Abstract
1. Background
2. Introduction to the Wnt Signaling Pathway and the Involvement of ncRNAs in Cancer
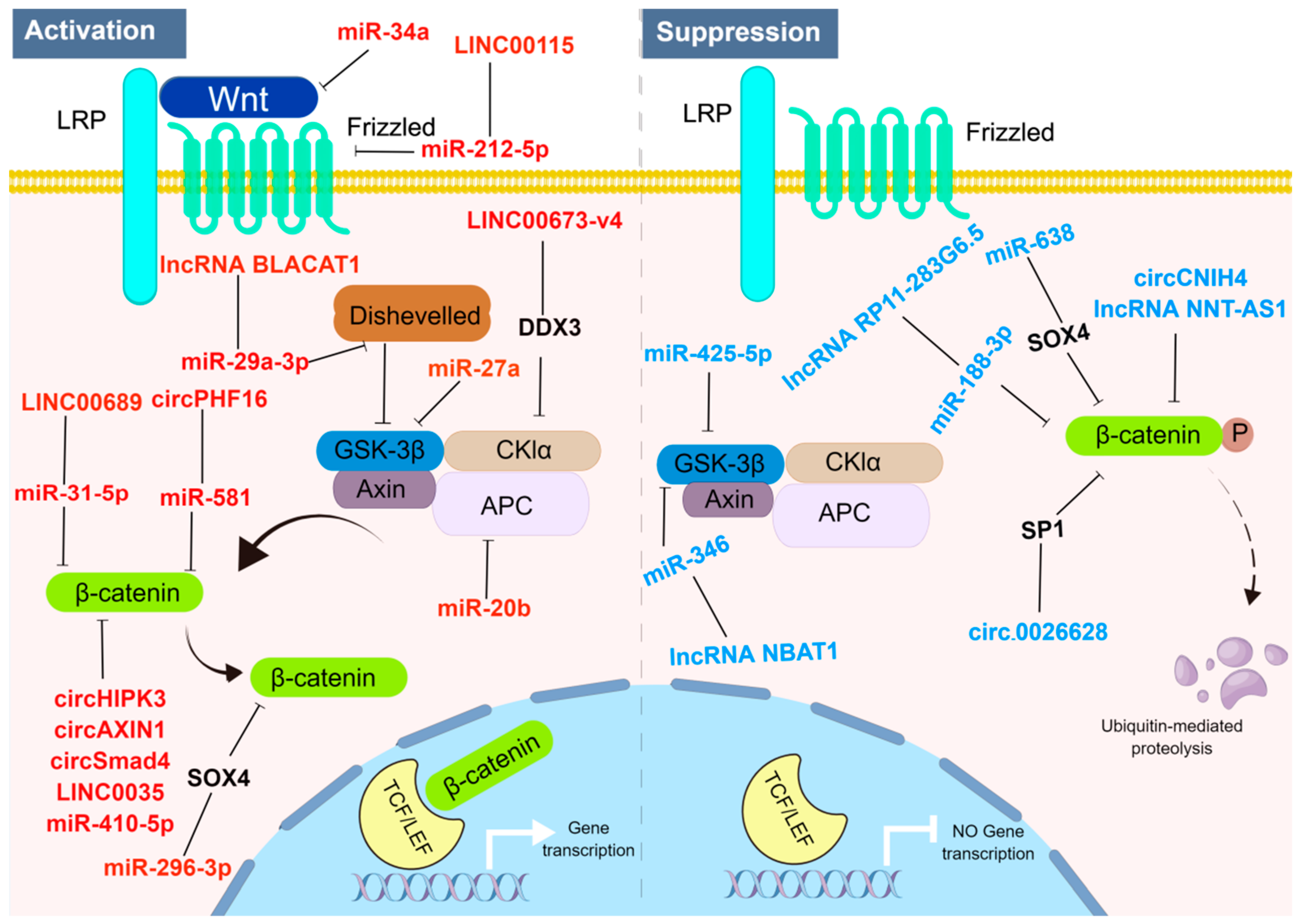
3. The Roles and Mechanisms of ncRNAs Involved in the Wnt/β-Catenin Signaling Pathway in Tumors
3.1. The Impact of the Interplay between ncRNAs and the Wnt/β-Catenin Signaling Pathway on Breast Cancer
3.2. The Effect of the Interaction between ncRNA and Wnt/β-Catenin Signaling Pathway on Lung Cancer
3.3. The Effect of the Interaction between ncRNA and Wnt/β-Catenin Signaling Pathway on Colorectal Cancer
3.4. The Effect of Interaction between ncRNA and Wnt/β-Catenin Signaling Pathway on Prostate Cancer
3.5. The Effect of the Interaction between ncRNA and Wnt/β-Catenin Signaling Pathway on Gastric Cancer
4. Perspective
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.; Treloar, A.E.; Lupien, M. Emergence of the Noncoding Cancer Genome: A Target of Genetic and Epigenetic Alterations. Cancer Discov. 2016, 6, 1215–1229. [Google Scholar] [CrossRef]
- Morel, D.; Jeffery, D.; Aspeslagh, S.; Almouzni, G.; Postel-Vinay, S. Combining epigenetic drugs with other therapies for solid tumours—Past lessons and future promise. Nat. Rev. Clin. Oncol. 2020, 17, 91–107. [Google Scholar] [CrossRef] [PubMed]
- van der Pol, Y.; Mouliere, F. Toward the Early Detection of Cancer by Decoding the Epigenetic and Environmental Fingerprints of Cell-Free DNA. Cancer Cell 2019, 36, 350–368. [Google Scholar] [CrossRef]
- Calses, P.C.; Crawford, J.J.; Lill, J.R.; Dey, A. Hippo Pathway in Cancer: Aberrant Regulation and Therapeutic Opportunities. Trends Cancer 2019, 5, 297–307. [Google Scholar] [CrossRef]
- Pópulo, H.; Lopes, J.M.; Soares, P. The mTOR signalling pathway in human cancer. Int. J. Mol. Sci. 2012, 13, 1886–1918. [Google Scholar] [CrossRef]
- Dhillon, A.S.; Hagan, S.; Rath, O.; Kolch, W. MAP kinase signalling pathways in cancer. Oncogene 2007, 26, 3279–3290. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, L.; Qu, Y. Targeting the β-catenin signaling for cancer therapy. Pharmacol. Res. 2020, 160, 104794. [Google Scholar] [CrossRef]
- Duchartre, Y.; Kim, Y.M.; Kahn, M. The Wnt signaling pathway in cancer. Crit. Rev. Oncol. Hematol. 2016, 99, 141–149. [Google Scholar] [CrossRef]
- Wang, B.; Tian, T.; Kalland, K.H.; Ke, X.; Qu, Y. Targeting Wnt/β-Catenin Signaling for Cancer Immunotherapy. Trends Pharmacol. Sci. 2018, 39, 648–658. [Google Scholar] [CrossRef]
- Yamamoto, D.; Oshima, H.; Wang, D.; Takeda, H.; Kita, K.; Lei, X.; Nakayama, M.; Murakami, K.; Ohama, T.; Takemura, H.; et al. Characterization of RNF43 frameshift mutations that drive Wnt ligand- and R-spondin-dependent colon cancer. J. Pathol. 2022, 257, 39–52. [Google Scholar] [CrossRef] [PubMed]
- Taheriazam, A.; Bayanzadeh, S.D.; Heydari Farahani, M.; Mojtabavi, S.; Zandieh, M.A.; Gholami, S.; Heydargoy, M.H.; Jamali Hondori, M.; Kangarloo, Z.; Behroozaghdam, M.; et al. Non-coding RNA-based therapeutics in cancer therapy: An emphasis on Wnt/β-catenin control. Eur. J. Pharmacol. 2023, 951, 175781. [Google Scholar] [CrossRef]
- Rahmani, F.; Avan, A.; Hashemy, S.I.; Hassanian, S.M. Role of Wnt/β-catenin signaling regulatory microRNAs in the pathogenesis of colorectal cancer. J. Cell. Physiol. 2018, 233, 811–817. [Google Scholar] [CrossRef]
- Spaan, I.; Raymakers, R.A.; van de Stolpe, A.; Peperzak, V. Wnt signaling in multiple myeloma: A central player in disease with therapeutic potential. J. Hematol. Oncol. 2018, 11, 67. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Weng, W.; Zhang, Q.; Wu, Y.; Ni, S.; Tan, C.; Xu, M.; Sun, H.; Liu, C.; Wei, P.; et al. The lncRNA NEAT1 activates Wnt/β-catenin signaling and promotes colorectal cancer progression via interacting with DDX5. J. Hematol. Oncol. 2018, 11, 113. [Google Scholar] [CrossRef] [PubMed]
- Esteller, M. Non-coding RNAs in human disease. Nat. Rev. Genet. 2011, 12, 861–874. [Google Scholar] [CrossRef] [PubMed]
- Esteller, M.; Pandolfi, P.P. The Epitranscriptome of Noncoding RNAs in Cancer. Cancer Discov. 2017, 7, 359–368. [Google Scholar] [CrossRef]
- Hombach, S.; Kretz, M. Non-coding RNAs: Classification, Biology and Functioning. Adv. Exp. Med. Biol. 2016, 937, 3–17. [Google Scholar] [CrossRef]
- Wei, L.; Wang, X.; Lv, L.; Liu, J.; Xing, H.; Song, Y.; Xie, M.; Lei, T.; Zhang, N.; Yang, M. The emerging role of microRNAs and long noncoding RNAs in drug resistance of hepatocellular carcinoma. Mol. Cancer 2019, 18, 147. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Hannon, G.J. MicroRNAs: Small RNAs with a big role in gene regulation. Nat. Rev. Genet. 2004, 5, 522–531. [Google Scholar] [CrossRef]
- Smith, A.J.; Sompel, K.M.; Elango, A.; Tennis, M.A. Non-Coding RNA and Frizzled Receptors in Cancer. Front. Mol. Biosci. 2021, 8, 712546. [Google Scholar] [CrossRef]
- Trzybulska, D.; Vergadi, E.; Tsatsanis, C. miRNA and Other Non-Coding RNAs as Promising Diagnostic Markers. eJIFCC 2018, 29, 221–226. [Google Scholar] [PubMed]
- Statello, L.; Guo, C.J.; Chen, L.L.; Huarte, M. Author Correction: Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 2021, 22, 159. [Google Scholar] [CrossRef] [PubMed]
- Larrea, E.; Sole, C.; Manterola, L.; Goicoechea, I.; Armesto, M.; Arestin, M.; Caffarel, M.M.; Araujo, A.M.; Araiz, M.; Fernandez-Mercado, M.; et al. New Concepts in Cancer Biomarkers: Circulating miRNAs in Liquid Biopsies. Int. J. Mol. Sci. 2016, 17, 627. [Google Scholar] [CrossRef]
- Zhang, X.; Yang, J. Role of Non-coding RNAs on the Radiotherapy Sensitivity and Resistance of Head and Neck Cancer: From Basic Research to Clinical Application. Front. Cell Dev. Biol. 2020, 8, 637435. [Google Scholar] [CrossRef] [PubMed]
- Slack, F.J.; Chinnaiyan, A.M. The Role of Non-coding RNAs in Oncology. Cell 2019, 179, 1033–1055. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Target recognition and regulatory functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef]
- Lagos-Quintana, M.; Rauhut, R.; Lendeckel, W.; Tuschl, T. Identification of novel genes coding for small expressed RNAs. Science 2001, 294, 853–858. [Google Scholar] [CrossRef]
- Lu, J.; Getz, G.; Miska, E.A.; Alvarez-Saavedra, E.; Lamb, J.; Peck, D.; Sweet-Cordero, A.; Ebert, B.L.; Mak, R.H.; Ferrando, A.A.; et al. MicroRNA expression profiles classify human cancers. Nature 2005, 435, 834–838. [Google Scholar] [CrossRef]
- Calin, G.A.; Croce, C.M. MicroRNA signatures in human cancers. Nat. Rev. Cancer 2006, 6, 857–866. [Google Scholar] [CrossRef]
- Zhao, W.; An, Y.; Liang, Y.; Xie, X.W. Role of HOTAIR long noncoding RNA in metastatic progression of lung cancer. Eur. Rev. Med. Pharmacol. Sci. 2014, 18, 1930–1936. [Google Scholar]
- Wang, N.; Yu, Y.; Xu, B.; Zhang, M.; Li, Q.; Miao, L. Pivotal prognostic and diagnostic role of the long non-coding RNA colon cancer-associated transcript 1 expression in human cancer. Mol. Med. Rep. 2019, 19, 771–782. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.J.; Dang, H.X.; Lim, D.A.; Feng, F.Y.; Maher, C.A. Long noncoding RNAs in cancer metastasis. Nat. Rev. Cancer 2021, 21, 446–460. [Google Scholar] [CrossRef]
- Sanger, H.L.; Klotz, G.; Riesner, D.; Gross, H.J.; Kleinschmidt, A.K. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc. Natl. Acad. Sci. USA 1976, 73, 3852–3856. [Google Scholar] [CrossRef] [PubMed]
- Cocquerelle, C.; Mascrez, B.; Hétuin, D.; Bailleul, B. Mis-splicing yields circular RNA molecules. FASEB J. 1993, 7, 155–160. [Google Scholar] [CrossRef]
- Alsaab, H.O. Pathological role of long non-coding (lnc) RNA in the regulation of Wnt/β-catenin signaling pathway during epithelial-mesenchymal transition (EMT). Pathol. Res. Pract. 2023, 248, 154566. [Google Scholar] [CrossRef]
- Anastasiadou, E.; Jacob, L.S.; Slack, F.J. Non-coding RNA networks in cancer. Nat. Rev. Cancer 2018, 18, 5–18. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.Z.; Cheng, T.T.; He, Q.J.; Lei, Z.Y.; Chi, J.; Tang, Z.; Liao, Q.X.; Zhang, H.; Zeng, L.S.; Cui, S.Z. LINC01133 as ceRNA inhibits gastric cancer progression by sponging miR-106a-3p to regulate APC expression and the Wnt/β-catenin pathway. Mol. Cancer 2018, 17, 126. [Google Scholar] [CrossRef] [PubMed]
- Nie, J.; Jiang, H.C.; Zhou, Y.C.; Jiang, B.; He, W.J.; Wang, Y.F.; Dong, J. MiR-125b regulates the proliferation and metastasis of triple negative breast cancer cells via the Wnt/β-catenin pathway and EMT. Biosci. Biotechnol. Biochem. 2019, 83, 1062–1071. [Google Scholar] [CrossRef]
- Liu, R.; Deng, P.; Zhang, Y.; Wang, Y.; Peng, C. Circ_0082182 promotes oncogenesis and metastasis of colorectal cancer in vitro and in vivo by sponging miR-411 and miR-1205 to activate the Wnt/β-catenin pathway. World J. Surg. Oncol. 2021, 19, 51. [Google Scholar] [CrossRef]
- Klaus, A.; Birchmeier, W. Wnt signalling and its impact on development and cancer. Nat. Rev. Cancer 2008, 8, 387–398. [Google Scholar] [CrossRef]
- Nusse, R.; Varmus, H.E. Many tumors induced by the mouse mammary tumor virus contain a provirus integrated in the same region of the host genome. Cell 1982, 31, 99–109. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Zhang, X.; Feng, X.; Fan, X.; Jin, Z. The crosstalk between microRNAs and the Wnt/β-catenin signaling pathway in cancer. Oncotarget 2017, 8, 14089–14106. [Google Scholar] [CrossRef] [PubMed]
- Habas, R.; Dawid, I.B. Dishevelled and Wnt signaling: Is the nucleus the final frontier? J. Biol. 2005, 4, 2. [Google Scholar] [CrossRef][Green Version]
- Kühl, M.; Sheldahl, L.C.; Park, M.; Miller, J.R.; Moon, R.T. The Wnt/Ca2+ pathway: A new vertebrate Wnt signaling pathway takes shape. Trends Genet. TIG 2000, 16, 279–283. [Google Scholar] [CrossRef] [PubMed]
- Mlodzik, M. Planar cell polarization: Do the same mechanisms regulate Drosophila tissue polarity and vertebrate gastrulation? Trends Genet. TIG 2002, 18, 564–571. [Google Scholar] [CrossRef] [PubMed]
- Nusse, R.; Clevers, H. Wnt/β-Catenin Signaling, Disease, and Emerging Therapeutic Modalities. Cell 2017, 169, 985–999. [Google Scholar] [CrossRef]
- Cao, M.Q.; You, A.B.; Zhu, X.D.; Zhang, W.; Zhang, Y.Y.; Zhang, S.Z.; Zhang, K.W.; Cai, H.; Shi, W.K.; Li, X.L.; et al. miR-182-5p promotes hepatocellular carcinoma progression by repressing FOXO3a. J. Hematol. Oncol. 2018, 11, 12. [Google Scholar] [CrossRef]
- Ge, X.; Wang, X. Role of Wnt canonical pathway in hematological malignancies. J. Hematol. Oncol. 2010, 3, 33. [Google Scholar] [CrossRef]
- Gajos-Michniewicz, A.; Czyz, M. WNT Signaling in Melanoma. Int. J. Mol. Sci. 2020, 21, 4852. [Google Scholar] [CrossRef] [PubMed]
- Farooqi, A.A.; Rakhmetova, V.S.; Kapanova, G.; Tashenova, G.; Tulebayeva, A.; Akhenbekova, A.; Ibekenov, O.; Turgambayeva, A.; Xu, B. Bufalin-Mediated Regulation of Cell Signaling Pathways in Different Cancers: Spotlight on JAK/STAT, Wnt/β-Catenin, mTOR, TRAIL/TRAIL-R, and Non-Coding RNAs. Molecules 2023, 28, 2231. [Google Scholar] [CrossRef]
- Lyu, H.; Zhang, J.; Wei, Q.; Huang, Y.; Zhang, R.; Xiao, S.; Guo, D.; Chen, X.-Z.; Zhou, C.; Tang, J. Identification of Wnt/β-Catenin- and Autophagy-Related lncRNA Signature for Predicting Immune Efficacy in Pancreatic Adenocarcinoma. Biology 2023, 12, 319. [Google Scholar] [CrossRef] [PubMed]
- Wan, H.; Lin, T.; Shan, M.; Lu, J.; Guo, Z. LINC00491 Facilitates Tumor Progression of Lung Adenocarcinoma via Wnt/β-Catenin-Signaling Pathway by Regulating MTSS1 Ubiquitination. Cells 2022, 11, 3737. [Google Scholar] [CrossRef] [PubMed]
- Monga, S.P. β-Catenin Signaling and Roles in Liver Homeostasis, Injury, and Tumorigenesis. Gastroenterology 2015, 148, 1294–1310. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, X. Targeting the Wnt/β-catenin signaling pathway in cancer. J. Hematol. Oncol. 2020, 13, 165. [Google Scholar] [CrossRef]
- Mafi, A.; Rismanchi, H.; Malek Mohammadi, M.; Hedayati, N.; Ghorbanhosseini, S.S.; Hosseini, S.A.; Gholinezhad, Y.; Mousavi Dehmordi, R.; Ghezelbash, B.; Zarepour, F.; et al. A spotlight on the interplay between Wnt/β-catenin signaling and circular RNAs in hepatocellular carcinoma progression. Front. Oncol. 2023, 13, 1224138. [Google Scholar] [CrossRef] [PubMed]
- Leung, D.H.L.; Phon, B.W.S.; Sivalingam, M.; Radhakrishnan, A.K.; Kamarudin, M.N.A. Regulation of EMT Markers, Extracellular Matrix, and Associated Signalling Pathways by Long Non-Coding RNAs in Glioblastoma Mesenchymal Transition: A Scoping Review. Biology 2023, 12, 818. [Google Scholar] [CrossRef]
- Tian, Y.; Lai, T.; Li, Z.; Mao, M.; Jin, Y.; Liu, Y.; Guo, R. Role of non-coding RNA intertwined with the Wnt/β-catenin signaling pathway in endometrial cancer. Mol. Med. Rep. 2023, 28, 150. [Google Scholar] [CrossRef]
- Zhou, P.; Liu, Y.; Wu, G.; Lu, K.; Zhao, T.; Yang, L. LincRNA PRNCR1 activates the Wnt/β-catenin pathway to drive the deterioration of hepatocellular carcinoma via regulating miR-411-3p/ZEB1 axis. Biotechnol. Genet. Eng. Rev. 2023. [Google Scholar] [CrossRef]
- van Ooyen, A.; Nusse, R. Structure and nucleotide sequence of the putative mammary oncogene int-1; proviral insertions leave the protein-encoding domain intact. Cell 1984, 39, 233–240. [Google Scholar] [CrossRef]
- Shi, J.; Li, F.; Luo, M.; Wei, J.; Liu, X. Distinct Roles of Wnt/β-Catenin Signaling in the Pathogenesis of Chronic Obstructive Pulmonary Disease and Idiopathic Pulmonary Fibrosis. Mediat. Inflamm. 2017, 2017, 3520581. [Google Scholar] [CrossRef]
- Zhan, T.; Rindtorff, N.; Boutros, M. Wnt signaling in cancer. Oncogene 2017, 36, 1461–1473. [Google Scholar] [CrossRef]
- Behrens, J.; von Kries, J.P.; Kühl, M.; Bruhn, L.; Wedlich, D.; Grosschedl, R.; Birchmeier, W. Functional interaction of beta-catenin with the transcription factor LEF-1. Nature 1996, 382, 638–642. [Google Scholar] [CrossRef] [PubMed]
- Peifer, M.; Pai, L.M.; Casey, M. Phosphorylation of the Drosophila adherens junction protein Armadillo: Roles for wingless signal and zeste-white 3 kinase. Dev. Biol. 1994, 166, 543–556. [Google Scholar] [CrossRef] [PubMed]
- Rubinfeld, B.; Albert, I.; Porfiri, E.; Fiol, C.; Munemitsu, S.; Polakis, P. Binding of GSK3beta to the APC-beta-catenin complex and regulation of complex assembly. Science 1996, 272, 1023–1026. [Google Scholar] [CrossRef]
- Yost, C.; Torres, M.; Miller, J.R.; Huang, E.; Kimelman, D.; Moon, R.T. The axis-inducing activity, stability, and subcellular distribution of beta-catenin is regulated in Xenopus embryos by glycogen synthase kinase 3. Genes Dev. 1996, 10, 1443–1454. [Google Scholar] [CrossRef] [PubMed]
- Damsky, W.E.; Curley, D.P.; Santhanakrishnan, M.; Rosenbaum, L.E.; Platt, J.T.; Gould Rothberg, B.E.; Taketo, M.M.; Dankort, D.; Rimm, D.L.; McMahon, M.; et al. β-catenin signaling controls metastasis in Braf-activated Pten-deficient melanomas. Cancer Cell 2011, 20, 741–754. [Google Scholar] [CrossRef]
- Khramtsov, A.I.; Khramtsova, G.F.; Tretiakova, M.; Huo, D.; Olopade, O.I.; Goss, K.H. Wnt/beta-catenin pathway activation is enriched in basal-like breast cancers and predicts poor outcome. Am. J. Pathol. 2010, 176, 2911–2920. [Google Scholar] [CrossRef]
- Kobayashi, M.; Honma, T.; Matsuda, Y.; Suzuki, Y.; Narisawa, R.; Ajioka, Y.; Asakura, H. Nuclear translocation of beta-catenin in colorectal cancer. Br. J. Cancer 2000, 82, 1689–1693. [Google Scholar] [CrossRef]
- Tao, J.; Calvisi, D.F.; Ranganathan, S.; Cigliano, A.; Zhou, L.; Singh, S.; Jiang, L.; Fan, B.; Terracciano, L.; Armeanu-Ebinger, S.; et al. Activation of β-catenin and Yap1 in human hepatoblastoma and induction of hepatocarcinogenesis in mice. Gastroenterology 2014, 147, 690–701. [Google Scholar] [CrossRef]
- Zhang, W.; Liu, Z. MiRNA-139-3p inhibits malignant progression in urothelial carcinoma of the bladder via targeting KIF18B and inactivating Wnt/beta-catenin pathway. Pharmacogenet. Genom. 2023, 33, 1–9. [Google Scholar] [CrossRef]
- Lu, C.; Jia, S.; Zhao, S.; Shao, X. MiR-342 regulates cell proliferation and apoptosis in hepatocellular carcinoma through Wnt/β-catenin signaling pathway. Cancer Biomark. Sect. A Dis. Markers 2019, 25, 115–126. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.L.; Cao, G.H.; Liu, Y.J.; Liu, C.H. Effect of LncRNA HOTAIR on the proliferation, apoptosis and drug resistance of Wilms tumor cells through Wnt/β-catenin signaling pathway. Chin. J. Oncol. 2021, 43, 769–774. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, C.; Duan, X.; Ren, F.; Li, S.; Jin, Z.; Wang, Y.; Feng, Y.; Liu, Z.; Chang, Z. CREPT/RPRD1B, a Recently Identified Novel Protein Highly Expressed in Tumors, Enhances the β-Catenin·TCF4 Transcriptional Activity in Response to Wnt Signaling. J. Biol. Chem. 2014, 289, 22589–22599. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Yang, X.; He, X.; Ma, W.; Wang, J.; Zhou, Q.; Li, M.; Yu, S. MicroRNA-449b-5p suppresses the growth and invasion of breast cancer cells via inhibiting CREPT-mediated Wnt/β-catenin signaling. Chem. Biol. Interact. 2019, 302, 74–82. [Google Scholar] [CrossRef]
- Tian, D.; Luo, L.; Wang, T.; Qiao, J. MiR-296-3p inhibits cell proliferation by the SOX4-Wnt/βcatenin pathway in triple-negative breast cancer. J. Biosci. 2021, 46, 98. [Google Scholar] [CrossRef]
- Xu, Q.; Zhang, Q.; Dong, M.; Yu, Y. MicroRNA-638 inhibits the progression of breast cancer through targeting HOXA9 and suppressing Wnt/β-cadherin pathway. World J. Surg. Oncol. 2021, 19, 247. [Google Scholar] [CrossRef]
- Yao, J.; Li, G.; Liu, M.; Yang, S.; Su, H.; Ye, C. lnc-MICAL2-1 sponges miR-25 to regulate DKK3 expression and inhibits activation of the Wnt/β-catenin signaling pathway in breast cancer. Int. J. Mol. Med. 2022, 49, 23. [Google Scholar] [CrossRef]
- Pei, J.; Zhang, S.; Yang, X.; Han, C.; Pan, Y.; Li, J.; Wang, Z.; Sun, C.; Zhang, J. Long non-coding RNA RP11-283G6.5 confines breast cancer development through modulating miR-188-3p/TMED3/Wnt/β-catenin signalling. RNA Biol. 2021, 18, 287–302. [Google Scholar] [CrossRef]
- Zhao, Z.; Han, X.; Nie, C.; Lin, S.; Wang, J.; Fang, H. Circ_0008784 activates Wnt/β-catenin pathway to affect the proliferation and apoptosis of triple-negative breast cancer cells. Pathol. Res. Pract. 2023, 241, 154185. [Google Scholar] [CrossRef]
- Tan, Z.; Zhao, L.; Huang, S.; Jiang, Q.; Wei, Y.; Wu, J.L.; Zhang, Z.; Li, Y. Small peptide LINC00511-133aa encoded by LINC00511 regulates breast cancer cell invasion and stemness through the Wnt/β-catenin pathway. Mol. Cell. Probes 2023, 69, 101913. [Google Scholar] [CrossRef] [PubMed]
- Jiang, B.; Zhu, H.; Tang, L.; Gao, T.; Zhou, Y.; Gong, F.; Tan, Y.; Xie, L.; Wu, X.; Li, Y. Apatinib Inhibits Stem Properties and Malignant Biological Behaviors of Breast Cancer Stem Cells by Blocking Wnt/β-catenin Signal Pathway through Downregulating LncRNA ROR. Anti-Cancer Agents Med. Chem. 2022, 22, 1723–1734. [Google Scholar] [CrossRef] [PubMed]
- Xin, G.; Hua, D.; Yingxu, S. Exosomes transfer non-coding RNA to regulate breast cancer drug resistance. Cancer Res. Prev. Treat. 2022, 49, 1071–1076. [Google Scholar] [CrossRef]
- Cheng, S.; Huang, Y.; Lou, C.; He, Y.; Zhang, Y.; Zhang, Q. FSTL1 enhances chemoresistance and maintains stemness in breast cancer cells via integrin β3/Wnt signaling under miR-137 regulation. Cancer Biol. Ther. 2019, 20, 328–337. [Google Scholar] [CrossRef]
- Kirby, T. Young non-smoker diagnosed with lung cancer. Lancet. Respir. Med. 2020, 8, 141–142. [Google Scholar] [CrossRef]
- Ma, Z.; Wang, Y.; He, B.; Cui, J.; Zhang, C.; Wang, H.; Feng, W.; Wang, B.; Wei, D.; Wu, Y.; et al. Expression of miR-590 in lung cancer and its correlation with prognosis. Oncol. Lett. 2018, 15, 1753–1757. [Google Scholar] [CrossRef]
- Xu, B.B.; Gu, Z.F.; Ma, M.; Wang, J.Y.; Wang, H.N. MicroRNA-590-5p suppresses the proliferation and invasion of non-small cell lung cancer by regulating GAB1. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 5954–5963. [Google Scholar] [CrossRef]
- Hao, X.; Su, A. MiR-590 suppresses the progression of non-small cell lung cancer by regulating YAP1 and Wnt/β-catenin signaling. Clin. Transl. Oncol. 2022, 24, 546–555. [Google Scholar] [CrossRef]
- Liu, S.; Yang, N.; Wang, L.; Wei, B.; Chen, J.; Gao, Y. lncRNA SNHG11 promotes lung cancer cell proliferation and migration via activation of Wnt/β-catenin signaling pathway. J. Cell. Physiol. 2020, 235, 7541–7553. [Google Scholar] [CrossRef]
- Lin, H.; Shangguan, Z.; Zhu, M.; Bao, L.; Zhang, Q.; Pan, S. lncRNA FLVCR1-AS1 silencing inhibits lung cancer cell proliferation, migration, and invasion by inhibiting the activity of the Wnt/β-catenin signaling pathway. J. Cell. Biochem. 2019, 120, 10625–10632. [Google Scholar] [CrossRef]
- Chen, T.; Feng, G.; Xing, Z.; Gao, X. Circ-EIF3I facilitates proliferation, migration, and invasion of lung cancer via regulating the activity of Wnt/β-catenin pathway through the miR-1253/NOVA2 axis. Thorac. Cancer 2022, 13, 3133–3144. [Google Scholar] [CrossRef]
- Ning, M.Y.; Cheng, Z.L.; Zhao, J. MicroRNA-448 targets SATB1 to reverse the cisplatin resistance in lung cancer via mediating Wnt/β-catenin signalling pathway. J. Biochem. 2020, 168, 41–51. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Li, X.; Cai, C.; Hong, C.; Zhang, B. MicroRNA-32 and MicroRNA-548a Promote the Drug Sensitivity of Non-Small Cell Lung Cancer Cells to Cisplatin by Targeting ROBO1 and Inhibiting the Activation of Wnt/β-Catenin Axis. Cancer Manag. Res. 2021, 13, 3005–3016. [Google Scholar] [CrossRef]
- Li, X.; Zheng, H. LncRNA SNHG1 influences cell proliferation, migration, invasion, and apoptosis of non-small cell lung cancer cells via the miR-361-3p/FRAT1 axis. Thorac. Cancer 2020, 11, 295–304. [Google Scholar] [CrossRef]
- Shi, S.L.; Zhang, Z.H. Long non-coding RNA SNHG1 contributes to cisplatin resistance in non-small cell lung cancer by regulating miR-140-5p/Wnt/β-catenin pathway. Neoplasma 2019, 66, 756–765. [Google Scholar] [CrossRef]
- Lu, M.; Xiong, H.; Xia, Z.K.; Liu, B.; Wu, F.; Zhang, H.X.; Hu, C.H.; Liu, P. circRACGAP1 promotes non-small cell lung cancer proliferation by regulating miR-144-5p/CDKL1 signaling pathway. Cancer Gene Ther. 2021, 28, 197–211. [Google Scholar] [CrossRef] [PubMed]
- Ye, Y.; Zhao, L.; Li, Q.; Xi, C.; Li, Y.; Li, Z. circ_0007385 served as competing endogenous RNA for miR-519d-3p to suppress malignant behaviors and cisplatin resistance of non-small cell lung cancer cells. Thorac. Cancer 2020, 11, 2196–2208. [Google Scholar] [CrossRef]
- Parizadeh, S.M.; Jafarzadeh-Esfehani, R.; Fazilat-Panah, D.; Hassanian, S.M.; Shahidsales, S.; Khazaei, M.; Parizadeh, S.M.R.; Ghayour-Mobarhan, M.; Ferns, G.A.; Avan, A. The potential therapeutic and prognostic impacts of the c-MET/HGF signaling pathway in colorectal cancer. IUBMB Life 2019, 71, 802–811. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Tu, J.; Liu, C.; Wang, L.; Yuan, X. MicroRNA-621 functions as a metastasis suppressor in colorectal cancer by directly targeting LEF1 and suppressing Wnt/β-catenin signaling. Life Sci. 2022, 308, 120941. [Google Scholar] [CrossRef]
- Han, T.; Gao, M.; Wang, X.; Li, W.; Zhuo, J.; Qu, Z.; Chen, Y. LINC00665 activates Wnt/β-catenin signaling pathway to facilitate tumor progression of colorectal cancer via upregulating CTNNB1. Exp. Mol. Pathol. 2021, 120, 104639. [Google Scholar] [CrossRef]
- Lv, L.; Huang, B.; Yi, L.; Zhang, L. Long non-coding RNA SNHG4 enhances RNF14 mRNA stability to promote the progression of colorectal cancer by recruiting TAF15 protein. Apoptosis 2022, 28, 414–431. [Google Scholar] [CrossRef]
- Zhang, X.; Yao, J.; Shi, H.; Gao, B.; Zhou, H.; Zhang, Y.; Zhao, D.; Gao, S.; Wang, C.; Zhang, L. Hsa_circ_0026628 promotes the development of colorectal cancer by targeting SP1 to activate the Wnt/β-catenin pathway. Cell Death Dis. 2021, 12, 802. [Google Scholar] [CrossRef]
- Du, Y.L.; Liang, Y.; Shi, G.Q.; Cao, Y.; Qiu, J.; Yuan, L.; Yong, Z.; Liu, L.; Li, J. LINC00689 participates in proliferation, chemoresistance and metastasis via miR-31-5p/YAP/β-catenin axis in colorectal cancer. Exp. Cell Res. 2020, 395, 112176. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Wang, L.; Sun, Q.; Chen, R.; Zhang, C.; Yang, P.; Tan, Y.; Peng, C.; Wang, T.; Jin, C.; et al. Hsa_circ_0001666 suppresses the progression of colorectal cancer through the miR-576-5p/PCDH10 axis. Clin. Transl. Med. 2021, 11, e565. [Google Scholar] [CrossRef]
- Tani, H.; Torimura, M. Identification of short-lived long non-coding RNAs as surrogate indicators for chemical stress response. Biochem. Biophys. Res. Commun. 2013, 439, 547–551. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Lin, H.; Kang, L.; Huang, P.; Huang, J.; Cai, J.; Xian, Z.; Zhu, P.; Huang, M.; Wang, L.; et al. Aberrant expression of long noncoding RNA SNHG15 correlates with liver metastasis and poor survival in colorectal cancer. J. Cell. Physiol. 2019, 234, 7032–7039. [Google Scholar] [CrossRef]
- Sun, X.; Bai, Y.; Yang, C.; Hu, S.; Hou, Z.; Wang, G. Long noncoding RNA SNHG15 enhances the development of colorectal carcinoma via functioning as a ceRNA through miR-141/SIRT1/Wnt/β-catenin axis. Artif. Cells Nanomed. Biotechnol. 2019, 47, 2536–2544. [Google Scholar] [CrossRef]
- Li, M.; Bian, Z.; Jin, G.; Zhang, J.; Yao, S.; Feng, Y.; Wang, X.; Yin, Y.; Fei, B.; You, Q.; et al. LncRNA-SNHG15 enhances cell proliferation in colorectal cancer by inhibiting miR-338-3p. Cancer Med. 2019, 8, 2404–2413. [Google Scholar] [CrossRef]
- Saeinasab, M.; Bahrami, A.R.; González, J.; Marchese, F.P.; Martinez, D.; Mowla, S.J.; Matin, M.M.; Huarte, M. SNHG15 is a bifunctional MYC-regulated noncoding locus encoding a lncRNA that promotes cell proliferation, invasion and drug resistance in colorectal cancer by interacting with AIF. J. Exp. Clin. Cancer Res. CR 2019, 38, 172. [Google Scholar] [CrossRef]
- Jiang, H.; Li, T.; Qu, Y.; Wang, X.; Li, B.; Song, J.; Sun, X.; Tang, Y.; Wan, J.; Yu, Y.; et al. Long non-coding RNA SNHG15 interacts with and stabilizes transcription factor Slug and promotes colon cancer progression. Cancer Lett. 2018, 425, 78–87. [Google Scholar] [CrossRef] [PubMed]
- Olatubosun, M.O.; Abubakar, M.B.; Batiha, G.E.; Malami, I.; Ibrahim, K.G.; Abubakar, B.; Bello, M.B.; Alexiou, A.; Imam, M.U. LncRNA SNHG15: A potential therapeutic target in the treatment of colorectal cancer. Chem. Biol. Drug Des. 2023, 101, 1138–1150. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer Statistics, 2021. CA Cancer J. Clin. 2021, 71, 7–33. [Google Scholar] [CrossRef]
- Basu, S.; Majumder, S.; Bhowal, A.; Ghosh, A.; Naskar, S.; Nandy, S.; Mukherjee, S.; Sinha, R.K.; Basu, K.; Karmakar, D.; et al. A study of molecular signals deregulating mismatch repair genes in prostate cancer compared to benign prostatic hyperplasia. PLoS ONE 2015, 10, e0125560. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Zhang, G.; Wei, M.; Lu, X.; Fu, H.; Feng, F.; Wang, S.; Lu, W.; Wu, N.; Lu, Z.; et al. The tumor suppressing effects of QKI-5 in prostate cancer: A novel diagnostic and prognostic protein. Cancer Biol. Ther. 2014, 15, 108–118. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Xie, S.; Liu, J.; Li, T.; Wang, W.; Xie, Z. MicroRNA-4429 suppresses proliferation of prostate cancer cells by targeting distal-less homeobox 1 and inactivating the Wnt/β-catenin pathway. BMC Urol. 2021, 21, 40. [Google Scholar] [CrossRef]
- Dai, J.; Yuan, G.; Li, Y.; Zhou, H. MicroRNA-596 is epigenetically inactivated and suppresses prostatic cancer cell growth and migration via regulating Wnt/β-catenin signaling. Clin. Transl. Oncol. 2021, 23, 1394–1404. [Google Scholar] [CrossRef]
- Fan, L.; Wang, Y.; Huo, W.; Wang, W.H. MicroRNA-301a-3p overexpression promotes cell invasion and proliferation by targeting runt-related transcription factor 3 in prostate cancer. Mol. Med. Rep. 2019, 20, 3755–3763. [Google Scholar] [CrossRef] [PubMed]
- Xing, R. miR-3648 Promotes Prostate Cancer Cell Proliferation by Inhibiting Adenomatous Polyposis Coli 2. J. Nanosci. Nanotechnol. 2019, 19, 7526–7531. [Google Scholar] [CrossRef]
- Ding, L.; Lin, Y.; Chen, X.; Wang, R.; Lu, H.; Wang, H.; Luo, W.; Lu, Z.; Xia, L.; Zhou, X.; et al. circPHF16 suppresses prostate cancer metastasis via modulating miR-581/RNF128/Wnt/β-catenin pathway. Cell. Signal. 2023, 102, 110557. [Google Scholar] [CrossRef]
- Wang, X.; Wang, R.; Wu, Z.; Bai, P. Circular RNA ITCH suppressed prostate cancer progression by increasing HOXB13 expression via spongy miR-17-5p. Cancer Cell Int. 2019, 19, 328. [Google Scholar] [CrossRef]
- Han, Y.; Hu, H.; Zhou, J. Knockdown of LncRNA SNHG7 inhibited epithelial-mesenchymal transition in prostate cancer though miR-324-3p/WNT2B axis in vitro. Pathol. Res. Pract. 2019, 215, 152537. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.C.; Song, L.L.; Liang, Q.; Hao, L.; Zhang, Z.G.; Han, C.H. Long noncoding RNA LEF1-AS1 silencing suppresses the initiation and development of prostate cancer by acting as a molecular sponge of miR-330-5p via LEF1 repression. J. Cell. Physiol. 2019, 234, 12727–12744. [Google Scholar] [CrossRef]
- Wang, L.; Wang, L.; Wang, Q.; Yosefi, B.; Wei, S.; Wang, X.; Shen, D. The function of long noncoding RNA HOTAIRM1 in the progression of prostate cancer cells. Andrologia 2021, 53, e13897. [Google Scholar] [CrossRef]
- Rahmani, F.; Safavi, P.; Fathollahpour, A.; Tanhaye Kalate Sabz, F.; Tajzadeh, P.; Arefnezhad, M.; Ferns, G.A.; Hassanian, S.M.; Avan, A. The interplay between non-coding RNAs and Wnt/ß-catenin signaling pathway in urinary tract cancers: From tumorigenesis to metastasis. EXCLI J. 2022, 21, 1273–1284. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Yang, Y.; Ren, L.; Yang, J.; Wang, B.; Xing, T.; Chen, H.; Chen, M. miR-15a-3p Suppresses Prostate Cancer Cell Proliferation and Invasion by Targeting SLC39A7 Via Downregulating Wnt/β-Catenin Signaling Pathway. Cancer Biother. Radiopharm. 2019, 34, 472–479. [Google Scholar] [CrossRef] [PubMed]
- Dong, B.; Xu, G.C.; Liu, S.T.; Liu, T.; Geng, B. MiR-34a affects G2 arrest in prostate cancer PC3 cells via Wnt pathway and inhibits cell growth and migration. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 8349–8358. [Google Scholar] [CrossRef]
- Wang, M.; Yin, C.; Wu, Z.; Wang, X.; Lin, Q.; Jiang, X.; Du, H.; Lang, C.; Peng, X.; Dai, Y. The long transcript of lncRNA TMPO-AS1 promotes bone metastases of prostate cancer by regulating the CSNK2A1/DDX3X complex in Wnt/β-catenin signaling. Cell Death Discov. 2023, 9, 287. [Google Scholar] [CrossRef]
- Liu, S.; Wang, Q.; Liu, Y.; Xia, Z.Y. miR-425-5p suppresses tumorigenesis and DDP resistance in human-prostate cancer by targeting GSK3β and inactivating the Wnt/β-catenin signaling pathway. J. Biosci. 2019, 44, 102. [Google Scholar] [CrossRef]
- Mirzaei, S.; Paskeh, M.D.A.; Okina, E.; Gholami, M.H.; Hushmandi, K.; Hashemi, M.; Kalu, A.; Zarrabi, A.; Nabavi, N.; Rabiee, N.; et al. Molecular Landscape of LncRNAs in Prostate Cancer: A focus on pathways and therapeutic targets for intervention. J. Exp. Clin. Cancer Res. CR 2022, 41, 214. [Google Scholar] [CrossRef]
- Venerito, M.; Vasapolli, R.; Rokkas, T.; Malfertheiner, P. Gastric cancer: Epidemiology, prevention, and therapy. Helicobacter 2018, 23 (Suppl. S1), e12518. [Google Scholar] [CrossRef]
- Rawla, P.; Barsouk, A. Epidemiology of gastric cancer: Global trends, risk factors and prevention. Przegląd Gastroenterol. 2019, 14, 26–38. [Google Scholar] [CrossRef] [PubMed]
- Smyth, E.C.; Nilsson, M.; Grabsch, H.I.; van Grieken, N.C.; Lordick, F. Gastric cancer. Lancet 2020, 396, 635–648. [Google Scholar] [CrossRef] [PubMed]
- Shi, W.J.; Gao, J.B. Molecular mechanisms of chemoresistance in gastric cancer. World J. Gastrointest. Oncol. 2016, 8, 673–681. [Google Scholar] [CrossRef]
- Akhavanfar, R.; Shafagh, S.G.; Mohammadpour, B.; Farahmand, Y.; Lotfalizadeh, M.H.; Kookli, K.; Adili, A.; Siri, G.; Eshagh Hosseini, S.M. A comprehensive insight into the correlation between ncRNAs and the Wnt/β-catenin signalling pathway in gastric cancer pathogenesis. Cell Commun. Signal. CCS 2023, 21, 166. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Zhang, X.; Lin, H.; Deng, S.; Qin, Y.; He, J.; Hu, F.; Zhu, X.; Feng, X.; Wang, J.; et al. Dual activation of Hedgehog and Wnt/β-catenin signaling pathway caused by downregulation of SUFU targeted by miRNA-150 in human gastric cancer. Aging 2021, 13, 10749–10769. [Google Scholar] [CrossRef]
- Ashrafizadeh, M.; Rafiei, H.; Mohammadinejad, R.; Farkhondeh, T.; Samarghandian, S. Wnt-regulating microRNAs role in gastric cancer malignancy. Life Sci. 2020, 250, 117547. [Google Scholar] [CrossRef]
- Chen, J.Q.; Huang, Z.P.; Li, H.F.; Ou, Y.L.; Huo, F.; Hu, L.K. MicroRNA-520f-3p inhibits proliferation of gastric cancer cells via targeting SOX9 and thereby inactivating Wnt signaling. Sci. Rep. 2020, 10, 6197. [Google Scholar] [CrossRef]
- Fang, X.; Pan, A. MiR-507 inhibits the progression of gastric carcinoma via targeting CBX4-mediated activation of Wnt/β-catenin and HIF-1α pathways. Clin. Transl. Oncol. 2022, 24, 2021–2028. [Google Scholar] [CrossRef]
- Li, J.; Sun, J.; Liu, Z.; Zeng, Z.; Ouyang, S.; Zhang, Z.; Ma, M.; Kang, W. The Roles of Non-Coding RNAs in Radiotherapy of Gastrointestinal Carcinoma. Front. Cell Dev. Biol. 2022, 10, 862563. [Google Scholar] [CrossRef]
- Liu, W.G.; Xu, Q. Upregulation of circHIPK3 promotes the progression of gastric cancer via Wnt/β-catenin pathway and indicates a poor prognosis. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 7905–7912. [Google Scholar] [CrossRef]
- Wang, L.; Li, B.; Yi, X.; Xiao, X.; Zheng, Q.; Ma, L. Circ_SMAD4 promotes gastric carcinogenesis by activating wnt/β-catenin pathway. Cell Prolif. 2021, 54, e12981. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Xu, Y.; Zhang, X.; Deng, S.; Yuan, Y.; Luo, X.; Hossain, M.T.; Zhu, X.; Du, K.; Hu, F.; et al. A novel protein AXIN1-295aa encoded by circAXIN1 activates the Wnt/β-catenin signaling pathway to promote gastric cancer progression. Mol. Cancer 2021, 20, 158. [Google Scholar] [CrossRef] [PubMed]
- Luan, P.B.; Sun, X.M.; Yao, J. LINC00355 inhibits apoptosis and promotes proliferation of gastric cancer cells by regulating Wnt/β-catenin signaling pathway. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 8377–8383. [Google Scholar] [CrossRef]
- Peng, Y.; Zhang, X.; Lin, H.; Deng, S.; Qin, Y.; Yuan, Y.; Feng, X.; Wang, J.; Chen, W.; Hu, F.; et al. SUFU mediates EMT and Wnt/β-catenin signaling pathway activation promoted by miRNA-324-5p in human gastric cancer. Cell Cycle 2020, 19, 2720–2733. [Google Scholar] [CrossRef]
- Shi, Q.; Zhou, C.; Xie, R.; Li, M.; Shen, P.; Lu, Y.; Ma, S. CircCNIH4 inhibits gastric cancer progression via regulating DKK2 and FRZB expression and Wnt/β-catenin pathway. J. Biol. Res. 2021, 28, 19. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhang, K.; Hou, Y. Long non-coding RNA NNT-AS1 knockdown represses the progression of gastric cancer via modulating the miR-142-5p/SOX4/Wnt/β-catenin signaling pathway. Mol. Med. Rep. 2020, 22, 687–696. [Google Scholar] [CrossRef]
- Yang, C.; Han, S. The circular RNA circ0005654 interacts with specificity protein 1 via microRNA-363 sequestration to promote gastric cancer progression. Bioengineered 2021, 12, 6305–6317. [Google Scholar] [CrossRef]
- Chen, Y.; Liu, H.; Zou, J.; Cao, G.; Li, Y.; Xing, C.; Wu, J. Exosomal circ_0091741 promotes gastric cancer cell autophagy and chemoresistance via the miR-330-3p/TRIM14/Dvl2/Wnt/β-catenin axis. Hum. Cell 2023, 36, 258–275. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.G.; Li, X.B.; Yin, R.H.; Li, X.F. lncRNA VIM-AS1 promotes cell proliferation, metastasis and epithelial-mesenchymal transition by activating the Wnt/β-catenin pathway in gastric cancer. Mol. Med. Rep. 2020, 22, 4567–4578. [Google Scholar] [CrossRef]
- Xu, Z.; Ran, J.; Gong, K.; Hou, Y.; Li, J.; Guo, Y. LncRNA SUMO1P3 regulates the invasion, migration and cell cycle of gastric cancer cells through Wnt/β-catenin signaling pathway. J. Recept. Signal Transduct. Res. 2021, 41, 574–581. [Google Scholar] [CrossRef]
- Zhou, W.; Ding, X.; Jin, P.; Li, P. miR-6838-5p Affects Cell Growth, Migration, and Invasion by Targeting GPRIN3 via the Wnt/β-Catenin Signaling Pathway in Gastric Cancer. Pathobiol. J. Immunopathol. Mol. Cell. Biol. 2020, 87, 327–337. [Google Scholar] [CrossRef]
- Chen, B.; Dragomir, M.P.; Yang, C.; Li, Q.; Horst, D.; Calin, G.A. Targeting non-coding RNAs to overcome cancer therapy resistance. Signal Transduct. Target. Ther. 2022, 7, 121. [Google Scholar] [CrossRef] [PubMed]
- Zhao, E.; Lan, Y.; Quan, F.; Zhu, X.; Suru, A.; Wan, L.; Xu, J.; Hu, J. Identification of a Six-lncRNA Signature With Prognostic Value for Breast Cancer Patients. Front. Genet. 2020, 11, 673. [Google Scholar] [CrossRef]
| Cancer Type | ncRNAs | Expression | Mechanisms | Functions to Wnt Pathway | PMID |
|---|---|---|---|---|---|
| Breast cancer | miR-125b | Up | Wnt/β-catenin | Activation | 30950326 |
| miR-296-3p | Down | SOX4 | Suppression | 34785625 | |
| miR-96-5p | Down | CTNND1 | Suppression | 31913290 | |
| miR-9501 | Up | β-catenin | Suppression | 32373971 | |
| miR-548c-5p | Down | Wnt1 | Suppression | 32329856 | |
| miR-516a-3p | Down | Pygopus2 | Suppression | 31273950 | |
| miR-454-3p | Up | RPRD1A | Activation | 30809286 | |
| miR-449b-5p | Down | CREPT | Suppression | 30738779 | |
| miR-429 | Down | Wnt/β-catenin | Suppression | 32961031 | |
| miR-34a | Up | Wnt3/Wnt1 | Activation | 30779084 | |
| miR-340-5p | Down | LGR5 | Suppression | 30300682 | |
| miR-296 | Down | FGFR1 | Suppression | 31841196 | |
| miR-27a | Up | GSK-3β | Activation | 33025840 | |
| miR-216a | Down | Wnt/β-catenin | Suppression | 30864744 | |
| miR-193b | Down | c-Met | Suppression | 34863149 | |
| miR-190 | Down | SOX9 | Activation | 30658681 | |
| miR-135 | Down | Wnt/β-catenin | Suppression | 35730603 | |
| miR-130a-3p | Down | NRARP | Suppression | 35797350 | |
| miR-124-3p.1 | Up | Axin1 | Activation | 32125723 | |
| lncRNA MICAL2-1 | Down | miR-25/DKK3 | Suppression | 34970696 | |
| lncRNA RP11-283G6.5 | Down | miR-188-3p/TMED3 | Suppression | 34416888 | |
| lncRNA RUSC1-AS-N | Up | Wnt1/β-catenin | Activation | 30569097 | |
| lncRNA RBM5-AS1 | Up | Wnt/β-catenin | Activation | 35110544 | |
| lncRNA HOTTIP | Up | miR-148a-3p/WNT1 | Activation | 32307830 | |
| lncRNA HOTTIP | Up | Wnt/β-catenin | Activation | 30676763 | |
| lncRNA H19 | Up | miR-340-3p/YWHAZ | Activation | 30676763 | |
| lncRNA C5orf66-AS1 | Up | miR-149-5p/CTCF/CTNNB1 | Activation | 35499320 | |
| lncRNA ASMTL-AS1 | Down | miR-1228-3p/SOX17 | Suppression | 34006305 | |
| lncRNA LUCAT1 | Up | miR-5582-3p/TCF7L2 | Activation | 31300015 | |
| lncRNA CCAT1 | Up | miR-204/211/miR-148a/152/ANXA2 | Activation | 31695775 | |
| LINC01287 | Up | Wnt/β-catenin | Activation | 31173295 | |
| LINC01234 | Up | miR-525-5p/MEIS2 | Activation | 34173712 | |
| circ_0008784 | Up | miR-506-3p/CTNNB1 | Activation | 36436315 | |
| circ-ITCH | Down | miR-214/miR-17 | Activation | 30509108 | |
| circARL8B | Up | miR-653-5p/HMGA2 | Activation | 34050452 | |
| circABCC4 | Up | miR-154-5p | Suppression | 34050452 | |
| Lung cancer | miR-590 | Down | YAP1 | Suppression | 35031966 |
| miR-448 | Down | SATB1 | Activation | 32525527 | |
| miR-489-3p | Up | USP48 | Activation | 35413838 | |
| miR-421 | Up | HOPX | Activation | 31115507 | |
| miR-23B | Down | RUNX2 | Suppression | 32495614 | |
| miR-20b | Down | APC | Activation | 31894264 | |
| miR-1b-19p | Down | MYPT3 | Suppression | 33964297 | |
| miR-147b | Down | RPS15A | Suppression | 31665807 | |
| miR-103 | Up | KLF7 | Activation | 32582959 | |
| miR-100 | Down | HOXA1 | Suppression | 32364673 | |
| miR-520a | Up | RRM2 | Suppression | 33859925 | |
| lncRNA SNHG11 | Up | miR-4436a/CTNNB1 | Activation | 32239719 | |
| lncRNA FLVCR1-AS1 | Up | Wnt/β-catenin | Suppression | 30697812 | |
| lncRNA-SNHG7 | Down | miR-181/cbx7 | Suppression | 32201260 | |
| lncRNASEH1-AS1 | Up | miR-516a-5p/FOXK1 | Activation | 35166053 | |
| lncRNA SNHG20 | Up | miR-197/TCF/LEF1 | Activation | 31957836 | |
| lncRNA PVT1 | Up | miR-361-3p/SOX9 | Activation | 32197208 | |
| lncRNA JPX | Up | miR-33a-5p/Twist1 | Activation | 32197208 | |
| lncRNA HJURP | Up | β-catenin | Suppression | 31115012 | |
| LncRNA DSCAM-AS1 | Up | miR-577/HMGB1 | Activation | 32386483 | |
| lncRNA AWPPH | Up | Wnt/β-catenin | Activation | 32386483 | |
| LncDBH-AS1 | Down | miR-155/AXIN1 | Activation | 33506901 | |
| LINC01006 | Up | miR-129-2-3p/CTNNB1 | Activation | 33753463 | |
| LINC00942 | Up | miR-5006-5p/FZD1 | Activation | 34253104 | |
| LINC00669 | Up | Wnt/β-catenin | Activation | 36621836 | |
| LINC00326 | Up | miR-657/DKK2 | Suppression | 36747258 | |
| LINC00673-v4 | Up | DDX3/CK1ε | Activation | 31235588 | |
| circ-EIF3I | Up | miR-1253/NOVA2 | Activation | 36193788 | |
| has_circ_0017109 | Up | miR-671-5p/FZD4 | Activation | 36434577 | |
| has_circ_0001946 | Up | miR-135a-5p/SIRT1 | Activation | 30841451 | |
| hsa_circ_0066903 | Down | miR-3681-3p/miR-3909/GSK3B | Suppression | 35821283 | |
| hsa_circ_0007059 | Down | miR-378 | Suppression | 31351967 | |
| has_circ_0006427 | Down | miR-6783-3p/DKK1 | Suppression | 30470570 | |
| circ-ZNF124 | Up | miR-498/YES1 | Suppression | 33186139 | |
| circVAPA | Up | miR-876-5p/WNT5a | Activation | 33619796 | |
| circ-PGC | Up | miR-2-532p/FOXR3 | Activation | 34494941 | |
| circ_0067934 | Up | miR-1182/KLF8 | Activation | 32768951 | |
| Colorectal cancer | miR-621 | Down | LEF1 | Suppression | 36087740 |
| miR-576-5p | Up | Wnt5a | Activation | 33300054 | |
| miR-532-3p | Down | ETS3/TGM1 | Suppression | 31570702 | |
| miR-501-3p | Up | APC | Activation | 31364752 | |
| miR-381 | Down | SPIN1 | Activation | 34753384 | |
| miR-377-3p | Down | ZEB2/XIAP | Suppression | 32220639 | |
| miR-30-5p | Down | USP2 | Suppression | 30338942 | |
| miR-19a-3p | Up | FOXF2 | Suppression | 32103872 | |
| miR-188 | Up | FOXL1 | Activation | 37305399 | |
| miR-183-5p | Up | RCN2 | Suppression | 30896885 | |
| miR-144-3p | Down | BCL6 | Suppression | 32206063 | |
| miR-103/107 | Up | Axin2 | Activation | 31273221 | |
| miR-6125 | Down | YTHDF2 | Activation | 34709763 | |
| miR-520e | Down | AEG-1 | Suppression | 31574178 | |
| LINC00665 | Up | miR-214-3p/CTNNB1 | Activation | 33865827 | |
| lncRNA TUG1 | Up | miR-542-3p/TRIB2 | Activation | 34030715 | |
| lncRNA PART1 | Up | miR-150-5p/miR-520h/CTNNB1 | Activation | 31669140 | |
| lncRNA NEAT1 | Up | miR-486-5p/NR4A1 | Activation | 33337350 | |
| lncRNA NEAT1 | Up | miR-34a/SIRT1 | Activation | 30312725 | |
| lncRNA HCG18 | Up | miR-1271/MTDH | Activation | 31854468 | |
| lncRNA ADAMTS9-AS1 | Down | Wnt/β-catenin | Suppression | 32889785 | |
| LINC01315 | Up | Wnt/β-catenin | Activation | 35322763 | |
| LINC00963-v2/-v3 | Down | miR-143/miR-217/miR-512/APC/Axin | Suppression | 36804476 | |
| LINC00365 | Up | CDK1 | Activation | 31544991 | |
| circ_0082182 | Up | miR-411/miR-1205 | Activation | 33596920 | |
| hsa_circ_0026628 | Up | miR-346/SP1 | Activation | 34420031 | |
| hsa_circ_0068464 | Up | miR-383 | Activation | 35168468 | |
| hsa_circ_0009361 | Down | miR-582/APC2 | Suppression | 31109967 | |
| hsa_circ_0005615 | Up | miR-149-5p/TNKS | Activation | 32393760 | |
| hsa_circ_0005075 | Up | Wnt/β-catenin | Activation | 31081084 | |
| circRASSF2 | Up | miR-195-5p/FZD4 | Activation | 33929991 | |
| circPTK2 | Up | miR-136-5p/YTHDF1 | Activation | 34974791 | |
| circ-IGF1R | Up | miR-362-5p/HMGB3 | Activation | 36542208 | |
| circIFT80 | Up | miR-142/miR-568/miR-634/CTNNB1 | Activation | 35783013 | |
| circAGFG1 | Up | miR-4262/miR-185-5/pYY1/CTNNB1 | Activation | 32681092 | |
| circ-ACAP2 | Up | miR-143-3p/FZD4 | Activation | 34085707 | |
| circ_0026344 | Down | miR-183 | Suppression | 31608699 | |
| Prostate cancer | miR-4429 | Down | DLX1 | Suppression | 33740948 |
| miR-596 | Down | β-catenin | Suppression | 33387246 | |
| miR-15a-3p | Down | SLC39A7 | Suppression | 31135177 | |
| miR-34a | Down | Wnt1 | Suppression | 32894541 | |
| miR-425-5p | Down | GSK3β | Suppression | 31502580 | |
| miR-653-5p | Down | SOX30 | Suppression | 31889959 | |
| miR-95-3p | Up | DKK3 | Activation | 30779066 | |
| lncRNA SOX2-OT | Up | miR-452-5p/HMGB3 | Suppression | 32407168 | |
| lncRNA SNHG12 | Up | miR-195 | Activation | 30945357 | |
| lncRNA HOTTIP | Up | Wnt/β-catenin | Suppression | 30809864 | |
| LINC00115 | Up | miR-212-5p/FZD5 | Activation | 34697900 | |
| circPHF16 | Down | miR-581/RNF128 | Suppression | 36503162 | |
| Gastric cancer | miRNA-150 | Up | SUFU | Activation | 33848981 |
| miR-520f-3p | Down | SOX9 | Suppression | 32277152 | |
| miR-507 | Down | CBX4 | Suppression | 35819589 | |
| miR-324-5p | Up | SUFU | Activation | 33017570 | |
| miR-6838-5p | Down | GPRIN3 | Suppression | 33254176 | |
| miR-192/-215 | Up | APC | Activation | 32091625 | |
| miR-188-5p | Up | PTEN | Activation | 31138169 | |
| miR-195-5p | Down | YAP | Activation | 31378888 | |
| miR-381/miR-489 | Down | CUL4B | Suppression | 30483755 | |
| miR-675 | Up | PITX1 | Activation | 31260797 | |
| LINC00355 | Up | Wnt/β-catenin | Activation | 32894544 | |
| lncRNA NNT-AS1 | Up | miR-142-5p/SOX4 | Activation | 32468065 | |
| lncRNA VIM-AS1 | Up | miR-8052/FDZ1 | Activation | 33173977 | |
| lncRNA SUMO1P3 | Up | Wnt/β-catenin | Activation | 33179980 | |
| LINC01225 | Up | Wnt/β-catenin | Activation | 31460694 | |
| LINC01503 | Up | Wnt/β-catenin | Activation | 32207034 | |
| lncRNA H19 | Up | β-catenin | Activation | 34348271 | |
| lncRNA MIR4435-2HG | Up | DSP | Activation | 31484163 | |
| lncRNA NCK1-AS1 | Up | miR-22-3p/BCL9 | Activation | 33974352 | |
| lncRNA SNHG11 | Up | miR-483-3p/miR-1276/CTNNB1/ATG12 | Activation | 33068778 | |
| lncRNA ZEB2-AS1 | Up | Wnt/β-catenin | Activation | 30635820 | |
| lncRNA ZFAS1 | Up | miR-200b/Wnt1 | Activation | 30999814 | |
| LOC100505817 | Down | Wnt/β-catenin | Activation | 34385891 | |
| LOC285194 | Down | Wnt/β-catenin | Suppression | 31991056 | |
| circ0005654 | Up | miR-363/sp1 | Activation | 34499009 | |
| circ_0091741 | Up | miR-330-3p/TRIM14 | Activation | 36323918 | |
| circ-SFMBT2 | Up | miR-885-3p/CHD7 | Activation | 34387601 | |
| cir-ITCH | Down | miR-17 | Suppression | 33060778 | |
| hsa_circ_0001649 | Down | miR-20a/ERK | Suppression | 32212290 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, X.; Du, Y.; Luo, L.; Xu, X.; Xiong, S.; Yang, X.; Guo, L.; Liang, T. Deciphering the Enigmatic Influence: Non-Coding RNAs Orchestrating Wnt/β-Catenin Signaling Pathway in Tumor Progression. Int. J. Mol. Sci. 2023, 24, 13909. https://doi.org/10.3390/ijms241813909
Yang X, Du Y, Luo L, Xu X, Xiong S, Yang X, Guo L, Liang T. Deciphering the Enigmatic Influence: Non-Coding RNAs Orchestrating Wnt/β-Catenin Signaling Pathway in Tumor Progression. International Journal of Molecular Sciences. 2023; 24(18):13909. https://doi.org/10.3390/ijms241813909
Chicago/Turabian StyleYang, Xinbing, Yajing Du, Lulu Luo, Xinru Xu, Shizheng Xiong, Xueni Yang, Li Guo, and Tingming Liang. 2023. "Deciphering the Enigmatic Influence: Non-Coding RNAs Orchestrating Wnt/β-Catenin Signaling Pathway in Tumor Progression" International Journal of Molecular Sciences 24, no. 18: 13909. https://doi.org/10.3390/ijms241813909
APA StyleYang, X., Du, Y., Luo, L., Xu, X., Xiong, S., Yang, X., Guo, L., & Liang, T. (2023). Deciphering the Enigmatic Influence: Non-Coding RNAs Orchestrating Wnt/β-Catenin Signaling Pathway in Tumor Progression. International Journal of Molecular Sciences, 24(18), 13909. https://doi.org/10.3390/ijms241813909





