Endoplasmic Reticulum Stress-Related Ten-Biomarker Risk Classifier for Survival Evaluation in Epithelial Ovarian Cancer and TRPM2: A Potential Therapeutic Target of Ovarian Cancer
Abstract
:1. Introduction
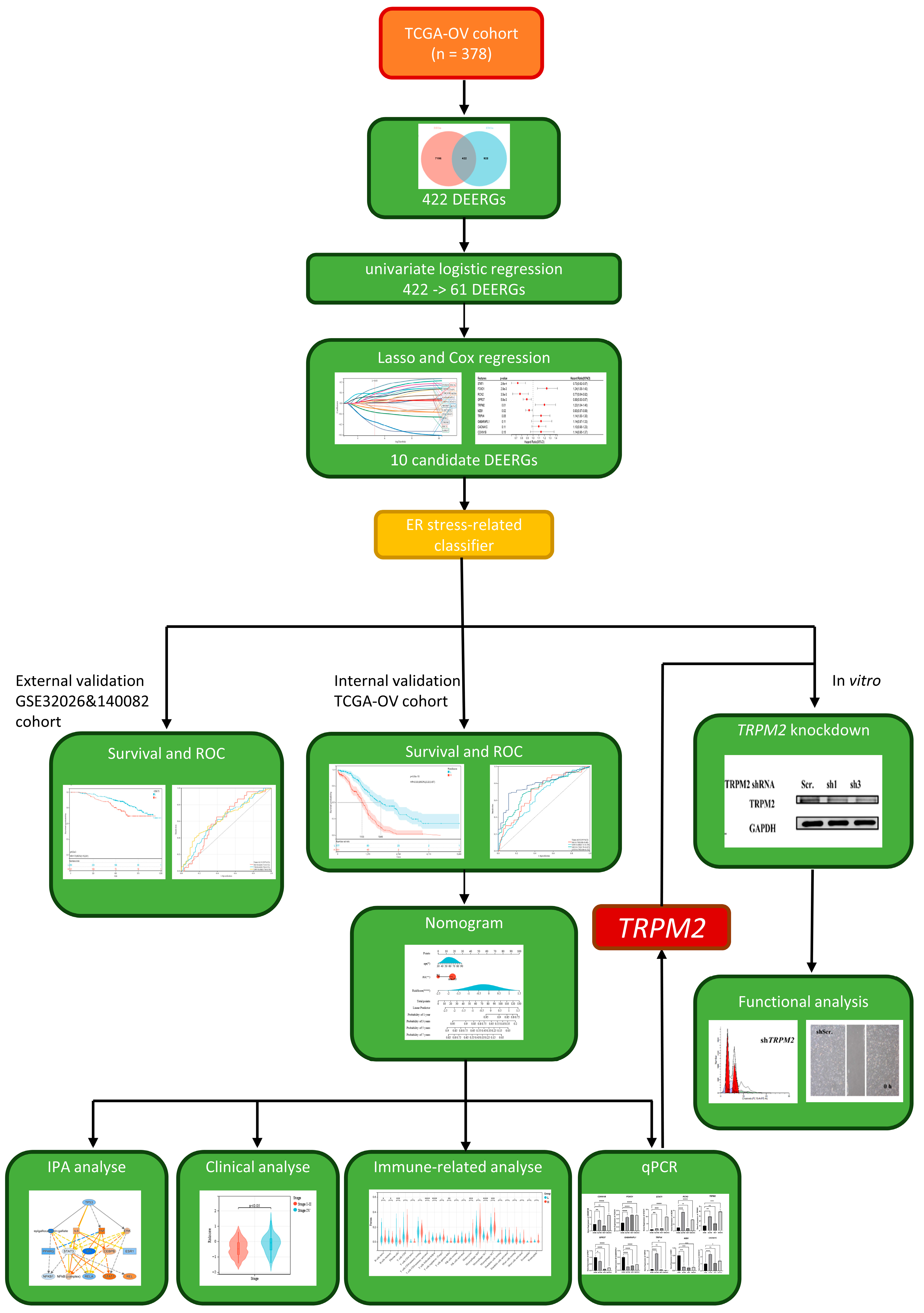
2. Results
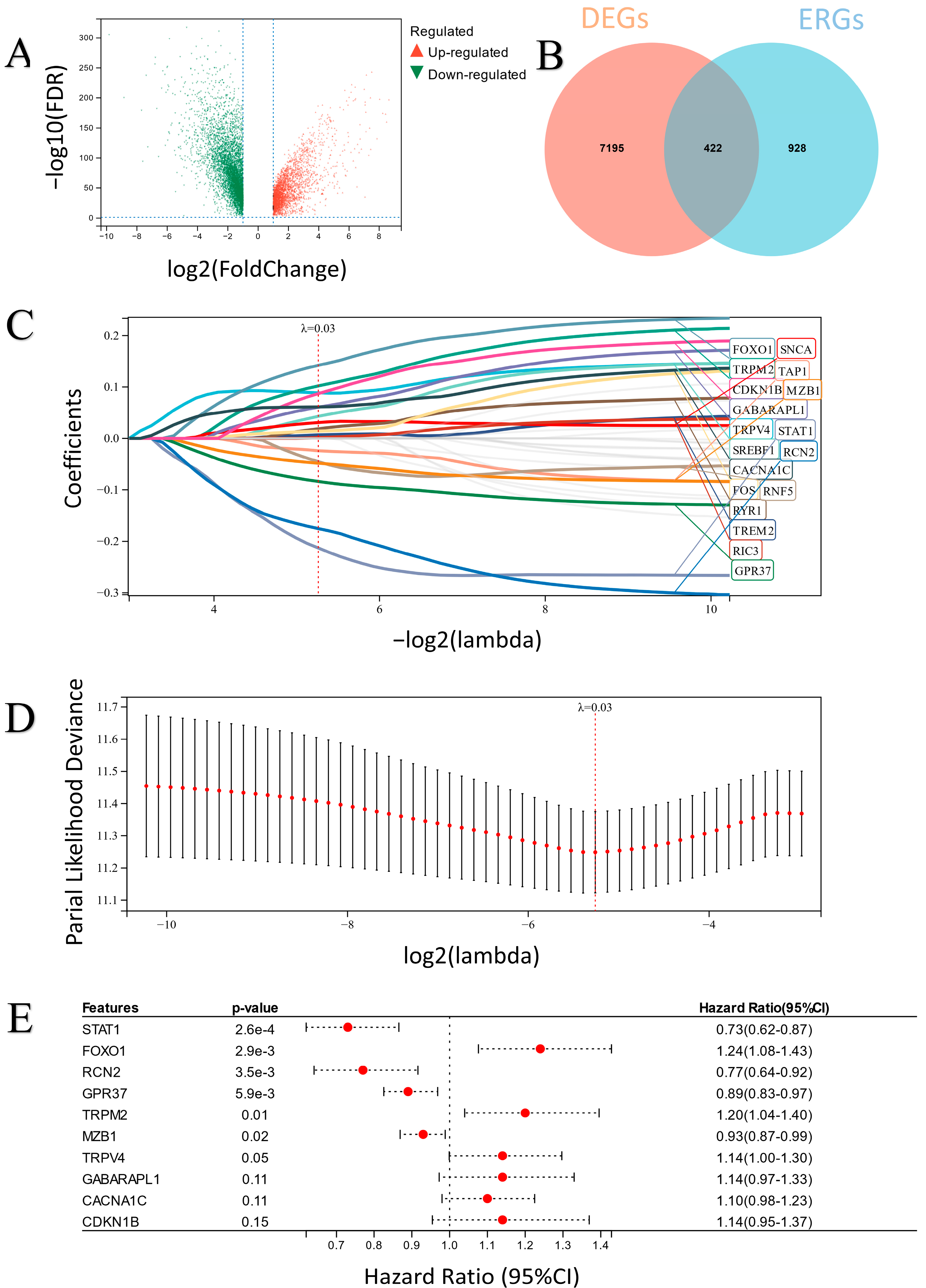
2.1. The Characteristics of the Differentially Expressed ERGs (DEERGs)
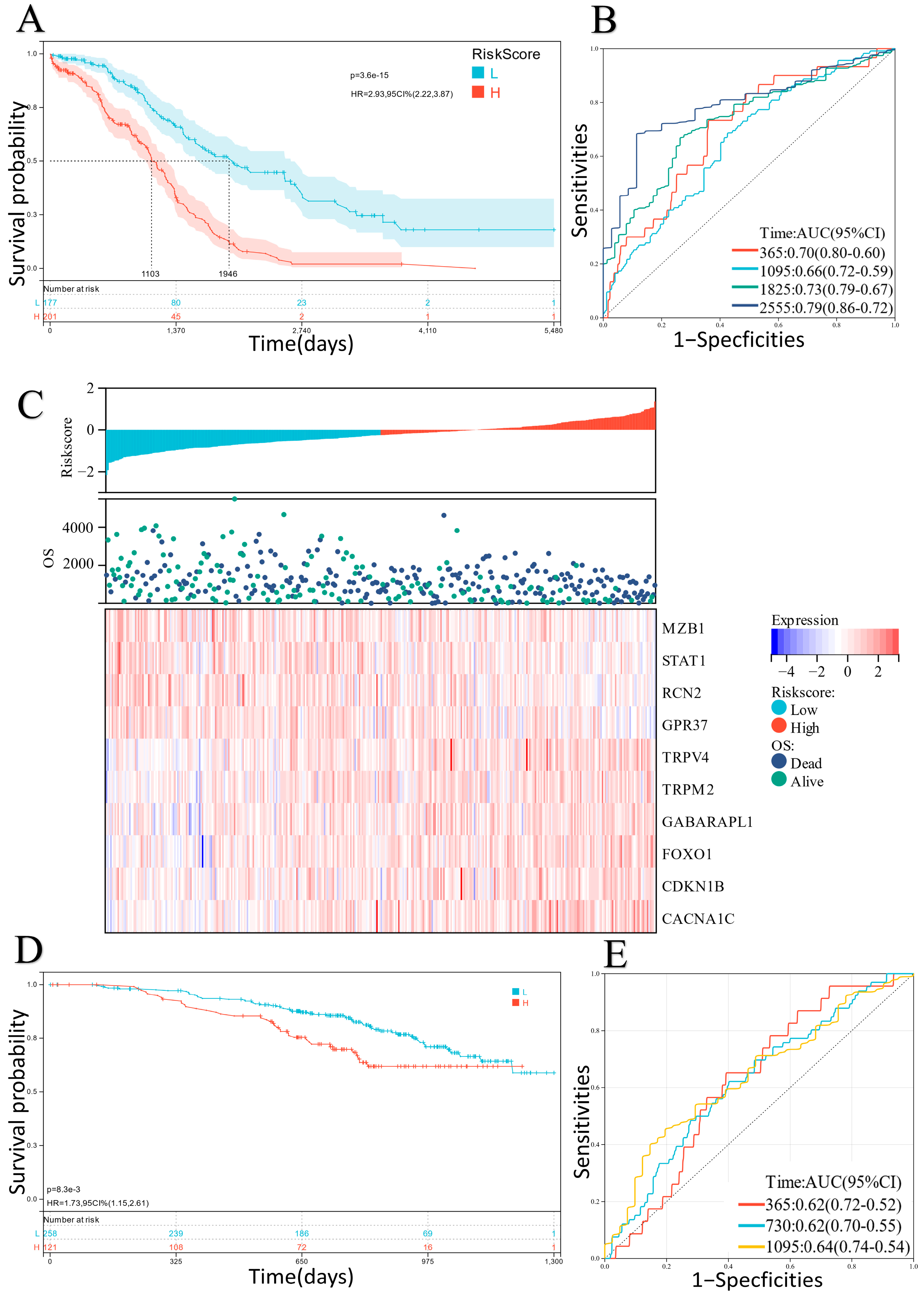
2.2. Construction and Verification of the ER Stress-Related Classifier
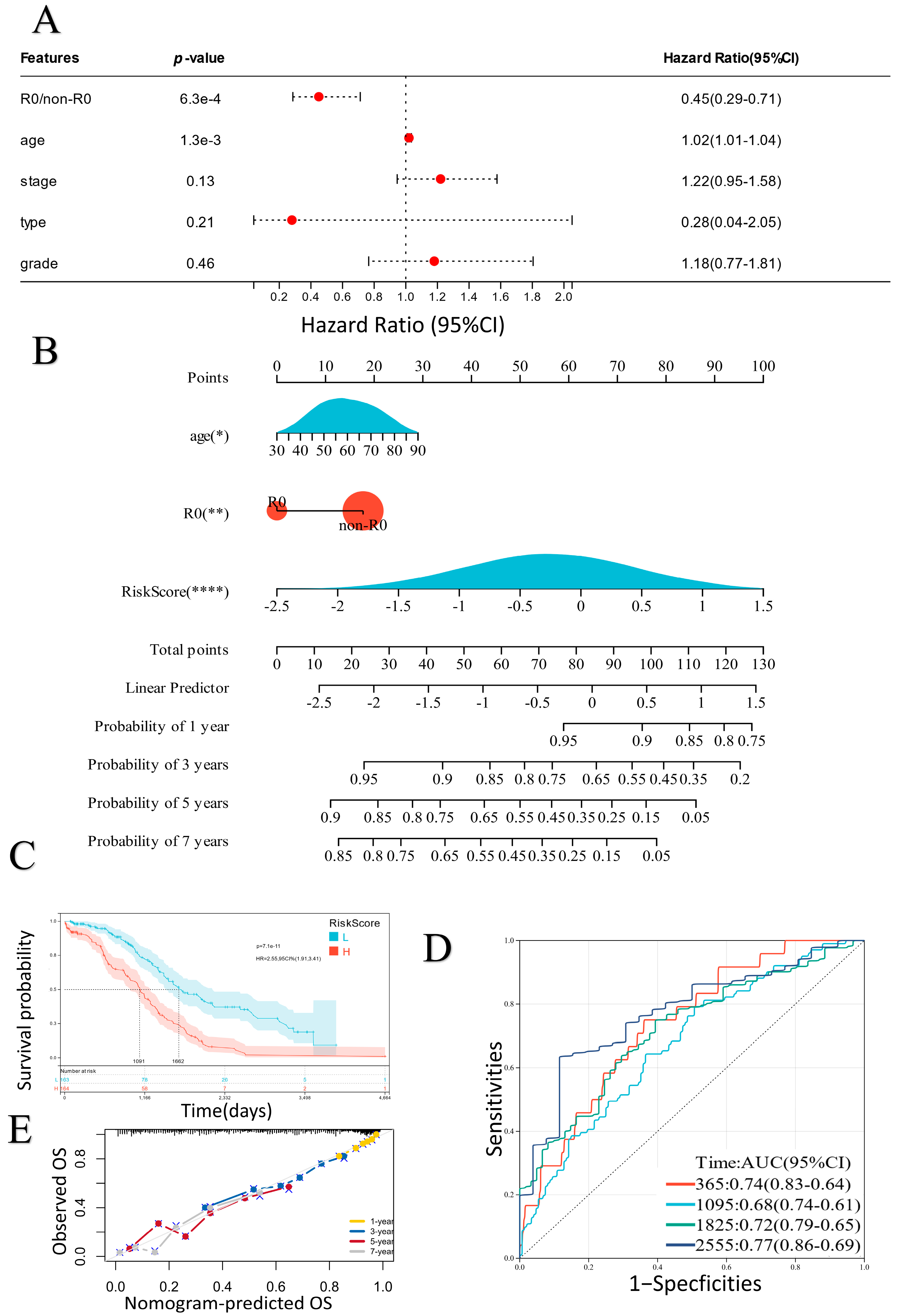
2.3. Establishment of a Prognostic Nomogram
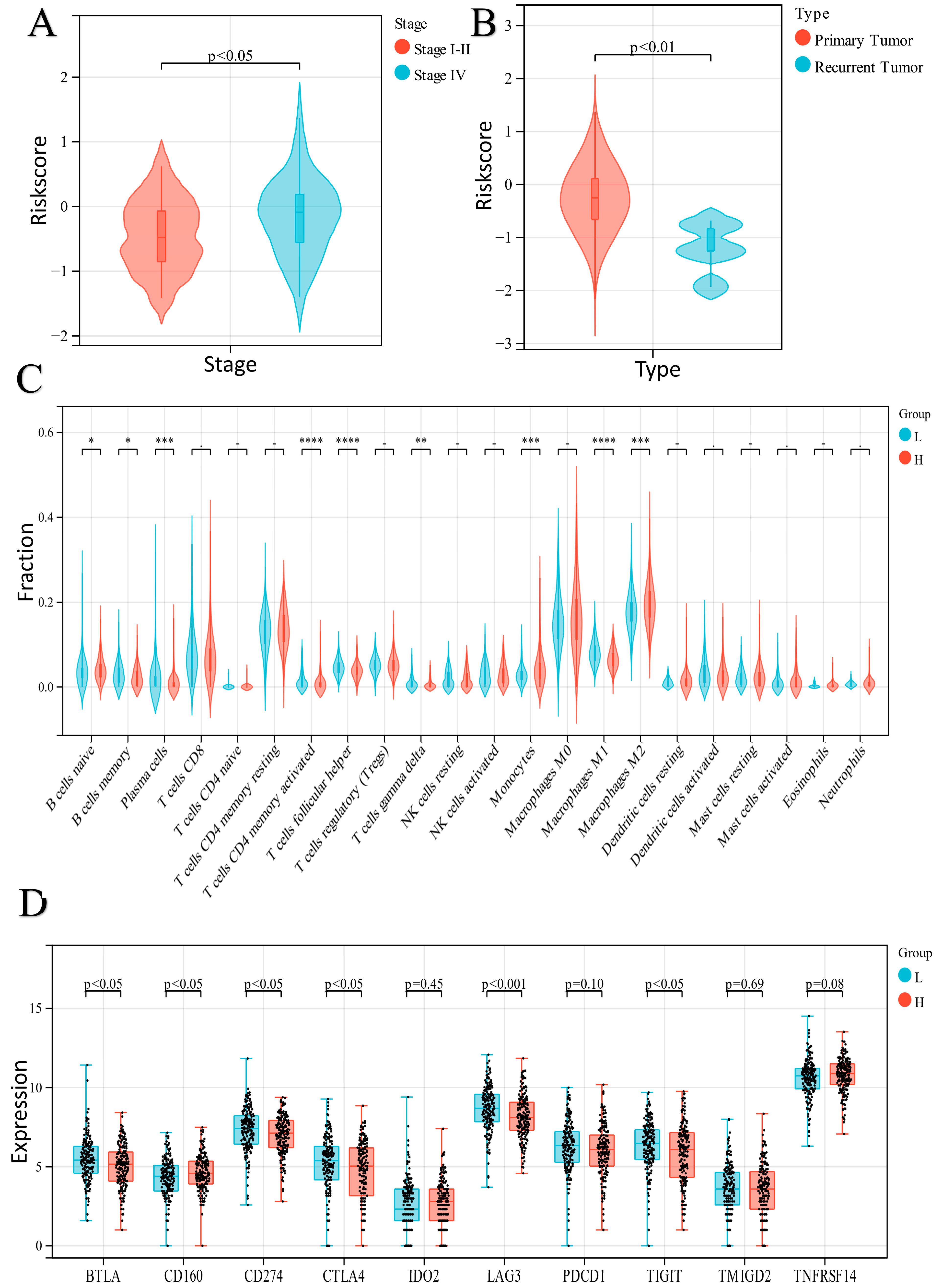
2.4. Exploration of the Relationship between the ER Stress-Related Classifier and Clinical Parameters
2.5. Analysis of Immune Cell Infiltration in the Two Groups
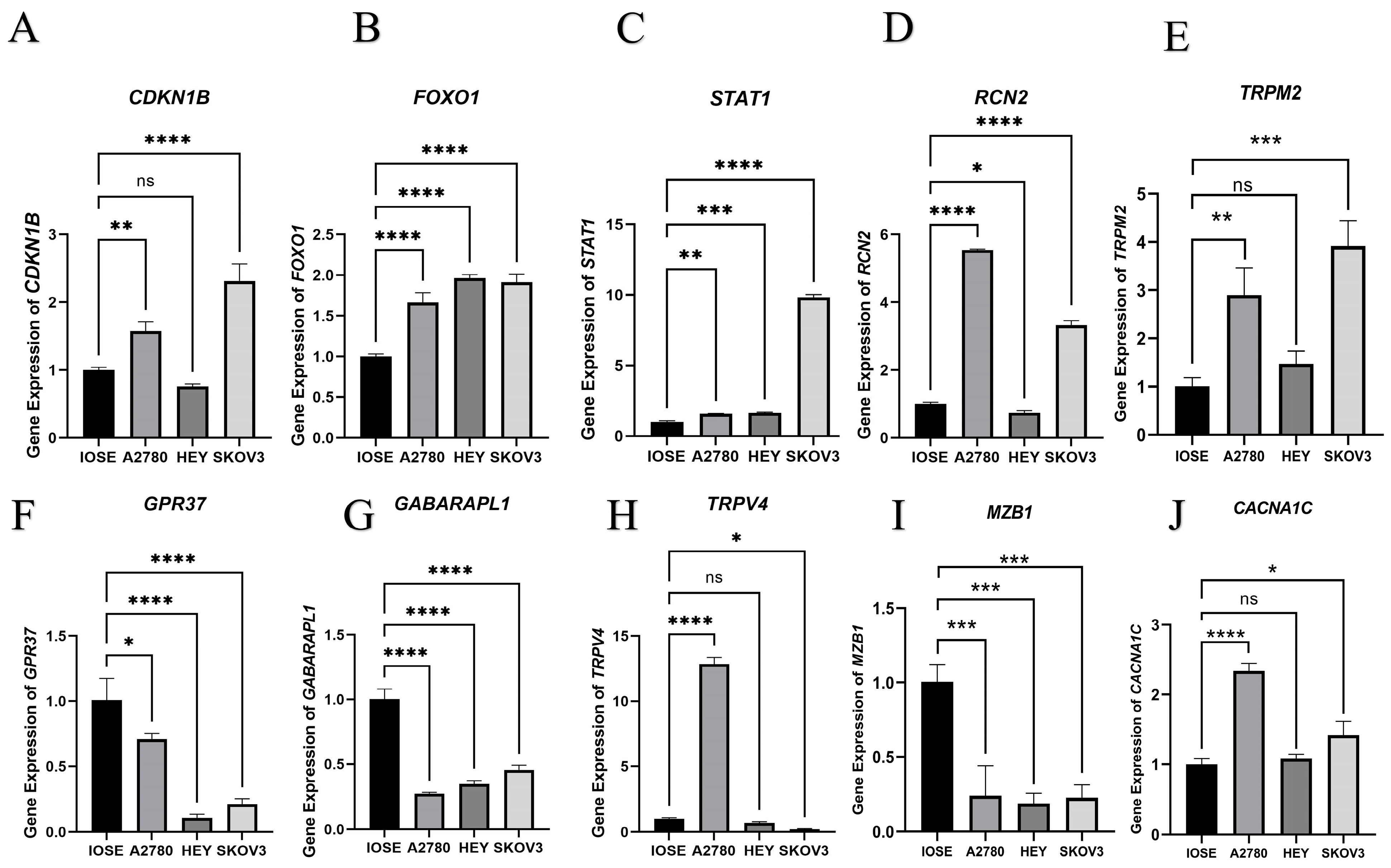
2.6. qPCR Assay of 10 Candidate DEERGs
2.7. Knockdown of TRPM2 Inhibited Ovarian Cancer Cell Apoptosis, Invasion, and Migration
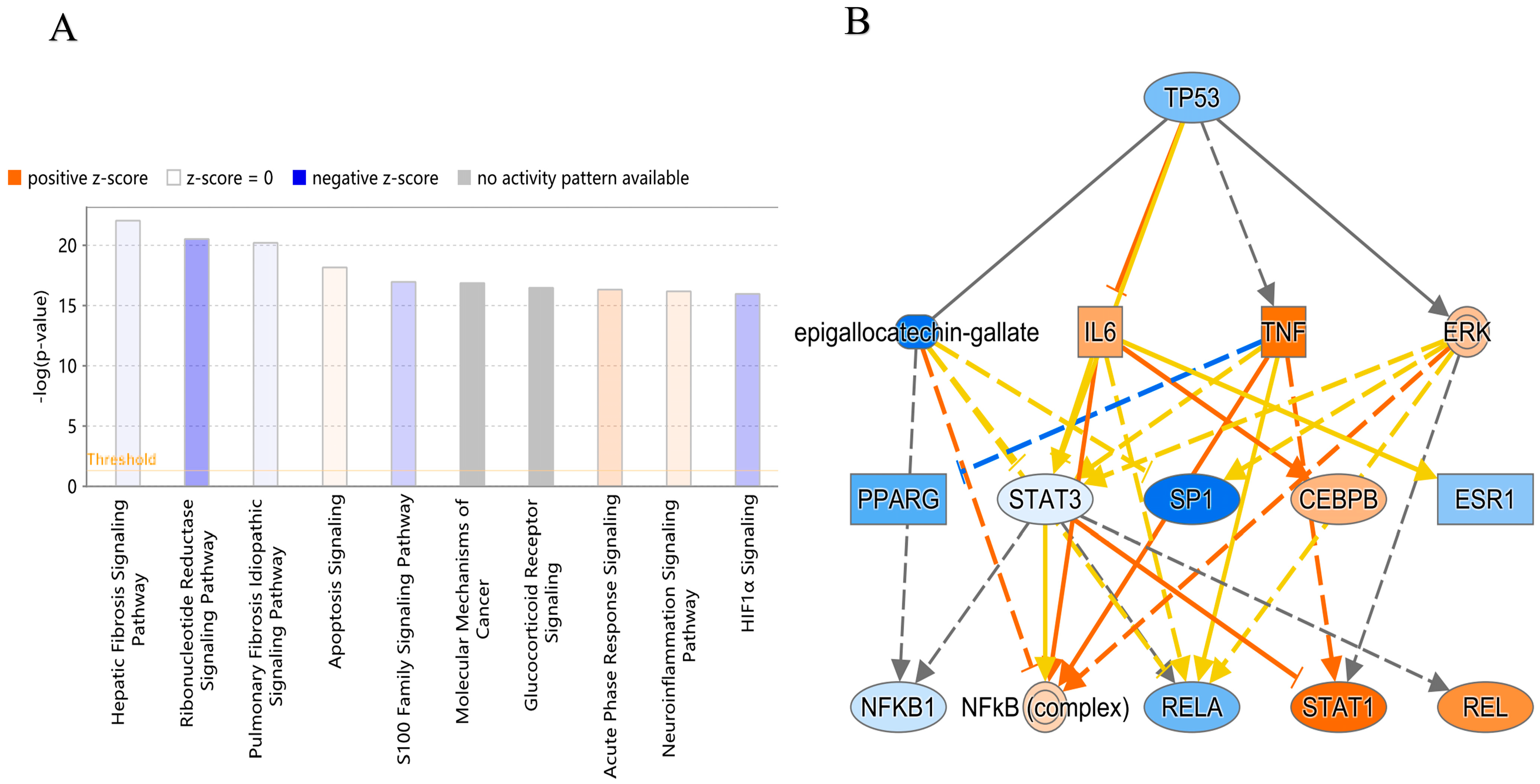
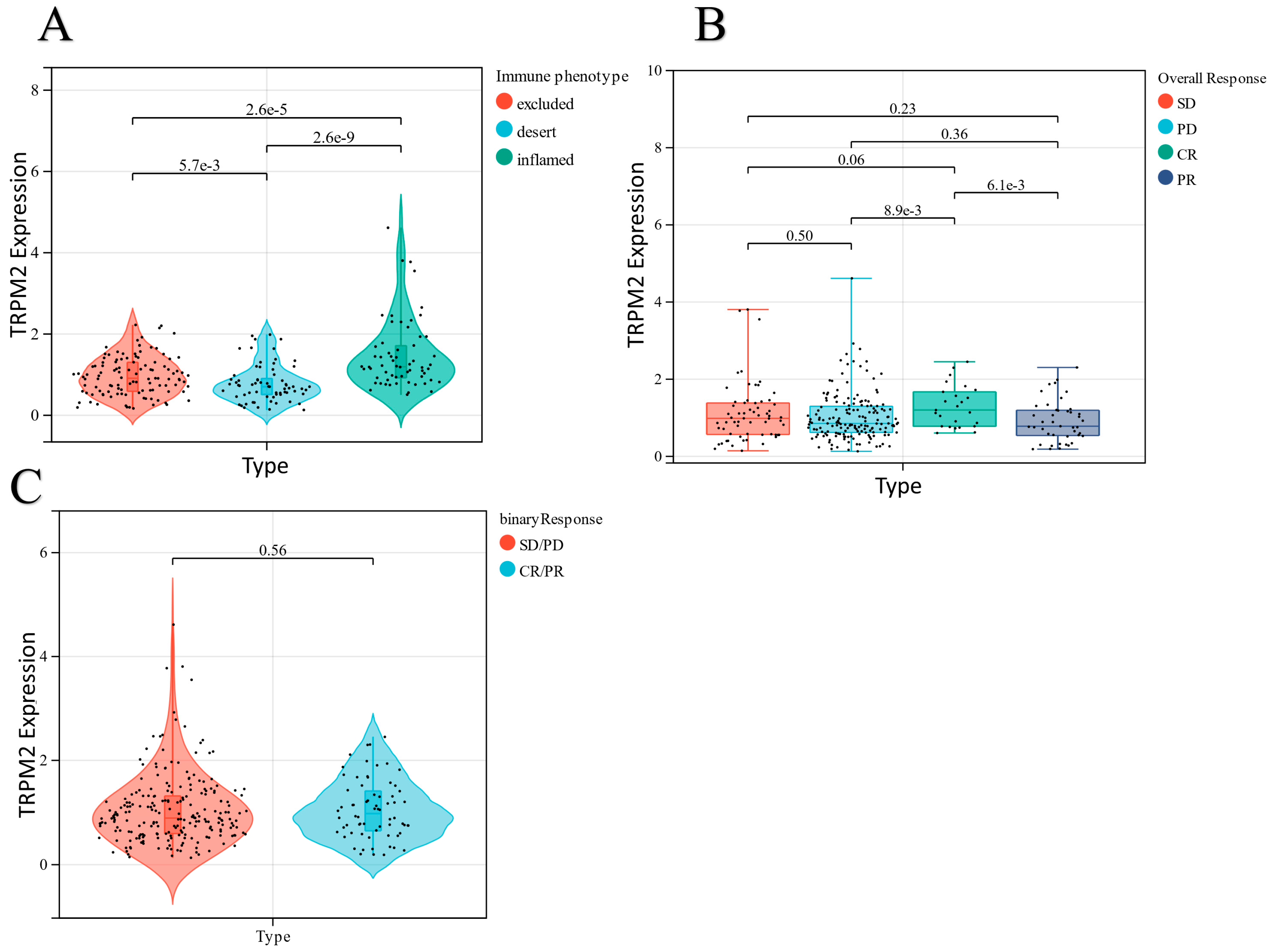
3. Discussion
4. Materials and Methods
4.1. Data Collection
4.2. Development of an ER Stress-Related Classifier Related to Prognosis
4.3. Survival Analysis
4.4. Determination of a Nomogram
4.5. Levels of Infiltration of Immune Cells
4.6. qPCR Assay
4.7. Cell Line
4.8. shRNA Knockdown of TRPM2 In Vitro
4.9. CCK8 Cell Proliferation Assay
4.10. Cell Cycle Assay
4.11. Cell Migration and Invasion Assays
4.12. ROS Measurements
4.13. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Yoon, H.; Lee, S. Fatty Acid Metabolism in Ovarian Cancer: Therapeutic Implications. Int. J. Mol. Sci. 2022, 23, 2170. [Google Scholar] [CrossRef] [PubMed]
- Cruz, I.N.; Coley, H.M.; Kramer, H.B.; Madhuri, T.K.; Safuwan, N.A.; Angelino, A.R.; Yang, M. Proteomics Analysis of Ovarian Cancer Cell Lines and Tissues Reveals Drug Resistance-associated Proteins. Cancer Genom. Proteom. 2017, 14, 35–51. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Cheng, S.; Jin, Y.; Zhao, Y.; Wang, Y. Roles of CA125 in diagnosis, prediction, and oncogenesis of ovarian cancer. Biochim. Biophys. Acta Rev. Cancer 2021, 1875, 188503. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Cubillos-Ruiz, J.R. Endoplasmic reticulum stress signals in the tumour and its microenvironment. Nat. Rev. Cancer 2021, 21, 71–88. [Google Scholar] [CrossRef]
- Xu, X.; Tang, Y.; Zhu, J.; Luo, J. Endoplasmic Reticulum Stress-Related Four-Biomarker Risk Classifier for Survival Evaluation in Esophageal Cancer. J. Oncol. 2022, 2022, 5860671. [Google Scholar] [CrossRef] [PubMed]
- Joo, J.H.; Liao, G.; Collins, J.B.; Grissom, S.F.; Jetten, A.M. Farnesol-induced apoptosis in human lung carcinoma cells is coupled to the endoplasmic reticulum stress response. Cancer Res. 2007, 67, 7929–7936. [Google Scholar] [CrossRef] [PubMed]
- Moenner, M.; Pluquet, O.; Bouchecareilh, M.; Chevet, E. Integrated endoplasmic reticulum stress responses in cancer. Cancer Res. 2007, 67, 10631–10634. [Google Scholar] [CrossRef]
- Wang, W.; Wang, M.; Xiao, Y.; Wang, Y.; Ma, L.; Guo, L.; Wu, X.; Lin, X.; Zhang, P. USP35 mitigates endoplasmic reticulum stress-induced apoptosis by stabilizing RRBP1 in non-small cell lung cancer. Mol. Oncol. 2022, 16, 1572–1590. [Google Scholar] [CrossRef]
- Samanta, S.; Tamura, S.; Dubeau, L.; Mhawech-Fauceglia, P.; Miyagi, Y.; Kato, H.; Lieberman, R.; Buckanovich, R.J.; Lin, Y.G.; Neamati, N. Clinicopathological significance of endoplasmic reticulum stress proteins in ovarian carcinoma. Sci. Rep. 2020, 10, 2160. [Google Scholar] [CrossRef]
- Xu, J.; Bi, G.; Luo, Q.; Liu, Y.; Liu, T.; Li, L.; Zeng, Q.; Wang, Q.; Wang, Y.; Yu, J.; et al. PHLDA1 Modulates the Endoplasmic Reticulum Stress Response and is required for Resistance to Oxidative Stress-induced Cell Death in Human Ovarian Cancer Cells. J. Cancer 2021, 12, 5486–5493. [Google Scholar] [CrossRef]
- Bi, F.; Jiang, Z.; Park, W.; Hartwich, T.M.P.; Ge, Z.; Chong, K.Y.; Yang, K.; Morrison, M.J.; Kim, D.; Kim, J.; et al. A Benzenesulfonamide-Based Mitochondrial Uncoupler Induces Endoplasmic Reticulum Stress and Immunogenic Cell Death in Epithelial Ovarian Cancer. Mol. Cancer Ther. 2021, 20, 2398–2409. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Gong, X.; Hu, T.; Chen, Y. Two novel prognostic models for ovarian cancer respectively based on ferroptosis and necroptosis. BMC Cancer 2022, 22, 74. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Zheng, M.; Wang, S.; Gao, L.; Gou, R.; Liu, O.; Dong, H.; Li, X.; Lin, B. Identification of a five-gene signature of the RGS gene family with prognostic value in ovarian cancer. Genomics 2021, 113, 2134–2144. [Google Scholar] [CrossRef] [PubMed]
- Bao, L.; Festa, F.; Freet, C.S.; Lee, J.P.; Hirschler-Laszkiewicz, I.M.; Chen, S.J.; Keefer, K.A.; Wang, H.G.; Patterson, A.D.; Cheung, J.Y.; et al. The Human Transient Receptor Potential Melastatin 2 Ion Channel Modulates ROS Through Nrf2. Sci. Rep. 2019, 9, 14132. [Google Scholar] [CrossRef] [PubMed]
- James, N.E.; Woodman, M.; Ribeiro, J.R. Prognostic immunologic signatures in epithelial ovarian cancer. Oncogene 2022, 41, 1389–1396. [Google Scholar] [CrossRef] [PubMed]
- Bao, M.; Zhang, L.; Hu, Y. Novel gene signatures for prognosis prediction in ovarian cancer. J. Cell. Mol. Med. 2020, 24, 9972–9984. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Wu, Y.; Wang, S.; Wu, J.; Wu, X.; Zheng, Z. A risk model of gene signatures for predicting platinum response and survival in ovarian cancer. J. Ovarian Res. 2022, 15, 39. [Google Scholar] [CrossRef]
- Wang, H.; Liu, J.; Yang, J.; Wang, Z.; Zhang, Z.; Peng, J.; Wang, Y.; Hong, L. A novel tumor mutational burden-based risk model predicts prognosis and correlates with immune infiltration in ovarian cancer. Front. Immunol. 2022, 13, 943389. [Google Scholar] [CrossRef]
- Beretta, G.L.; Corno, C.; Zaffaroni, N.; Perego, P. Role of FoxO Proteins in Cellular Response to Antitumor Agents. Cancers 2019, 11, 90. [Google Scholar] [CrossRef]
- Oakes, S.A. Endoplasmic Reticulum Stress Signaling in Cancer Cells. Am. J. Pathol. 2020, 190, 934–946. [Google Scholar] [CrossRef]
- Yang, J.Y.; Hung, M.C. Deciphering the role of forkhead transcription factors in cancer therapy. Curr. Drug Targets 2011, 12, 1284–1290. [Google Scholar] [CrossRef] [PubMed]
- Christian, M.; Lam, E.W.; Wilson, M.S.; Brosens, J.J. FOXO transcription factors and their role in disorders of the female reproductive tract. Curr. Drug Targets 2011, 12, 1291–1302. [Google Scholar] [CrossRef] [PubMed]
- Brosens, J.J.; Wilson, M.S.; Lam, E.W. FOXO transcription factors: From cell fate decisions to regulation of human female reproduction. Adv. Exp. Med. Biol. 2009, 665, 227–241. [Google Scholar] [CrossRef] [PubMed]
- Song, M.; Bode, A.M.; Dong, Z.; Lee, M.H. AKT as a Therapeutic Target for Cancer. Cancer Res. 2019, 79, 1019–1031. [Google Scholar] [CrossRef] [PubMed]
- White, J.P.; Cibelli, M.; Urban, L.; Nilius, B.; McGeown, J.G.; Nagy, I. TRPV4: Molecular Conductor of a Diverse Orchestra. Physiol. Rev. 2016, 96, 911–973. [Google Scholar] [CrossRef] [PubMed]
- So, C.L.; Milevskiy, M.J.G.; Monteith, G.R. Transient receptor potential cation channel subfamily V and breast cancer. Lab. Investig. 2020, 100, 199–206. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Liu, Z. STAT1 in cancer: Friend or foe? Discov. Med. 2017, 24, 19–29. [Google Scholar]
- Honoré, B. The rapidly expanding CREC protein family: Members, localization, function, and role in disease. Bioessays 2009, 31, 262–277. [Google Scholar] [CrossRef]
- Cai, R.; Wang, P.; Zhao, X.; Lu, X.; Deng, R.; Wang, X.; Hong, C.; Lin, J. Reticulocalbin3: A Ca2+ homeostasis regulator that promotes esophageal squamous cell carcinoma progression and cisplatin resistance. Cancer Sci. 2022, 113, 3593–3607. [Google Scholar] [CrossRef]
- Biswas, S.K.; Mantovani, A. Macrophage plasticity and interaction with lymphocyte subsets: Cancer as a paradigm. Nat. Immunol. 2010, 11, 889–896. [Google Scholar] [CrossRef]
- Yoshitomi, H.; Ueno, H. Shared and distinct roles of T peripheral helper and T follicular helper cells in human diseases. Cell. Mol. Immunol. 2021, 18, 523–527. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Mezzadra, R.; Schumacher, T.N. Regulation and Function of the PD-L1 Checkpoint. Immunity 2018, 48, 434–452. [Google Scholar] [CrossRef] [PubMed]
- Lane, D.J.; Mills, T.M.; Shafie, N.H.; Merlot, A.M.; Saleh Moussa, R.; Kalinowski, D.S.; Kovacevic, Z.; Richardson, D.R. Expanding horizons in iron chelation and the treatment of cancer: Role of iron in the regulation of ER stress and the epithelial-mesenchymal transition. Biochim. Biophys. Acta 2014, 1845, 166–181. [Google Scholar] [CrossRef] [PubMed]
- Aubrey, B.J.; Strasser, A.; Kelly, G.L. Tumor-Suppressor Functions of the TP53 Pathway. Cold Spring Harb. Perspect. Med. 2016, 6, a026062. [Google Scholar] [CrossRef] [PubMed]
- Fliegert, R.; Bauche, A.; Wolf Pérez, A.M.; Watt, J.M.; Rozewitz, M.D.; Winzer, R.; Janus, M.; Gu, F.; Rosche, A.; Harneit, A.; et al. 2′-Deoxyadenosine 5′-diphosphoribose is an endogenous TRPM2 superagonist. Nat. Chem. Biol. 2017, 13, 1036–1044. [Google Scholar] [CrossRef] [PubMed]
- Malko, P.; Jiang, L.H. TRPM2 channel-mediated cell death: An important mechanism linking oxidative stress-inducing pathological factors to associated pathological conditions. Redox Biol. 2020, 37, 101755. [Google Scholar] [CrossRef] [PubMed]
- Miller, B.A. TRPM2 in Cancer. Cell Calcium 2019, 80, 8–17. [Google Scholar] [CrossRef]
- Sommerfeld, L.; Finkernagel, F.; Jansen, J.M.; Wagner, U.; Nist, A.; Stiewe, T.; Müller-Brüsselbach, S.; Sokol, A.M.; Graumann, J.; Reinartz, S.; et al. The multicellular signalling network of ovarian cancer metastases. Clin. Transl. Med. 2021, 11, e633. [Google Scholar] [CrossRef]
- Newman, A.M.; Liu, C.L.; Green, M.R.; Gentles, A.J.; Feng, W.; Xu, Y.; Hoang, C.D.; Diehn, M.; Alizadeh, A.A. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods 2015, 12, 453–457. [Google Scholar] [CrossRef]

| Gene | Sequence (5′- > 3′) | |
|---|---|---|
| FOXO1 | F primer | TCGTCATAATCTGTCCCTACACA |
| R primer | CGGCTTCGGCTCTTAGCAAA | |
| TRPM2 | F primer | TCCCCGCCGAGTACATACTG |
| R primer | GTCTGCTCCGATATGAACTTCTC | |
| RCN2 | F primer | TTCAGGTCCCGGTTTGAGTCT |
| R primer | TCAAGCCTGCCATCGTTATCT | |
| MZB1 | F primer | AGTTGGTCTACACGGATGTCC |
| R primer | CTTGGTCCACTTCTCGAACTC | |
| GPR37 | F primer | ATGTCGCGGCTACTGCTTC |
| R primer | GCAGAACGTCTCTTGCAGAAT | |
| GABARAPL1 | F primer | ATGAAGTTCCAGTACAAGGAGGA |
| R primer | GCTTTTGGAGCCTTCTCTACAAT | |
| CDKN1B | F primer | ATCACAAACCCCTAGAGGGCA |
| R primer | GGGTCTGTAGTAGAACTCGGG | |
| STAT1 | F primer | CAGCTTGACTCAAAATTCCTGGA |
| R primer | TGAAGATTACGCTTGCTTTTCCT | |
| TRPV4 | F primer | TCCACCCTATATGAGTCCTCGG |
| R primer | TAGGTGCCGTAGTCAAACAGT | |
| CACNA1C | F primer | TGATTCCAACGCCACCAATTC |
| R primer | GAGGAGTCCATAGGCGATTACT | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, M.; Wang, Y.; Xu, S.; Huang, S.; Wu, M.; Chen, G.; Wang, Y. Endoplasmic Reticulum Stress-Related Ten-Biomarker Risk Classifier for Survival Evaluation in Epithelial Ovarian Cancer and TRPM2: A Potential Therapeutic Target of Ovarian Cancer. Int. J. Mol. Sci. 2023, 24, 14010. https://doi.org/10.3390/ijms241814010
Zhang M, Wang Y, Xu S, Huang S, Wu M, Chen G, Wang Y. Endoplasmic Reticulum Stress-Related Ten-Biomarker Risk Classifier for Survival Evaluation in Epithelial Ovarian Cancer and TRPM2: A Potential Therapeutic Target of Ovarian Cancer. International Journal of Molecular Sciences. 2023; 24(18):14010. https://doi.org/10.3390/ijms241814010
Chicago/Turabian StyleZhang, Minghai, Yingjie Wang, Shilin Xu, Shan Huang, Meixuan Wu, Guangquan Chen, and Yu Wang. 2023. "Endoplasmic Reticulum Stress-Related Ten-Biomarker Risk Classifier for Survival Evaluation in Epithelial Ovarian Cancer and TRPM2: A Potential Therapeutic Target of Ovarian Cancer" International Journal of Molecular Sciences 24, no. 18: 14010. https://doi.org/10.3390/ijms241814010





