Genome-Wide Analysis of MBF1 Family Genes in Five Solanaceous Plants and Functional Analysis of SlER24 in Salt Stress
Abstract
:1. Introduction
2. Results
2.1. Identification of MBF1 Genes in Five Solanaceae Species
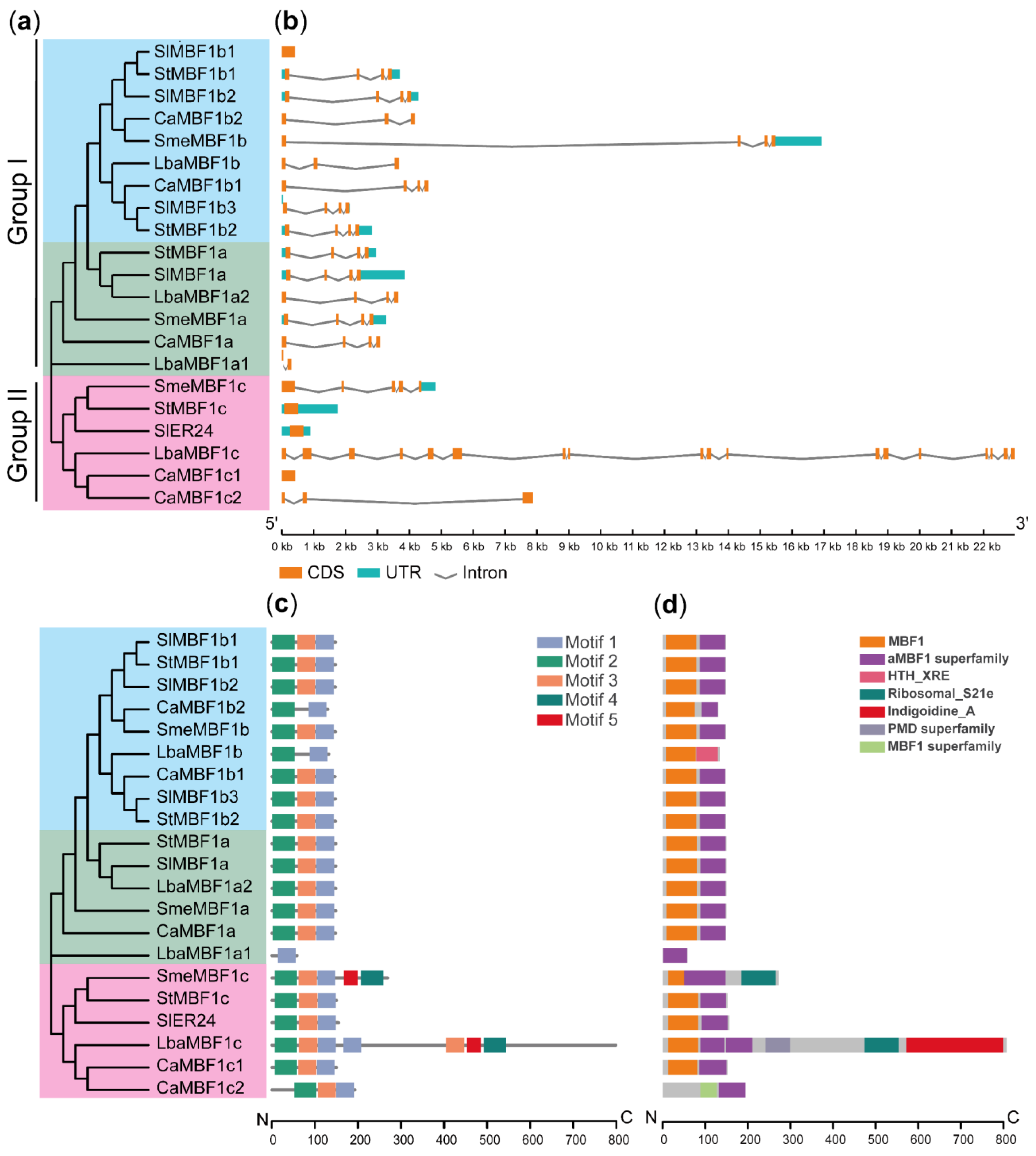
2.2. The Classification, Gene Structure, Motif Composition, and Conserved Domain of MBF1 Genes in Five Solanaceae Plants
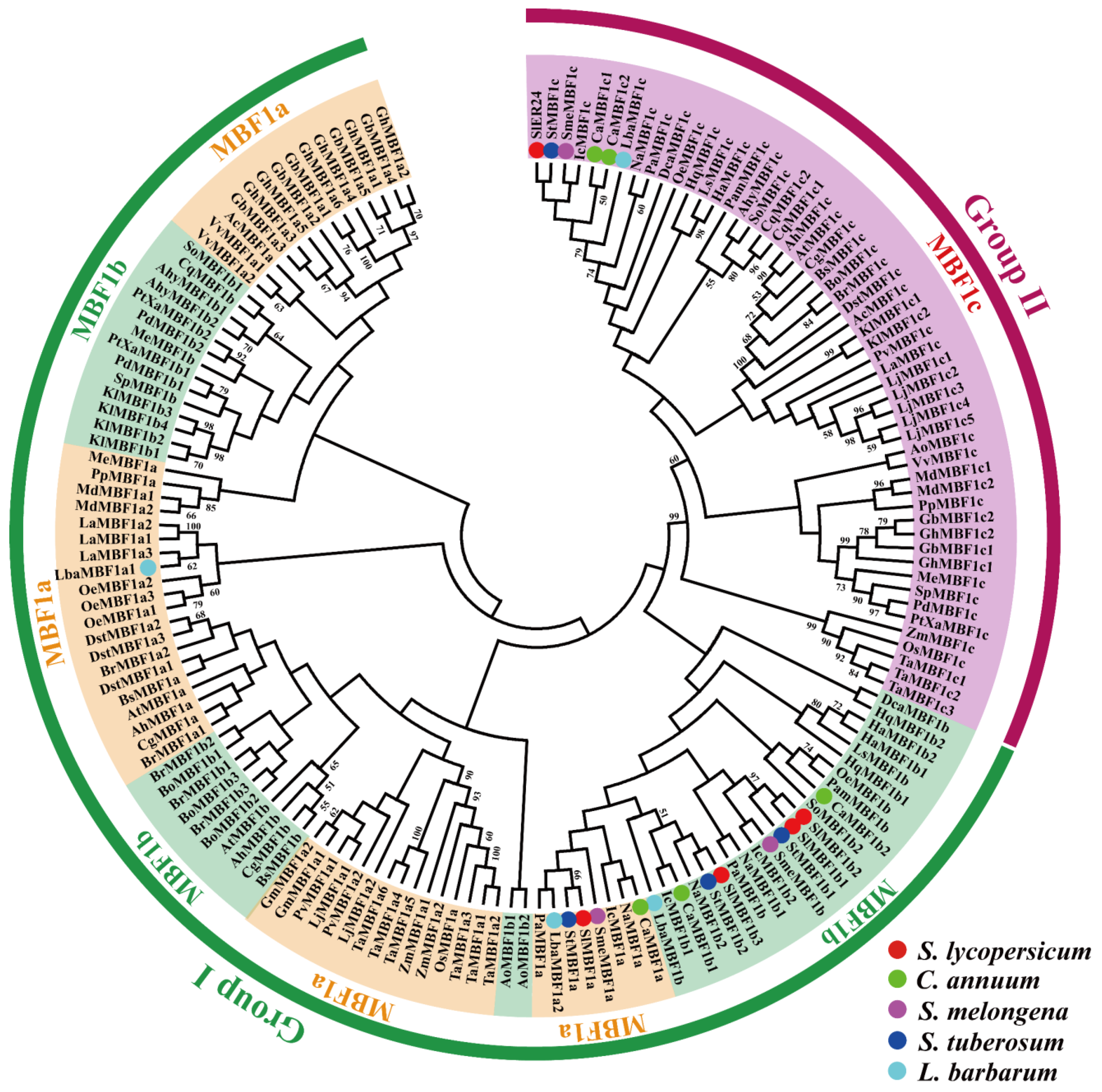
2.3. Phylogenetic Analysis of the MBF1 Family
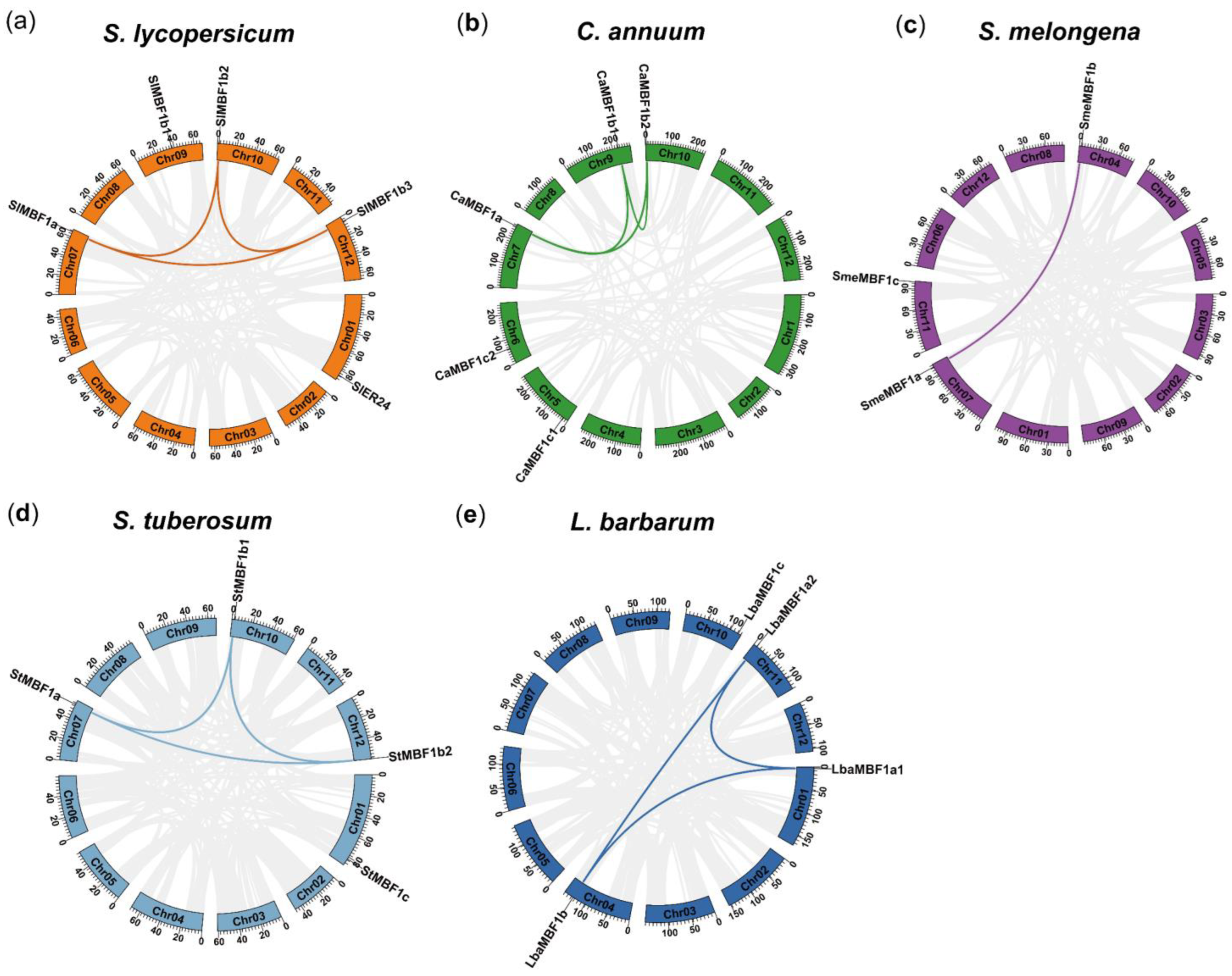
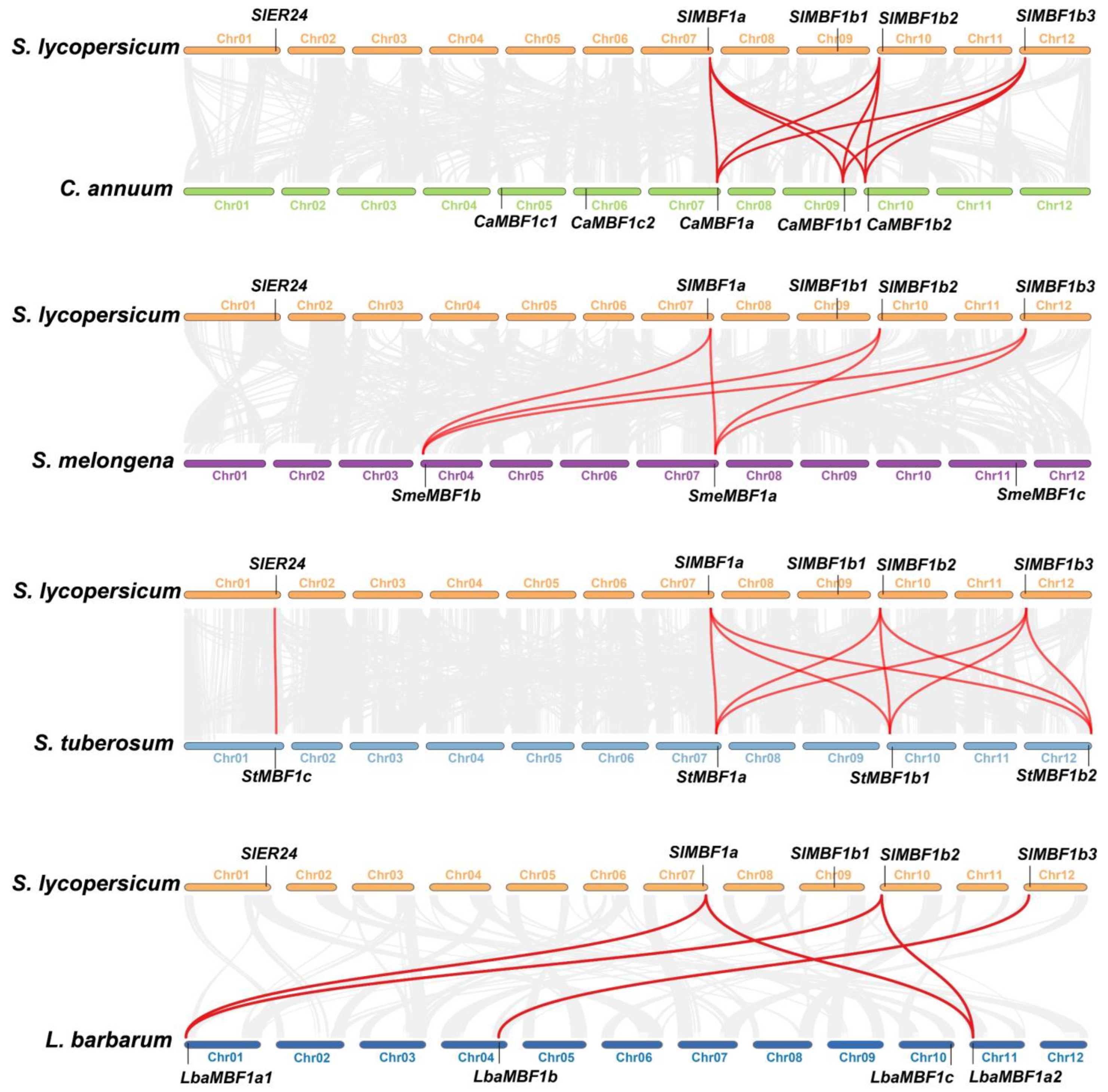
2.4. Chromosomal Location, Collinearity, and Gene Duplication Events Analysis of MBF1 Family in Five Solanaceae Plants
2.5. Subcellular Localization Analysis of MBF1 Family in Five Solanaceae Plants
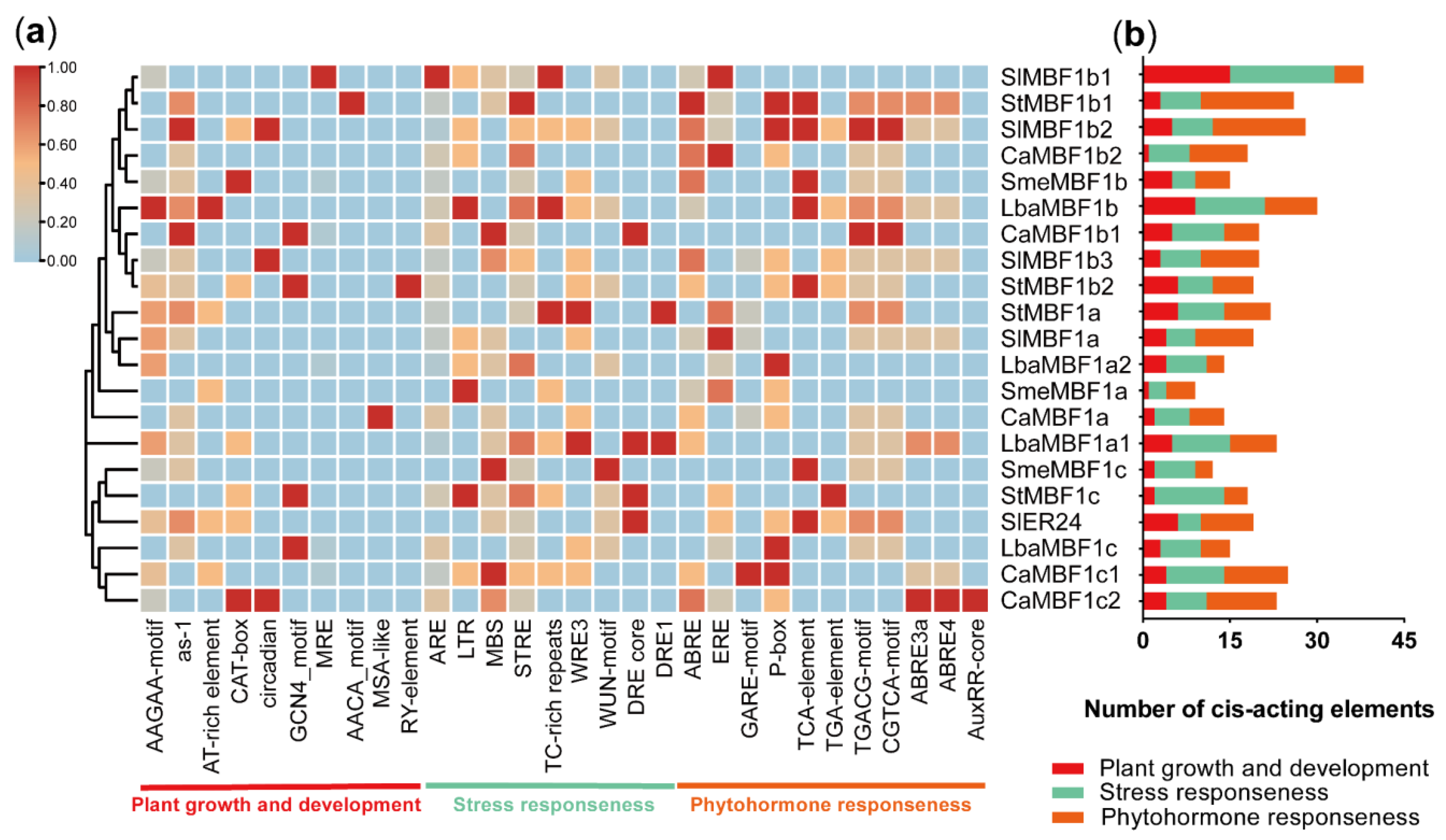
2.6. Analysis of cis-Elements in MBF1 Family of Five Solanaceae Plants
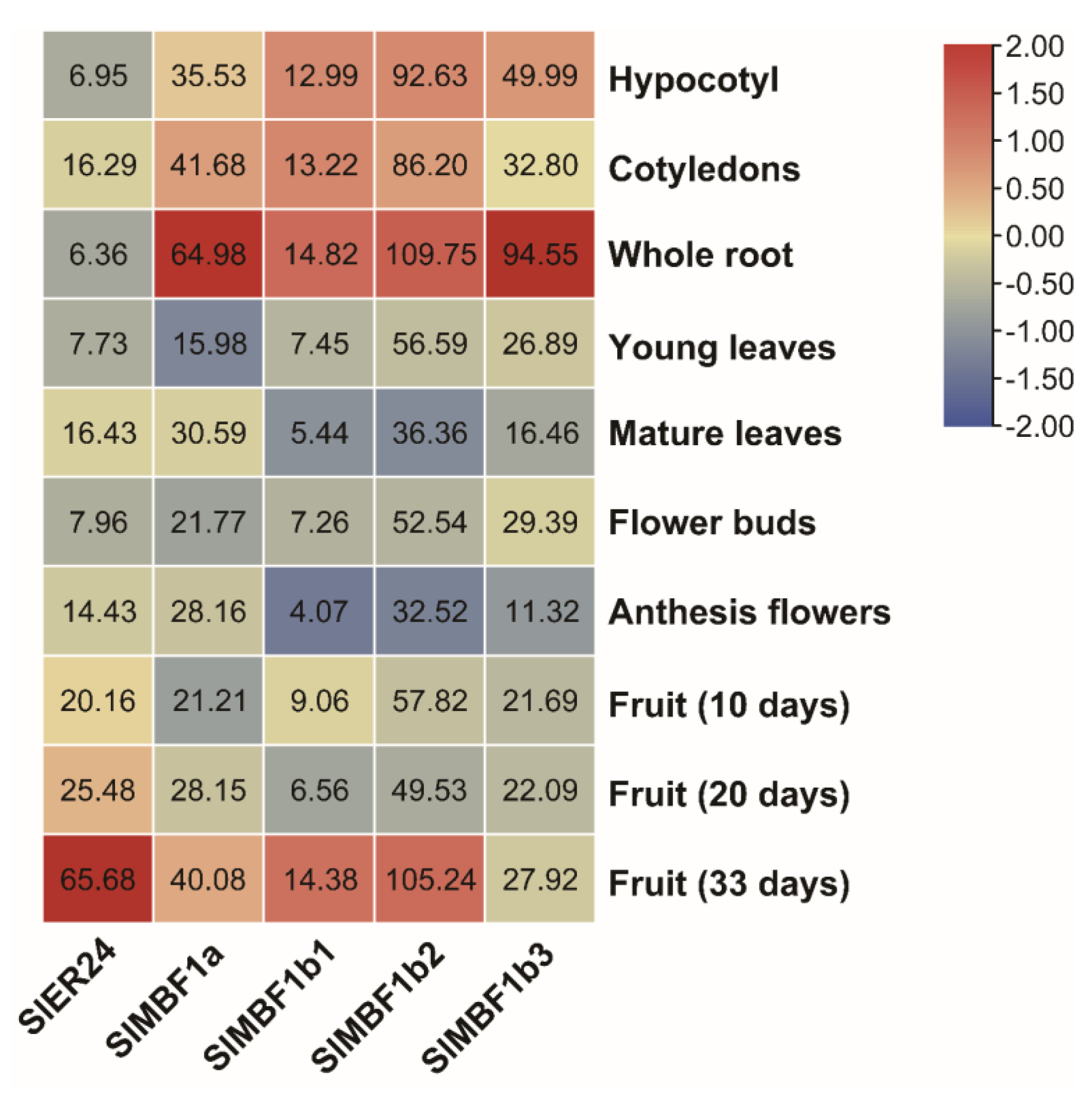
2.7. Analysis of the Expression Patterns of SlMBF1s in Tissue and Organ
2.8. Expression Patterns of SlMBF1s under NaCl, PEG, ABA, and Ethrel Treatments
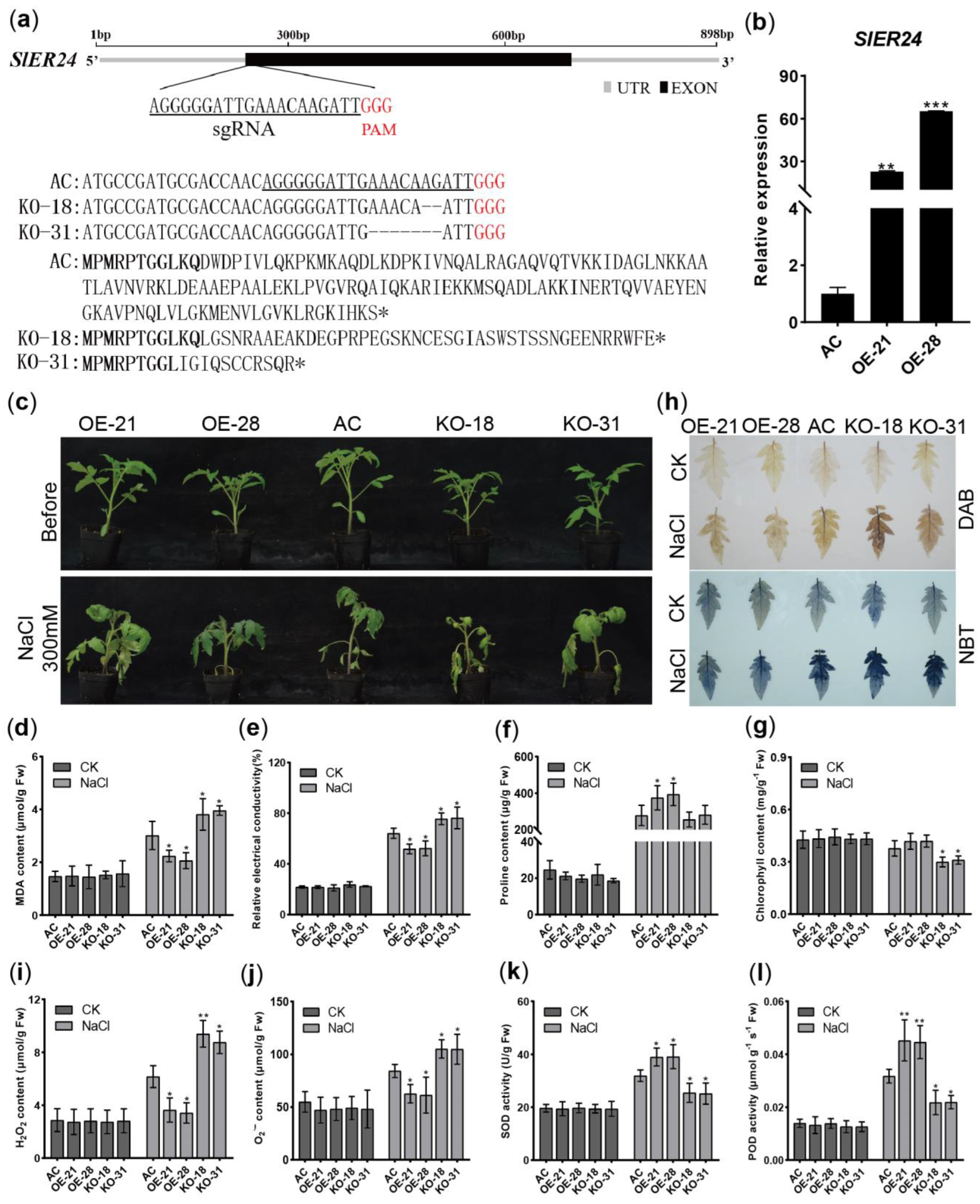
2.9. Overexpression of SlER24 Increases Salt Tolerance in Tomato
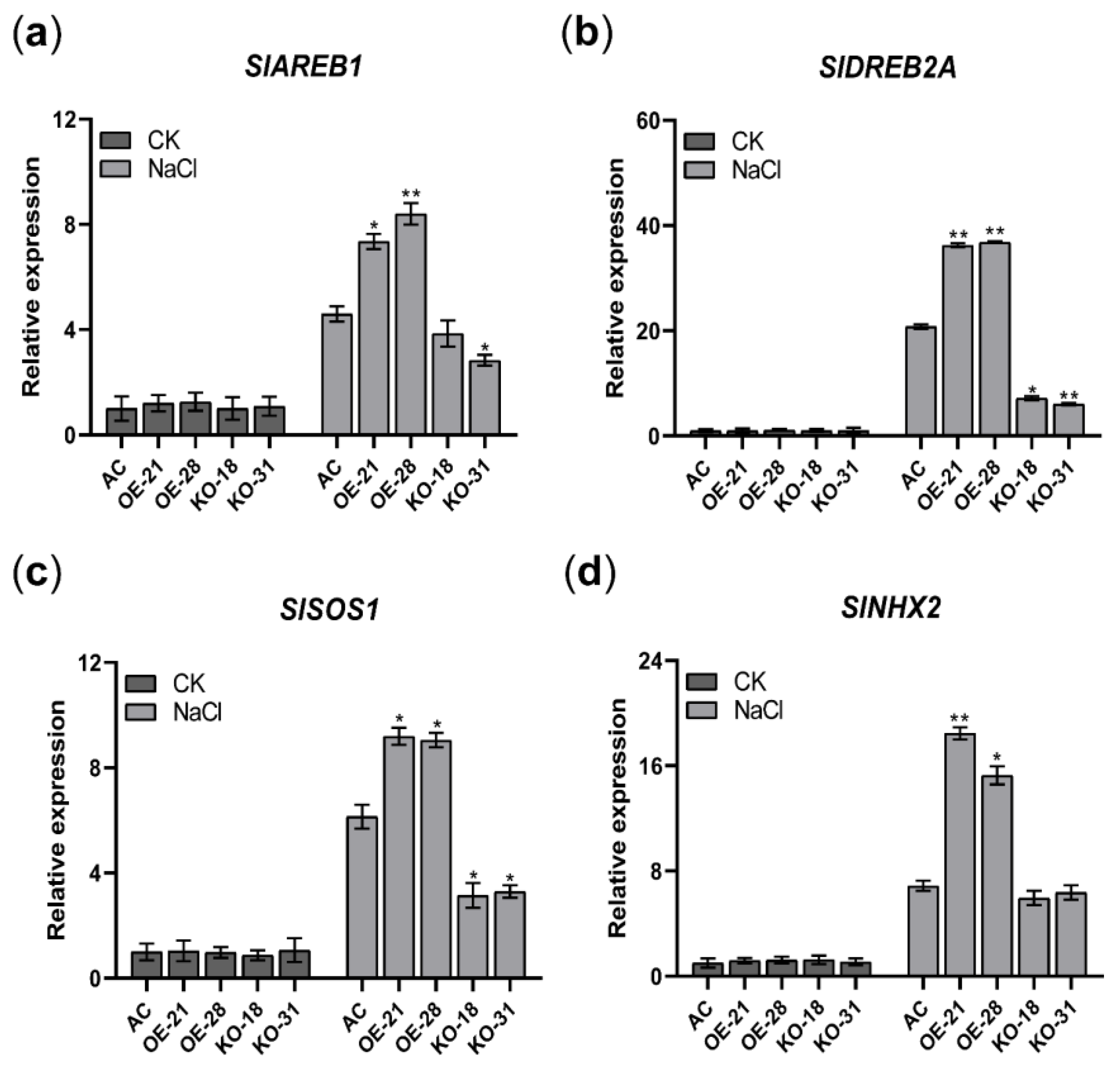
2.10. Overexpression of SlER24 Regulates the Expression of Salt Stress-Related Genes
3. Discussion
4. Materials and Methods
4.1. Identification of MBF1 Genes in Five Solanaceae Species
4.2. The Classification, Gene Structure, Motif Composition, and Conserved Domain of MBF1 Genes in Five Solanaceae Plants
4.3. Phylogenetic Analysis of the MBF1 Family
4.4. Chromosomal Location, Collinearity, and Gene Duplication Events Analysis of MBF1 Family in Five Solanaceae Plants
4.5. Subcellular Localization Analysis of MBF1 Family in Five Solanaceae Plants
4.6. Analysis of cis-Elements in MBF1 Family of Solanaceae
4.7. Analysis of the Expression Patterns of SlMBF1s in Tissue and Organ
4.8. Plant Materials and Treatments for qRT-PCR Analysis
4.9. Overexpression and CRISPR/Cas9 Vector Construction and Tomato Transformation
4.10. Salt Stress Treatments
4.11. Chlorophyll, Proline, H2O2, and O2− Content Determination
4.12. Determination of Antioxidant Enzyme Activities
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| qRT-PCR | Quantitative reverse transcription polymerase chain reaction |
| WGD | Whole-genome duplication |
| TD | Tandem duplication |
| PD | Proximal duplication |
| TRD | Transposed duplication |
| DSD | Dispersed duplication |
| FPKM | Fragments per kilobase of transcript per million fragments mapped |
| MW | Molecular weight |
| pI | Isoelectric point |
| HMM | Hidden Markov Model |
| MS | Murashige and Skoog medium |
| NCBI | National Center for Biotechnology Information |
| NJ | Neighbor-joining |
References
- Jaimes-Miranda, F.; Montes, R.A.C. The plant MBF1 protein family: A bridge between stress and transcription. J. Exp. Bot. 2020, 71, 1782–1791. [Google Scholar] [CrossRef]
- Hui, W.K.; Zheng, H.; Fan, J.T.; Wang, J.Y.; Saba, T.; Wang, K.; Wu, J.J.; Wu, H.; Zhong, Y.; Chen, G.; et al. Genome-wide characterization of the MBF1 gene family and its expression pattern in different tissues and stresses in Zanthoxylum armatum. BMC Genom. 2022, 23, 652. [Google Scholar] [CrossRef] [PubMed]
- Aravind, L.; Anantharaman, V.; Balaji, S.; Babu, M.M.; Iyer, L.M. The many faces of the helix-turn-helix domain: Transcription regulation and beyond. FEMS Microbiol. Rev. 2005, 29, 231–262. [Google Scholar] [CrossRef] [PubMed]
- Takemaru, K.; Li, F.Q.; Ueda, H.; Hirose, S. Multiprotein bridging factor 1 (MBF1) is an evolutionarily conserved transcriptional coactivator that connects a regulatory factor and TATA element-binding protein. Proc. Natl. Acad. Sci. USA 1997, 94, 7251–7256. [Google Scholar] [CrossRef] [PubMed]
- Takemaru, K.; Harashima, S.; Ueda, H.; Hirose, S. Yeast coactivator MBF1 mediates GCN4-dependent transcriptional activation. Mol. Cell. Biol. 1998, 18, 4971–4976. [Google Scholar] [CrossRef] [PubMed]
- Li, F.Q.; Ueda, H.; Hirose, S. Mediators of activation of fushi-tarazu gene transcription by Bmftz-F1. Mol. Cell. Biol. 1994, 14, 3013–3021. [Google Scholar] [CrossRef] [PubMed]
- Kabe, Y.; Goto, M.; Shima, D.; Imai, T.; Wada, T.; Morohashi, K.; Shirakawa, M.; Hirose, S.; Handa, H. The role of human MBF1 as a transcriptional coactivator. J. Biol. Chem. 1999, 274, 34196–34202. [Google Scholar] [CrossRef]
- Wang, J.; Zhou, J.; Yang, Q.; Grayhack, E.J. Multi-protein bridging factor 1(Mbf1), Rps3 and Asc1 prevent stalled ribosomes from frameshifting. eLife 2018, 7, e39637. [Google Scholar] [CrossRef]
- Baltz, A.G.; Munschauer, M.; Schwanhausser, B.; Vasile, A.; Murakawa, Y.; Schueler, M.; Youngs, N.; Penfold-Brown, D.; Drew, K.; Milek, M.; et al. The mRNA-bound proteome and its global occupancy profile on protein-coding transcripts. Mol. Cell. 2012, 46, 674–690. [Google Scholar] [CrossRef]
- Klass, D.M.; Scheibe, M.; Butter, F.; Hogan, G.J.; Mann, M.; Brown, P.O. Quantitative proteomic analysis reveals concurrent RNA-protein interactions and identifies new RNA-binding proteins in Saccharomyces cerevisiae. Genome Res. 2013, 23, 1028–1038. [Google Scholar] [CrossRef]
- Blombach, F.; Launay, H.; Snijders, A.P.; Zorraquino, V.; Wu, H.; de Koning, B.; Brouns, S.J.; Ettema, T.J.; Camilloni, C.; Cavalli, A.; et al. Archaeal MBF1 binds to 30S and 70S ribosomes via its helix-turn-helix domain. Biochem. J. 2014, 462, 373–384. [Google Scholar] [CrossRef] [PubMed]
- Zhao, N.; Li, C.; Yan, Y.; Wang, H.; Wang, L.; Jiang, J.; Chen, S.; Chen, F. The transcriptional coactivator CmMBF1c is required for waterlogging tolerance in Chrysanthemum morifolium. Hortic. Res. 2022, 9, uhac215. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, N.; Bajad, S.; Shuman, J.; Shulaev, V.; Mittler, R. The transcriptional co-activator MBF1c is a key regulator of thermotolerance in Arabidopsis thaliana. J. Biol. Chem. 2008, 283, 9269–9275. [Google Scholar] [CrossRef] [PubMed]
- Kim, G.D.; Cho, Y.H.; Yoo, S.D. Regulatory functions of evolutionarily conserved AN1/A20-like Zinc finger family proteins in Arabidopsis stress responses under high temperature. Biochem. Biophys. Res. Commun. 2015, 457, 213–220. [Google Scholar] [CrossRef]
- Dezar, C.A.; Gago, G.M.; Gonzalez, D.H.; Chan, R.L. Hahb-4, a sunflower homeobox-leucine zipper gene, is a developmental regulator and confers drought tolerance to Arabidopsis thaliana plants. Transgenic Res. 2005, 14, 429–440. [Google Scholar] [CrossRef]
- Zanetti, M.E.; Chan, R.L.; Godoy, A.V.; Gonzalez, D.H.; Casalongue, C.A. Homeodomain-leucine zipper proteins interact with a plant homologue of the transcriptional co-activator multiprotein bridging factor 1. J. Biochem. Mol. Biol. 2004, 37, 320–324. [Google Scholar] [CrossRef]
- Tian, X.; Qin, Z.; Zhao, Y.; Wen, J.; Lan, T.; Zhang, L.; Wang, F.; Qin, D.; Yu, K.; Zhao, A.; et al. Stress granule-associated TaMBF1c confers thermotolerance through regulating specific mRNA translation in wheat (Triticum aestivum). New Phytol. 2022, 233, 1719–1731. [Google Scholar] [CrossRef]
- Suzuki, N.; Sejima, H.; Tam, R.; Schlauch, K.; Mittler, R. Identification of the MBF1 heat-response regulon of Arabidopsis thaliana. Plant J. 2011, 66, 844–851. [Google Scholar] [CrossRef]
- Wang, Y.Q.; Xia, D.N.; Li, W.Q.; Cao, X.Y.; Ma, F.; Wang, Q.Q.; Zhan, X.Q.; Hu, T.X. Overexpression of a tomato AP2/ERF transcription factor SlERF.B1 increases sensitivity to salt and drought stresses. Sci. Hortic. 2022, 304, 111332. [Google Scholar] [CrossRef]
- Julkowska, M.M.; Testerink, C. Tuning plant signaling and growth to survive salt. Trends Plant Sci. 2015, 20, 586–594. [Google Scholar] [CrossRef]
- Zhao, S.; Zhang, Q.; Liu, M.; Zhou, H.; Ma, C.; Wang, P. Regulation of plant responses to salt stress. Int. J. Mol. Sci. 2021, 22, 4609. [Google Scholar] [CrossRef]
- Arif, Y.; Singh, P.; Siddiqui, H.; Bajguz, A.; Hayat, S. Salinity induced physiological and biochemical changes in plants: An omic approach towards salt stress tolerance. Plant Physiol. Biochem. 2020, 156, 64–77. [Google Scholar] [CrossRef] [PubMed]
- Munns, R.; Tester, M. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 2008, 59, 651–681. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, A.A.; Taleisnik, E.L. Determination of reactive oxygen species in salt-stressed plant tissues. Methods Mol. Biol. 2012, 913, 225–236. [Google Scholar] [CrossRef] [PubMed]
- Alavilli, H.; Lee, H.; Park, M.; Lee, B.H. Antarctic moss multiprotein bridging factor 1c overexpression in Arabidopsis resulted in enhanced tolerance to salt stress. Front. Plant Sci. 2017, 8, 1206. [Google Scholar] [CrossRef]
- Zhao, Q.; He, L.; Wang, B.; Liu, Q.; Pan, Y.; Zhang, F.; Jiang, B.; Zhang, L. Overexpression of a multiprotein bridging factor 1 gene DgMBF1 improves the salinity tolerance of Chrysanthemum. Int. J. Mol. Sci. 2019, 20, 2453. [Google Scholar] [CrossRef]
- Qin, C.; Yu, C.; Shen, Y.; Fang, X.; Chen, L.; Min, J.; Cheng, J.; Zhao, S.; Xu, M.; Luo, Y.; et al. Whole-genome sequencing of cultivated and wild peppers provides insights into Capsicum domestication and specialization. Proc. Natl. Acad. Sci. USA 2014, 111, 5135–5140. [Google Scholar] [CrossRef]
- Cao, Y.L.; Li, Y.L.; Fan, Y.F.; Li, Z.; Yoshida, K.; Wang, J.Y.; Ma, X.K.; Wang, N.; Mitsuda, N.; Kotake, T.; et al. Wolfberry genomes and the evolution of Lycium (Solanaceae). Commun. Biol. 2021, 4, 671. [Google Scholar] [CrossRef]
- Tomato Genome Consortium. The tomato genome sequence provides insights into fleshy fruit evolution. Nature. 2012, 485, 635–641. [Google Scholar] [CrossRef]
- Diambra, L.A. Genome sequence and analysis of the tuber crop potato. Nature. 2011, 475, 189–194. [Google Scholar] [CrossRef]
- Wei, Q.Z.; Wang, J.L.; Wang, W.H.; Hu, T.H.; Hu, H.J.; Bao, C.L. A high-quality chromosome-level genome assembly reveals genetics for important traits in eggplant. Hortic. Res. 2020, 7, 153. [Google Scholar] [CrossRef] [PubMed]
- Vanneste, K.; Van de Peer, Y.; Maere, S. Inference of genome duplications from age distributions revisited. Mol. Biol. Evol. 2013, 30, 177–190. [Google Scholar] [CrossRef] [PubMed]
- Zou, L.; Yu, B.; Ma, X.L.; Cao, B.; Chen, G.; Chen, C.; Lei, J. Cloning and expression analysis of the BocMBF1c gene involved in heat tolerance in Chinese Kale. Int. J. Mol. Sci. 2019, 20, 5637. [Google Scholar] [CrossRef]
- Sasidharan, R.; Schippers, J.H.M.; Schmidt, R.R. Redox and low-oxygen stress: Signal integration and interplay. Plant Physiol. 2021, 186, 66–78. [Google Scholar] [CrossRef]
- Patel, M.K.; Pandey, S.; Burritt, D.J.; Tran, L.S.P. Plant responses to low-oxygen stress: Interplay between ROS and NO signaling pathways. Environ. Exp. Bot. 2019, 161, 134–142. [Google Scholar] [CrossRef]
- Tsuda, K.; Tsuji, T.; Hirose, S.; Yamazaki, K. Three Arabidopsis MBF1 homologs with distinct expression profiles play roles as transcriptional co-activators. Plant Cell Physiol. 2004, 45, 225–231. [Google Scholar] [CrossRef]
- Arce, D.P.; Godoy, A.V.; Tsuda, K.; Yamazaki, K.; Valle, E.M.; Iglesias, M.J.; Di Mauro, M.F.; Casalongue, C.A. The analysis of an Arabidopsis triple knock-down mutant reveals functions for MBF1 genes under oxidative stress conditions. J. Plant Physiol. 2010, 167, 194–200. [Google Scholar] [CrossRef]
- Hommel, M.; Khalil-Ahmad, Q.; Jaimes-Miranda, F.; Mila, I.; Pouzet, C.; Latche, A.; Pech, J.C.; Bouzayen, M.; Regad, F. Over-expression of a chimeric gene of the transcriptional co-activator MBF1 fused to the EAR repressor motif causes developmental alteration in Arabidopsis and tomato. Plant Sci. 2008, 175, 168–177. [Google Scholar] [CrossRef]
- Zhang, X.; Xu, Z.; Chen, L.; Ren, Z. Comprehensive analysis of multiprotein bridging factor 1 family genes and SlMBF1c negatively regulate the resistance to Botrytis cinerea in tomato. BMC Plant Biol. 2019, 19, 437. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.S.; Nie, S.; Liu, H.; Shi, T.L.; Tian, X.C.; Zhou, S.S.; Bao, Y.T.; Jia, K.H.; Guo, J.F.; Zhao, W.; et al. Chromosome-level genome assembly of a parent species of widely cultivated azaleas. Nat. Commun. 2020, 11, 5269. [Google Scholar] [CrossRef] [PubMed]
- Yin, Y.; Guo, C.; Shi, H.; Zhao, J.; Ma, F.; An, W.; He, X.; Luo, Q.; Cao, Y.; Zhan, X. Genome-wide comparative analysis of the R2R3-MYB gene family in five Solanaceae species and identification of members regulating carotenoid biosynthesis in wolfberry. Int. J. Mol. Sci. 2022, 23, 2259. [Google Scholar] [CrossRef] [PubMed]
- Huang, B.; Huang, Z.; Ma, R.; Chen, J.; Zhang, Z.; Yrjala, K. Genome-wide identification and analysis of the heat shock transcription factor family in moso bamboo (Phyllostachys edulis). Sci. Rep. 2021, 11, 16492. [Google Scholar] [CrossRef]
- Zegzouti, H.; Jones, B.; Frasse, P.; Marty, C.; Maitre, B.; Latch, A.; Pech, J.C.; Bouzayen, M. Ethylene-regulated gene expression in tomato fruit: Characterization of novel ethylene-responsive and ripening-related genes isolated by differential display. Plant J. 1999, 18, 589–600. [Google Scholar] [CrossRef]
- Tsuda, K.; Yamazaki, K. Structure and expression analysis of three subtypes of Arabidopsis MBF1 genes. Biochim. Biophys. Acta. 2004, 1680, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.L.; Chen, R.G.; Du, X.H.; Zhang, Z.; Yin, Y.X.; Gong, Z.H.; Wang, G.Y. Reduced tolerance to abiotic stress in transgenic Arabidopsis overexpressing a Capsicum annuum multiprotein bridging factor 1. Bmc Plant Biol. 2014, 14, 138. [Google Scholar] [CrossRef]
- Yan, Q.; Hou, H.M.; Singer, S.D.; Yan, X.X.; Guo, R.R.; Wang, X.P. The grape VvMBF1 gene improves drought stress tolerance in transgenic Arabidopsis thaliana. Plant Cell Tiss. Org. 2014, 118, 571–582. [Google Scholar] [CrossRef]
- Nakashima, K.; Yamaguchi-Shinozaki, K. ABA signaling in stress-response and seed development. Plant Cell Rep. 2013, 32, 959–970. [Google Scholar] [CrossRef]
- Yu, Z.P.; Duan, X.B.; Luo, L.; Dai, S.J.; Ding, Z.J.; Xia, G.M. How plant hormones mediate salt stress responses. Trends Plant Sci. 2020, 25, 1117–1130. [Google Scholar] [CrossRef]
- Riyazuddin, R.; Verma, R.; Singh, K.; Nisha, N.; Keisham, M.; Bhati, K.K.; Kim, S.T.; Gupta, R. Ethylene: A master regulator of salinity stress tolerance in plants. Biomolecules 2020, 10, 959. [Google Scholar] [CrossRef]
- Wang, Y.N.; Liu, C.; Li, K.X.; Sun, F.F.; Hu, H.Z.; Li, X.; Zhao, Y.K.; Han, C.Y.; Zhang, W.S.; Duan, Y.F.; et al. Arabidopsis EIN2 modulates stress response through abscisic acid response pathway. Plant Mol. Biol. 2007, 64, 633–644. [Google Scholar] [CrossRef] [PubMed]
- Di Mauro, M.F.; Iglesias, M.J.; Arce, D.P.; Valle, E.M.; Arnold, R.B.; Tsuda, K.; Yamazaki, K.; Casalongue, C.A.; Godoy, A.V. MBF1s regulate ABA-dependent germination of Arabidopsis seeds. Plant Signal. Behav. 2012, 7, 188–192. [Google Scholar] [CrossRef]
- Qin, D.; Wang, F.; Geng, X.; Zhang, L.; Yao, Y.; Ni, Z.; Peng, H.; Sun, Q. Overexpression of heat stress-responsive TaMBF1c, a wheat (Triticum aestivum L.) multiprotein bridging factor, confers heat tolerance in both yeast and rice. Plant Mol. Biol. 2015, 87, 31–45. [Google Scholar] [CrossRef]
- Mittler, R.; Zandalinas, S.I.; Fichman, Y.; Van Breusegem, F. Reactive oxygen species signalling in plant stress responses. Nat. Rev. Mol. Cell Biol. 2022, 23, 663–679. [Google Scholar] [CrossRef] [PubMed]
- Miller, G.; Suzuki, N.; Ciftci-Yilmaz, S.; Mittler, R. Reactive oxygen species homeostasis and signalling during drought and salinity stresses. Plant Cell Environ. 2010, 33, 453–467. [Google Scholar] [CrossRef]
- Nadarajah, K.K. ROS homeostasis in abiotic stress tolerance in plants. Int. J. Mol. Sci. 2020, 21, 5208. [Google Scholar] [CrossRef] [PubMed]
- Malakar, P.; Chattopadhyay, D. Adaptation of plants to salt stress: The role of the ion transporters. J. Plant Biochem Biot. 2021, 30, 668–683. [Google Scholar] [CrossRef]
- Liu, X.; Gao, T.; Liu, C.; Mao, K.; Gong, X.; Li, C.; Ma, F. Fruit crops combating drought: Physiological responses and regulatory pathways. Plant Physiol. 2023, 192, 1768–1784. [Google Scholar] [CrossRef]
- Hsieh, T.H.; Li, C.W.; Su, R.C.; Cheng, C.P.; Sanjaya; Tsai, Y. C.; Chan, M.T. A tomato bZIP transcription factor, SlAREB, is involved in water deficit and salt stress response. Planta 2010, 231, 1459–1473. [Google Scholar] [CrossRef]
- Orellana, S.; Yanez, M.; Espinoza, A.; Verdugo, I.; Gonzalez, E.; Ruiz-Lara, S.; Casaretto, J.A. The transcription factor SlAREB1 confers drought, salt stress tolerance and regulates biotic and abiotic stress-related genes in tomato. Plant Cell Environ. 2010, 33, 2191–2208. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Zhang, L.; Nie, L.; Zheng, Y.; Zhu, S.; Hou, J.; Li, R.; Chen, G.; Tang, X.; Wang, C.; et al. Genome-wide analysis of the DREB family genes and functional identification of the involvement of BrDREB2B in abiotic stress in wucai (Brassica campestris L.). BMC Genom. 2022, 23, 598. [Google Scholar] [CrossRef]
- Zhang, J.L.; Shi, H. Physiological and molecular mechanisms of plant salt tolerance. Photosynth. Res. 2013, 115, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Olias, R.; Eljakaoui, Z.; Pardo, J.M.; Belver, A. The Na(+)/H(+) exchanger SOS1 controls extrusion and distribution of Na(+) in tomato plants under salinity conditions. Plant Signal. Behav. 2009, 4, 973–976. [Google Scholar] [CrossRef]
- Huertas, R.; Rubio, L.; Cagnac, O.; Garcia-Sanchez, M.J.; Alche Jde, D.; Venema, K.; Fernandez, J.A.; Rodriguez-Rosales, M.P. The K+/H+ antiporter LeNHX2 increases salt tolerance by improving K+ homeostasis in transgenic tomato. Plant Cell Environ. 2013, 36, 2135–2149. [Google Scholar] [CrossRef] [PubMed]
- El-Gebali, S.; Mistry, J.; Bateman, A.; Eddy, S.R.; Luciani, A.; Potter, S.C.; Qureshi, M.; Richardson, L.J.; Salazar, G.A.; Smart, A.; et al. The Pfam protein families database in 2019. Nucleic Acids Res. 2019, 47, D427–D432. [Google Scholar] [CrossRef]
- Johnson, L.S.; Eddy, S.R.; Portugaly, E. Hidden Markov model speed heuristic and iterative HMM search procedure. BMC Bioinform. 2010, 11, 431. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Jin, J.P.; Guo, A.Y.; Zhang, H.; Luo, J.C.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.Y.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Lu, S.N.; Wang, J.Y.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; Gwadz, M.; Hurwitz, D.I.; Marchler, G.H.; Song, J.S.; et al. CDD/SPARCLE: The conserved domain database in 2020. Nucleic Acids Res. 2020, 48, D265–D268. [Google Scholar] [CrossRef]
- Chen, C.J.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.H.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Wang, Y.P.; Tang, H.B.; DeBarry, J.D.; Tan, X.; Li, J.P.; Wang, X.Y.; Lee, T.H.; Jin, H.Z.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [PubMed]
- Lescot, M.; Dehais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouze, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Fei, Z.J.; Joung, J.G.; Tang, X.M.; Zheng, Y.; Huang, M.Y.; Lee, J.M.; McQuinn, R.; Tieman, D.M.; Alba, R.; Klee, H.J.; et al. Tomato Functional Genomics Database: A comprehensive resource and analysis package for tomato functional genomics. Nucleic Acids Res. 2011, 39, D1156–D1163. [Google Scholar] [CrossRef] [PubMed]
- Hu, T.X.; Wang, Y.Q.; Wang, Q.Q.; Dang, N.N.; Wang, L.; Liu, C.C.; Zhu, J.H.; Zhan, X.Q. The tomato 2-oxoglutarate-dependent dioxygenase gene SlF3HL is critical for chilling stress tolerance. Hortic. Res. 2019, 6, 45. [Google Scholar] [CrossRef]
- Liu, G.Z.; Li, C.X.; Yu, H.Y.; Tao, P.W.; Yuan, L.; Ye, J.; Chen, W.F.; Wang, Y.; Ge, P.F.; Zhang, J.H.; et al. GREEN STRIPE, encoding methylated TOMATO AGAMOUS-LIKE 1, regulates chloroplast development and Chl synthesis in fruit. New Phytol. 2020, 228, 302–317. [Google Scholar] [CrossRef]
- McGrath, K.C.; Dombrecht, B.; Manners, J.M.; Schenk, P.M.; Edgar, C.I.; Maclean, D.J.; Scheible, W.R.; Udvardi, M.K.; Kazan, K. Repressor- and activator-type ethylene response factors functioning in jasmonate signaling and disease resistance identified via a genome-wide screen of Arabidopsis transcription factor gene expression. Plant Physiol. 2005, 139, 949–959. [Google Scholar] [CrossRef] [PubMed]


| Gene ID | Gene Name | Pfam ID | Description | TAIR BlastX |
|---|---|---|---|---|
| Solyc01g104740.3.1 | SlER24 | PF08523.13/PF01381.25 | MBF1/HTH_3 | AtMBF1c |
| Solyc07g062400.3.1 | SlMBF1a | PF08523.13/PF01381.25 | MBF1/HTH_3 | AtMBF1b |
| Solyc09g055470.1.1 | SlMBF1b1 | PF08523.13/PF01381.25 | MBF1/HTH_3 | AtMBF1b |
| Solyc10g007350.4.1 | SlMBF1b2 | PF08523.13/PF01381.25 | MBF1/HTH_3 | AtMBF1b |
| Solyc12g014290.2.1 | SlMBF1b3 | PF08523.13/PF01381.25 | MBF1/HTH_3 | AtMBF1b |
| CaDEM05G05830 | CaMBF1c1 | PF08523.13/PF01381.25 | MBF1/HTH_3 | AtMBF1c |
| CaDEM06G07520 | CaMBF1c2 | PF08523.13/PF01381.25 | MBF1/HTH_3 | AtMBF1c |
| CaDEM07G25960 | CaMBF1a | PF08523.13/PF01381.25 | MBF1/HTH_3 | AtMBF1a |
| CaDEM09G17680 | CaMBF1b1 | PF08523.13/PF01381.25 | MBF1/HTH_3 | AtMBF1b |
| CaDEM10G01710 | CaMBF1b2 | PF08523.13/PF01381.25 | MBF1/HTH_3 | AtMBF1b |
| Smechr0400261.1 | SmeMBF1b | PF08523.13/PF01381.25 | MBF1/HTH_3 | AtMBF1b |
| Smechr0702353.1 | SmeMBF1a | PF08523.13/PF01381.25 | MBF1/HTH_3 | AtMBF1b |
| Smechr1102702.1 | SmeMBF1c | PF08523.13/PF01381.25/PF01249.21 | MBF1/HTH_3/Ribosomal_S21e | AtMBF1c |
| Soltu.DM.01G043930.1 | StMBF1c | PF08523.13/PF01381.25 | MBF1/HTH_3 | AtMBF1c |
| Soltu.DM.07G023530.1 | StMBF1a | PF08523.13/PF01381.25 | MBF1/HTH_3 | AtMBF1b |
| Soltu.DM.10G002940.1 | StMBF1b1 | PF08523.13/PF01381.25 | MBF1/HTH_3 | AtMBF1b |
| Soltu.DM.12G030120.1 | StMBF1b2 | PF08523.13/PF01381.25 | MBF1/HTH_3 | AtMBF1b |
| Lba01g00195 | LbaMBF1a1 | PF01381.25 | HTH_3 | AtMBF1c |
| Lba04g02218 | LbaMBF1b | PF08523.13/PF01381.25 | MBF1/HTH_3 | AtMBF1b |
| Lba10g02214 | LbaMBF1c | PF08523.13/PF01381.25/PF01249.21/PF04227.15 | MBF1/HTH_3/Ribosomal_S21e/Indigoidine_A | AtMBF1c |
| Lba11g00477 | LbaMBF1a2 | PF08523.13/PF01381.25 | MBF1/HTH_3 | AtMBF1b |
| Gene ID | Gene Name | GDE | Ka | Ks | Ka/Ks |
|---|---|---|---|---|---|
| Solyc01g104740.3.1 | SlER24 | TRD | |||
| Solyc07g062400.3.1 | SlMBF1a | WGD | 0.04 | 0.77 | 0.05 |
| Solyc09g055470.1.1 | SlMBF1b1 | TRD | |||
| Solyc10g007350.4.1 | SlMBF1b2 | WGD | 0.04 | 1.09 | 0.04 |
| Solyc12g014290.2.1 | SlMBF1b3 | WGD | 0.03 | 0.88 | 0.03 |
| CaDEM05G05830 | CaMBF1c1 | TRD | |||
| CaDEM06G07520 | CaMBF1c2 | TRD | |||
| CaDEM07G25960 | CaMBF1a | WGD | 0.05 | 0.78 | 0.07 |
| CaDEM09G17680 | CaMBF1b1 | WGD | 0.10 | 0.97 | 0.10 |
| CaDEM10G01710 | CaMBF1b2 | WGD | 0.11 | 1.03 | 0.11 |
| Smechr0400261.1 | SmeMBF1b | WGD | 0.05 | 0.70 | 0.08 |
| Smechr0702353.1 | SmeMBF1a | WGD | |||
| Smechr1102702.1 | SmeMBF1c | TRD | |||
| Soltu.DM.01G043930.1 | StMBF1c | TRD | |||
| Soltu.DM.07G023530.1 | StMBF1a | WGD | 0.04 | 0.61 | 0.07 |
| Soltu.DM.10G002940.1 | StMBF1b1 | WGD | 0.04 | 0.99 | 0.04 |
| Soltu.DM.12G030120.1 | StMBF1b2 | WGD | 0.04 | 0.71 | 0.05 |
| Lba01g00195 | LbaMBF1a1 | WGD | 0.04 | 0.92 | 0.04 |
| Lba04g02218 | LbaMBF1b | WGD | 0.06 | 0.99 | 0.06 |
| Lba10g02214 | LbaMBF1c | TRD | |||
| Lba11g00477 | LbaMBF1a2 | WGD | 0.02 | 0.54 | 0.04 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xia, D.; Guan, L.; Yin, Y.; Wang, Y.; Shi, H.; Li, W.; Zhang, D.; Song, R.; Hu, T.; Zhan, X. Genome-Wide Analysis of MBF1 Family Genes in Five Solanaceous Plants and Functional Analysis of SlER24 in Salt Stress. Int. J. Mol. Sci. 2023, 24, 13965. https://doi.org/10.3390/ijms241813965
Xia D, Guan L, Yin Y, Wang Y, Shi H, Li W, Zhang D, Song R, Hu T, Zhan X. Genome-Wide Analysis of MBF1 Family Genes in Five Solanaceous Plants and Functional Analysis of SlER24 in Salt Stress. International Journal of Molecular Sciences. 2023; 24(18):13965. https://doi.org/10.3390/ijms241813965
Chicago/Turabian StyleXia, Dongnan, Lulu Guan, Yue Yin, Yixi Wang, Hongyan Shi, Wenyu Li, Dekai Zhang, Ran Song, Tixu Hu, and Xiangqiang Zhan. 2023. "Genome-Wide Analysis of MBF1 Family Genes in Five Solanaceous Plants and Functional Analysis of SlER24 in Salt Stress" International Journal of Molecular Sciences 24, no. 18: 13965. https://doi.org/10.3390/ijms241813965
APA StyleXia, D., Guan, L., Yin, Y., Wang, Y., Shi, H., Li, W., Zhang, D., Song, R., Hu, T., & Zhan, X. (2023). Genome-Wide Analysis of MBF1 Family Genes in Five Solanaceous Plants and Functional Analysis of SlER24 in Salt Stress. International Journal of Molecular Sciences, 24(18), 13965. https://doi.org/10.3390/ijms241813965







