CRISPR/Cas9-Mediated CtBP1 Gene Editing Enhances Chemosensitivity and Inhibits Metastatic Potential in Esophageal Squamous Cell Carcinoma Cells
Abstract
:1. Introduction
2. Results
2.1. Established PTX-Resistant ESCC Cell Lines
2.2. CtBP1 Is Notably Expressed in Esophageal Squamous Cell Carcinoma (ESCC) Cells
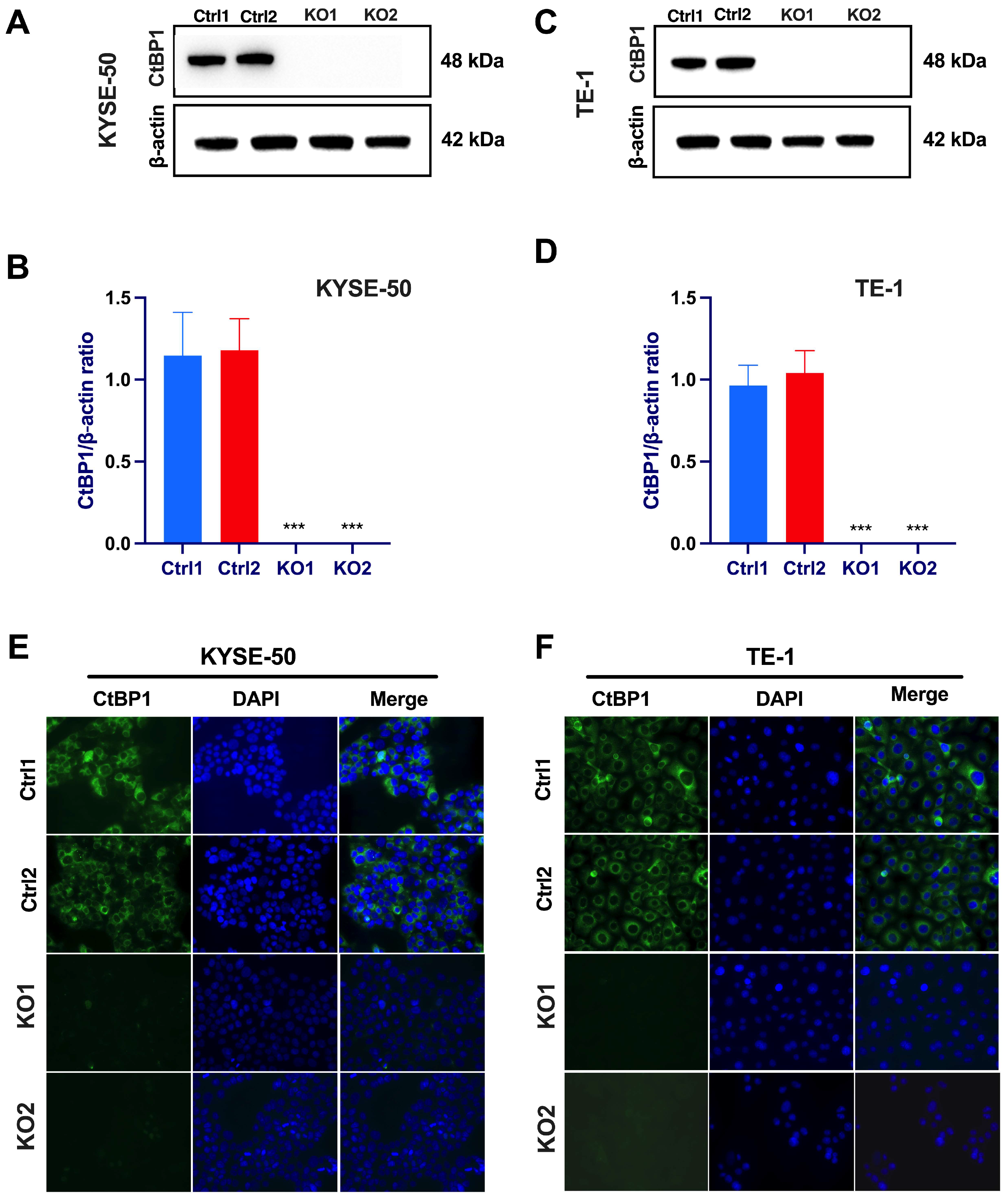
2.3. CRISPR/Cas9-Mediated Suppression of Oncogenic CtBP1 Expression in Paclitaxel-Resistant ESCC Cells
2.4. CtBP1 Knockout Promotes Paclitaxel Sensitivity and Inhibits Proliferation In Vitro
2.5. CtBP1 Expression Drives Cell Migration and Invasion in ESCC Cells
3. Discussion
4. Materials and Methods
4.1. Chemicals and Reagents
4.2. Cell Lines and Cell Culture
4.3. Antibodies
4.4. Guide RNA Design and Construction
4.5. Creation of CtBP1-KO Clonal ESCC Cell Lines Using a CRISPR-Directed Gene-Editing Approach
4.6. Immunofluorescence
4.7. Western Blotting
4.8. Cell Proliferation Assay
4.9. Cell Viability Assay
4.10. IC50 Calculation
4.11. Colony Formation Assay
4.12. Wound Healing Assay
4.13. Cell Migration Assa
4.14. Cell Invasion Assay
4.15. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Enzinger, P.C.; Mayer, R.J. Esophageal cancer. N. Engl. J. Med. 2003, 349, 2241–2252. [Google Scholar] [CrossRef]
- Zhao, J.; He, Y.T.; Zheng, R.S.; Zhang, S.W.; Chen, W.Q. Analysis of esophageal cancer time trends in China, 1989–2008. Asian Pac. J. Cancer Prev. 2012, 13, 4613–4617. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Wang, H.; Zeng, X.; Yin, P.; Zhu, J.; Chen, W.; Li, X.; Wang, L.; Wang, L.; Liu, Y.; et al. Mortality, morbidity, and risk factors in China and its provinces, 1990–2017: A systematic analysis for the Global Burden of Disease Study 2017. Lancet 2019, 394, 1145–1158. [Google Scholar] [CrossRef] [PubMed]
- Allemani, C.; Matsuda, T.; Di Carlo, V.; Harewood, R.; Matz, M.; Niksic, M.; Bonaventure, A.; Valkov, M.; Johnson, C.J.; Esteve, J.; et al. Global surveillance of trends in cancer survival 2000-14 (CONCORD-3): Analysis of individual records for 37,513,025 patients diagnosed with one of 18 cancers from 322 population-based registries in 71 countries. Lancet 2018, 391, 1023–1075. [Google Scholar] [CrossRef]
- Morgan, E.; Soerjomataram, I.; Rumgay, H.; Coleman, H.G.; Thrift, A.P.; Vignat, J.; Laversanne, M.; Ferlay, J.; Arnold, M. The Global Landscape of Esophageal Squamous Cell Carcinoma and Esophageal Adenocarcinoma Incidence and Mortality in 2020 and Projections to 2040: New Estimates from GLOBOCAN 2020. Gastroenterology 2022, 163, 649–658.e642. [Google Scholar] [CrossRef]
- Mariette, C.; Piessen, G.; Balon, J.M.; Van Seuningen, I.; Triboulet, J.P. Surgery alone in the curative treatment of localised oesophageal carcinoma. Eur. J. Surg. Oncol. 2004, 30, 869–876. [Google Scholar] [CrossRef]
- Roca, E.; Pennella, E.; Sardi, M.; Carraro, S.; Barugel, M.; Milano, C.; Fiorini, A.; Giglio, R.; Gonzalez, G.; Kneitschel, R.; et al. Combined intensive chemoradiotherapy for organ preservation in patients with resectable and non-resectable oesophageal cancer. Eur. J. Cancer 1996, 32A, 429–432. [Google Scholar] [CrossRef]
- Ilson, D.H.; van Hillegersberg, R. Management of Patients with Adenocarcinoma or Squamous Cancer of the Esophagus. Gastroenterology 2018, 154, 437–451. [Google Scholar] [CrossRef]
- Backemar, L.; Lagergren, P.; Djarv, T.; Johar, A.; Wikman, A.; Lagergren, J. Comorbidities and Risk of Complications After Surgery for Esophageal Cancer: A Nationwide Cohort Study in Sweden. World J. Surg. 2015, 39, 2282–2288. [Google Scholar] [CrossRef]
- Yip, C.; Landau, D.; Kozarski, R.; Ganeshan, B.; Thomas, R.; Michaelidou, A.; Goh, V. Primary esophageal cancer: Heterogeneity as potential prognostic biomarker in patients treated with definitive chemotherapy and radiation therapy. Radiology 2014, 270, 141–148. [Google Scholar] [CrossRef] [PubMed]
- Mayanagi, S.; Irino, T.; Kawakubo, H.; Kitagawa, Y. Neoadjuvant treatment strategy for locally advanced thoracic esophageal cancer. Ann. Gastroenterol. Surg. 2019, 3, 269–275. [Google Scholar] [CrossRef]
- Yang, H.; Liu, H.; Chen, Y.; Zhu, C.; Fang, W.; Yu, Z.; Mao, W.; Xiang, J.; Han, Y.; Chen, Z.; et al. Neoadjuvant Chemoradiotherapy Followed by Surgery Versus Surgery Alone for Locally Advanced Squamous Cell Carcinoma of the Esophagus (NEOCRTEC5010): A Phase III Multicenter, Randomized, Open-Label Clinical Trial. J. Clin. Oncol. 2018, 36, 2796–2803. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Liu, H.; Chen, Y.; Zhu, C.; Fang, W.; Yu, Z.; Mao, W.; Xiang, J.; Han, Y.; Chen, Z.; et al. Long-term Efficacy of Neoadjuvant Chemoradiotherapy Plus Surgery for the Treatment of Locally Advanced Esophageal Squamous Cell Carcinoma: The NEOCRTEC5010 Randomized Clinical Trial. JAMA Surg. 2021, 156, 721–729. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.; Dou, Y.; Zhou, K.; Huo, J.; Yang, T.; Qin, T.; Liu, W.; Wang, S.; Yang, D.; Chang, L.; et al. Targeting the Bcl-2 family and P-glycoprotein reverses paclitaxel resistance in human esophageal carcinoma cell line. Biomed. Pharmacother. 2017, 90, 897–905. [Google Scholar] [CrossRef]
- Bellesis, A.G.; Jecrois, A.M.; Hayes, J.A.; Schiffer, C.A.; Royer, W.E., Jr. Assembly of human C-terminal binding protein (CtBP) into tetramers. J. Biol. Chem. 2018, 293, 9101–9112. [Google Scholar] [CrossRef]
- Chen, X.; Zhang, Q.; Dang, X.; Song, T.; Wang, Y.; Yu, Z.; Zhang, S.; Fan, J.; Cong, F.; Zhang, W.; et al. Targeting the CtBP1-FOXM1 transcriptional complex with small molecules to overcome MDR1-mediated chemoresistance in osteosarcoma cancer stem cells. J. Cancer 2021, 12, 482–497. [Google Scholar] [CrossRef]
- Blevins, M.A.; Huang, M.; Zhao, R. The Role of CtBP1 in Oncogenic Processes and Its Potential as a Therapeutic Target. Mol. Cancer Ther. 2017, 16, 981–990. [Google Scholar] [CrossRef]
- Wang, R.; Asangani, I.A.; Chakravarthi, B.V.; Ateeq, B.; Lonigro, R.J.; Cao, Q.; Mani, R.S.; Camacho, D.F.; McGregor, N.; Schumann, T.E.; et al. Role of transcriptional corepressor CtBP1 in prostate cancer progression. Neoplasia 2012, 14, 905–914. [Google Scholar] [CrossRef]
- Chinnadurai, G. CtBP, an unconventional transcriptional corepressor in development and oncogenesis. Mol. Cell 2002, 9, 213–224. [Google Scholar] [CrossRef]
- Di, L.J.; Byun, J.S.; Wong, M.M.; Wakano, C.; Taylor, T.; Bilke, S.; Baek, S.; Hunter, K.; Yang, H.; Lee, M.; et al. Genome-wide profiles of CtBP link metabolism with genome stability and epithelial reprogramming in breast cancer. Nat. Commun. 2013, 4, 1449. [Google Scholar] [CrossRef] [PubMed]
- Birts, C.N.; Harding, R.; Soosaipillai, G.; Halder, T.; Azim-Araghi, A.; Darley, M.; Cutress, R.I.; Bateman, A.C.; Blaydes, J.P. Expression of CtBP family protein isoforms in breast cancer and their role in chemoresistance. Biol. Cell 2010, 103, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Jordan, M.A.; Wilson, L. Microtubules as a target for anticancer drugs. Nat. Rev. Cancer 2004, 4, 253–265. [Google Scholar] [CrossRef]
- Yang, B.; Guo, X.; Le, C.; Su, W.; Li, X.; Zhang, Y.; Yang, G.; Liang, W.; Zheng, Z.; Wu, J.; et al. Efficacy and Safety of Apatinib plus Neoadjuvant Chemotherapy for Locally Advanced Esophageal Squamous Cancer: A Phase II Trial. BioMed Res. Int. 2022, 2022, 4727407. [Google Scholar] [CrossRef]
- Liu, Y.; Ren, Z.; Yuan, L.; Xu, S.; Yao, Z.; Qiao, L.; Li, K. Paclitaxel plus cisplatin vs. 5-fluorouracil plus cisplatin as first-line treatment for patients with advanced squamous cell esophageal cancer. Am. J. Cancer Res. 2016, 6, 2345–2350. [Google Scholar] [PubMed]
- Akhtar, J.; Wang, Z.; Yu, C.; Zhang, Z.P.; Bi, M.M. STMN-1 gene: A predictor of survival in stage iia esophageal squamous cell carcinoma after Ivor-Lewis esophagectomy? Ann. Surg. Oncol. 2014, 21, 315–321. [Google Scholar] [CrossRef]
- Akhtar, J.; Wang, Z.; Jiang, W.P.; Bi, M.M.; Zhang, Z.P. Stathmin overexpression identifies high risk for lymphatic metastatic recurrence in pN0 esophageal squamous cell carcinoma patients. J. Gastroenterol. Hepatol. 2014, 29, 944–950. [Google Scholar] [CrossRef]
- Akhtar, J.; Wang, Z.; Zhang, Z.P.; Bi, M.M. Lentiviral-mediated RNA interference targeting stathmin1 gene in human gastric cancer cells inhibits proliferation in vitro and tumor growth in vivo. J. Transl. Med. 2013, 11, 212. [Google Scholar] [CrossRef]
- Akhtar, J.; Wang, Z.; Yu, C.; Zhang, Z.P. Effectiveness of local injection of lentivirus-delivered stathmin1 and stathmin1 shRNA in human gastric cancer xenograft mouse. J. Gastroenterol. Hepatol. 2014, 29, 1685–1691. [Google Scholar] [CrossRef]
- Habib, R.; Akhtar, J.; Taqi, M.; Yu, C.; Zhang, C. Lentiviral vector-mediated survivin shRNA delivery in gastric cancer cell lines significantly inhibits cell proliferation and tumor growth. Oncol. Rep. 2015, 34, 859–867. [Google Scholar] [CrossRef]
- Wang, S.; Akhtar, J.; Wang, Z. Anti-STMN1 therapy improves sensitivity to antimicrotubule drugs in esophageal squamous cell carcinoma. Tumour Biol. 2015, 36, 7797–7806. [Google Scholar] [CrossRef] [PubMed]
- Doudna, J.A.; Charpentier, E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science 2014, 346, 1258096. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; La Russa, M.; Qi, L.S. CRISPR/Cas9 in Genome Editing and Beyond. Annu. Rev. Biochem. 2016, 85, 227–264. [Google Scholar] [CrossRef] [PubMed]
- Karamouzis, M.V.; Gorgoulis, V.G.; Papavassiliou, A.G. Transcription factors and neoplasia: Vistas in novel drug design. Clin. Cancer Res. 2002, 8, 949–961. [Google Scholar]
- Vaiopoulos, A.G.; Kostakis, I.D.; Athanasoula, K.; Papavassiliou, A.G. Targeting transcription factor corepressors in tumor cells. Cell Mol. Life Sci. 2012, 69, 1745–1753. [Google Scholar] [CrossRef]
- Battaglia, S.; Maguire, O.; Campbell, M.J. Transcription factor co-repressors in cancer biology: Roles and targeting. Int. J. Cancer 2010, 126, 2511–2519. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Bi, Y.; Bi, H.; Diao, C.; Zhang, G.; Cheng, K.; Yang, Z. miR-137 suppresses the invasion and procedure of EMT of human breast cancer cell line MCF-7 through targeting CtBP1. Hum. Cell 2016, 29, 30–36. [Google Scholar] [CrossRef]
- Chinnadurai, G. The transcriptional corepressor CtBP: A foe of multiple tumor suppressors. Cancer Res. 2009, 69, 731–734. [Google Scholar] [CrossRef]
- Zhang, X.L.; Huang, C.X.; Zhang, J.; Inoue, A.; Zeng, S.E.; Xiao, S.J. CtBP1 is involved in epithelial-mesenchymal transition and is a potential therapeutic target for hepatocellular carcinoma. Oncol. Rep. 2013, 30, 809–814. [Google Scholar] [CrossRef]
- Jin, W.; Scotto, K.W.; Hait, W.N.; Yang, J.M. Involvement of CtBP1 in the transcriptional activation of the MDR1 gene in human multidrug resistant cancer cells. Biochem. Pharmacol. 2007, 74, 851–859. [Google Scholar] [CrossRef]
- Wang, C.; Guo, L.B.; Ma, J.Y.; Li, Y.M.; Liu, H.M. Establishment, and characterization of a paclitaxel-resistant human esophageal carcinoma cell line. Int. J. Oncol. 2013, 43, 1607–1617. [Google Scholar] [CrossRef]
- Chen, S.Y.; Hu, S.S.; Dong, Q.; Cai, J.X.; Zhang, W.P.; Sun, J.Y.; Wang, T.T.; Xie, J.; He, H.R.; Xing, J.F.; et al. Establishment of paclitaxel-resistant breast cancer cell line and nude mice models, and underlying multidrug resistance mechanisms in vitro and in vivo. Asian Pac. J. Cancer Prev. 2013, 14, 6135–6140. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhao, J.; Zhang, W.; Liu, G.; Yin, D.; Li, J.; Zhang, S.; Li, H. Establishment of paclitaxel-resistant cell line and the underlying mechanism on drug resistance. Int. J. Gynecol. Cancer 2012, 22, 1450–1456. [Google Scholar] [CrossRef] [PubMed]
- Nunes, M.; Silva, P.M.A.; Coelho, R.; Pinto, C.; Resende, A.; Bousbaa, H.; Almeida, G.M.; Ricardo, S. Generation of Two Paclitaxel-Resistant High-Grade Serous Carcinoma Cell Lines with Increased Expression of P-Glycoprotein. Front. Oncol. 2021, 11, 752127. [Google Scholar] [CrossRef] [PubMed]
- Aldonza, M.B.; Hong, J.Y.; Lee, S.K. Paclitaxel-resistant cancer cell-derived secretomes elicit ABCB1-associated docetaxel cross-resistance and escape from apoptosis through FOXO3a-driven glycolytic regulation. Exp. Mol. Med. 2017, 49, e286. [Google Scholar] [CrossRef]
- He, Y.; He, Z.; Lin, J.; Chen, C.; Chen, Y.; Liu, S. CtBP1/2 differentially regulate genomic stability and DNA repair pathway in high-grade serous ovarian cancer cell. Oncogenesis 2021, 10, 49. [Google Scholar] [CrossRef]
- Logan, C.M.; Menko, A.S. Microtubules: Evolving roles and critical cellular interactions. Exp. Biol. Med. 2019, 244, 1240–1254. [Google Scholar] [CrossRef]
- Orr, G.A.; Verdier-Pinard, P.; McDaid, H.; Horwitz, S.B. Mechanisms of Taxol resistance related to microtubules. Oncogene 2003, 22, 7280–7295. [Google Scholar] [CrossRef]
- Saiki, Y.; Yoshino, Y.; Fujimura, H.; Manabe, T.; Kudo, Y.; Shimada, M.; Mano, N.; Nakano, T.; Lee, Y.; Shimizu, S.; et al. DCK is frequently inactivated in acquired gemcitabine-resistant human cancer cells. Biochem. Biophys. Res. Commun. 2012, 421, 98–104. [Google Scholar] [CrossRef]
- Doench, J.G.; Fusi, N.; Sullender, M.; Hegde, M.; Vaimberg, E.W.; Donovan, K.F.; Smith, I.; Tothova, Z.; Wilen, C.; Orchard, R.; et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat. Biotechnol. 2016, 34, 184–191. [Google Scholar] [CrossRef]
- Hsu, P.D.; Scott, D.A.; Weinstein, J.A.; Ran, F.A.; Konermann, S.; Agarwala, V.; Li, Y.; Fine, E.J.; Wu, X.; Shalem, O.; et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 2013, 31, 827–832. [Google Scholar] [CrossRef] [PubMed]
- Sanjana, N.E.; Shalem, O.; Zhang, F. Improved vectors, and genome-wide libraries for CRISPR screening. Nat. Methods 2014, 11, 783–784. [Google Scholar] [CrossRef] [PubMed]
- Shalem, O.; Sanjana, N.E.; Hartenian, E.; Shi, X.; Scott, D.A.; Mikkelson, T.; Heckl, D.; Ebert, B.L.; Root, D.E.; Doench, J.G.; et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 2014, 343, 84–87. [Google Scholar] [CrossRef] [PubMed]
- Akhtar, J.; Han, Y.; Han, S.; Lin, W.; Cao, C.; Ge, R.; Babarinde, I.A.; Jia, Q.; Yuan, Y.; Chen, G.; et al. Bistable insulin response: The win-win solution for glycemic control. iScience 2022, 25, 105561. [Google Scholar] [CrossRef]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef]
- Yue, P.Y.; Leung, E.P.; Mak, N.K.; Wong, R.N. A simplified method for quantifying cell migration/wound healing in 96-well plates. J. Biomol. Screen. 2010, 15, 427–433. [Google Scholar] [CrossRef]




| Name | Sequence (5′–3′) |
|---|---|
| 1-sgRNAScramble | GCACTACCAGAGCTAACTCA |
| 2-sgRNAScramble | GTCCACCCTTATCTAGGCTA |
| 1-sgRNACtBP1 | AATCACTGAAGCCTGCGTCG |
| 2-sgRNACtBP1 | AGTTCTGGTCGGACCTTCGA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Akhtar, J.; Imran, M.; Wang, G. CRISPR/Cas9-Mediated CtBP1 Gene Editing Enhances Chemosensitivity and Inhibits Metastatic Potential in Esophageal Squamous Cell Carcinoma Cells. Int. J. Mol. Sci. 2023, 24, 14030. https://doi.org/10.3390/ijms241814030
Akhtar J, Imran M, Wang G. CRISPR/Cas9-Mediated CtBP1 Gene Editing Enhances Chemosensitivity and Inhibits Metastatic Potential in Esophageal Squamous Cell Carcinoma Cells. International Journal of Molecular Sciences. 2023; 24(18):14030. https://doi.org/10.3390/ijms241814030
Chicago/Turabian StyleAkhtar, Javed, Muhammad Imran, and Guanyu Wang. 2023. "CRISPR/Cas9-Mediated CtBP1 Gene Editing Enhances Chemosensitivity and Inhibits Metastatic Potential in Esophageal Squamous Cell Carcinoma Cells" International Journal of Molecular Sciences 24, no. 18: 14030. https://doi.org/10.3390/ijms241814030
APA StyleAkhtar, J., Imran, M., & Wang, G. (2023). CRISPR/Cas9-Mediated CtBP1 Gene Editing Enhances Chemosensitivity and Inhibits Metastatic Potential in Esophageal Squamous Cell Carcinoma Cells. International Journal of Molecular Sciences, 24(18), 14030. https://doi.org/10.3390/ijms241814030








