Identification of P450 Candidates Associated with the Biosynthesis of Physalin-Class Compounds in Physalis angulata
Abstract
:1. Introduction

2. Results
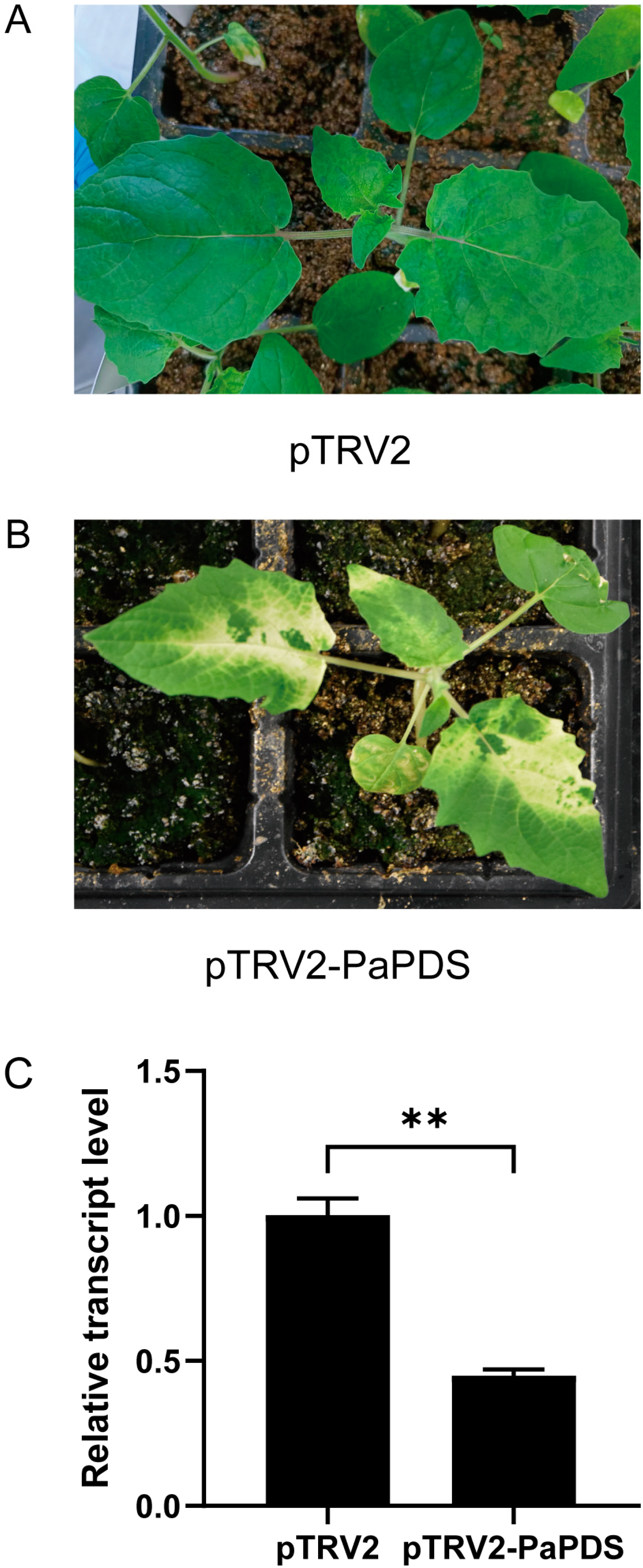
2.1. Confirmation of the VIGS System for P. angulata
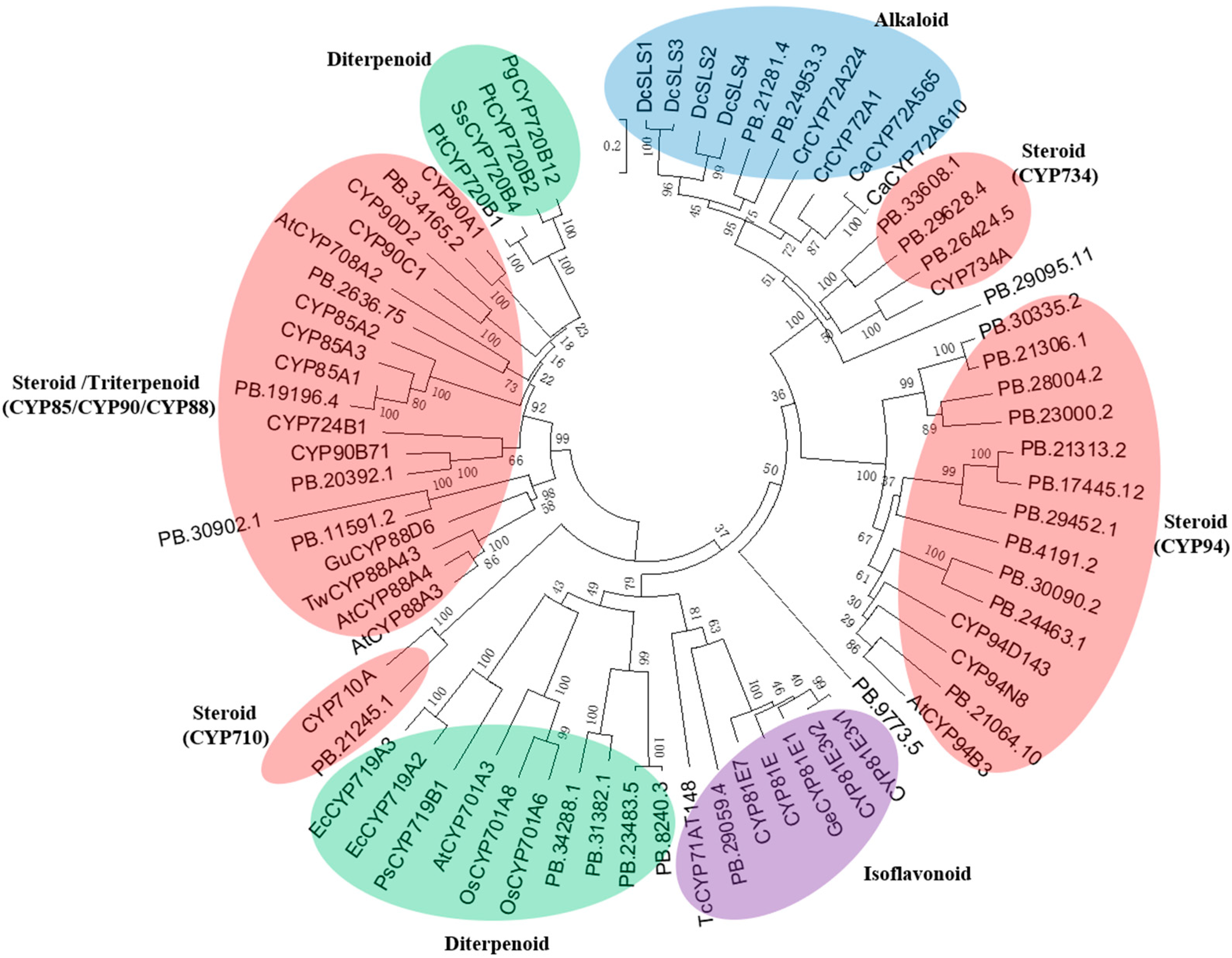
2.2. Identification of the P450 Candidates Associated with the Biosynthesis of Physalin-Class Compounds
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Chemicals
4.2. Phylogenetic Analysis of the P. angulata P450 Candidates
4.3. Transient Suppression of the P. angulata P450 Candidates
4.4. Real-Time PCR Analysis
4.5. Phytochemical Analysis
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sun, C.P.; Kutateladze, A.G.; Zhao, F.; Chen, L.X.; Qiu, F. A novel withanolide with an unprecedented carbon skeleton from Physalis angulata. Org. Biomol. Chem. 2017, 15, 1110–1114. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.; He, J.X.; Hu, H.X.; Zhang, K.; Wang, X.N.; Zhao, B.B.; Lou, H.X.; Ren, D.M.; Shen, T. Withanolides from the genus Physalis: A review on their phytochemical and pharmacological aspects. J. Pharm. Pharmacol. 2020, 72, 649–669. [Google Scholar] [CrossRef] [PubMed]
- Makino, B.; Kawai, M.; Kito, K.; Yamamura, H.; Butsugan, Y. New Physalins Possessing an Additional Carbon-Carbon Bond from Physalis-Alkekengi Var Francheti. Tetrahedron 1995, 51, 12529–12538. [Google Scholar] [CrossRef]
- Lima Mda, S.; Evangelista, A.F.; Santos, G.G.; Ribeiro, I.M.; Tomassini, T.C.; Pereira Soares, M.B.; Villarreal, C.F. Antinociceptive properties of physalins from Physalis angulata. J. Nat. Prod. 2014, 77, 2397–2403. [Google Scholar] [CrossRef]
- Wang, A.; Wang, S.; Zhou, F.; Li, P.; Wang, Y.; Gan, L.; Lin, L. Physalin B induces cell cycle arrest and triggers apoptosis in breast cancer cells through modulating p53-dependent apoptotic pathway. Biomed. Pharmacother. 2018, 101, 334–341. [Google Scholar] [CrossRef]
- Ding, N.; Lu, Y.; Cui, H.; Ma, Q.; Qiu, D.; Wei, X.; Dou, C.; Cao, N. Physalin D inhibits RANKL-induced osteoclastogenesis and bone loss via regulating calcium signaling. BMB Rep. 2020, 53, 154–159. [Google Scholar] [CrossRef]
- Pal, S.; Rastogi, S.; Nagegowda, D.A.; Gupta, M.M.; Shasany, A.K.; Chanotiya, C.S. RNAi of Sterol Methyl Transferase1 Reveals its Direct Role in Diverting Intermediates Towards Withanolide/Phytosterol Biosynthesis in Withania somnifera. Plant Cell Physiol. 2019, 60, 672–686. [Google Scholar] [CrossRef]
- Sonawane, P.D.; Pollier, J.; Panda, S.; Szymanski, J.; Massalha, H.; Yona, M.; Unger, T.; Malitsky, S.; Arendt, P.; Pauwels, L.; et al. Plant cholesterol biosynthetic pathway overlaps with phytosterol metabolism. Nat. Plants 2016, 3, 16205. [Google Scholar] [CrossRef]
- Knoch, E.; Sugawara, S.; Mori, T.; Poulsen, C.; Fukushima, A.; Harholt, J.; Fujimoto, Y.; Umemoto, N.; Saito, K. Third DWF1 paralog in Solanaceae, sterol Delta(24)-isomerase, branches withanolide biosynthesis from the general phytosterol pathway. Proc. Natl. Acad. Sci. USA 2018, 115, E8096–E8103. [Google Scholar] [CrossRef]
- Yang, J.; Tian, J.; Yang, Y.; Zhu, Y.; Li, C.; Zhang, Y. RNAi of Sterol Delta24-Isomerase Implicated Its Involvement in Physalin Biosynthesis in Physalis angulata L. Front. Plant Sci. 2022, 13, 850711. [Google Scholar] [CrossRef]
- Yang, J. Isolation and Functional Analysis of Key Enzymes Involved in the Biosynthesis of the Anti-Cancer Compound Physalins; Shanghai University: Shanghai, China, 2022. [Google Scholar]
- Rasool, S.; Mohamed, R. Plant cytochrome P450s: Nomenclature and involvement in natural product biosynthesis. Protoplasma 2016, 253, 1197–1209. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Xu, Q.; Chen, C.; Song, C.; Xu, Y.; Xiang, Y.; Feng, Y.; Ouyang, H.; Zhang, Y.; Jiang, H. The rapid discovery and identification of physalins in the calyx of Physalis alkekengi L. var. franchetii (Mast.) Makino using ultra-high performance liquid chromatography-quadrupole time of flight tandem mass spectrometry together with a novel three-step data mining strategy. J. Chromatogr. A 2014, 1361, 139–152. [Google Scholar] [CrossRef] [PubMed]
- Huang, C. The Use of UPLC-MS/MS in Rapid Characterization of Physalins in Ph. alkekengi L. var. franchetii and in Quality Evaluation of the Herb; Huazhong University of Science and Technology: Wuhan, China, 2014. [Google Scholar]
- He, H.; Zang, L.H.; Feng, Y.S.; Wang, J.; Liu, W.W.; Chen, L.X.; Kang, N.; Tashiro, S.; Onodera, S.; Qiu, F.; et al. Physalin A induces apoptotic cell death and protective autophagy in HT1080 human fibrosarcoma cells. J. Nat. Prod. 2013, 76, 880–888. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Yang, Y.; Tian, J.; Wu, Y.; An, F.; Li, C.; Zhang, Y. 22R- but not 22S-hydroxycholesterol is recruited for diosgenin biosynthesis. Plant J. Cell Mol. Biol. 2022, 109, 940–951. [Google Scholar] [CrossRef]
- Christ, B.; Xu, C.; Xu, M.; Li, F.S.; Wada, N.; Mitchell, A.J.; Han, X.L.; Wen, M.L.; Fujita, M.; Weng, J.K. Repeated evolution of cytochrome P450-mediated spiroketal steroid biosynthesis in plants. Nat. Commun. 2019, 10, 3206. [Google Scholar] [CrossRef]
- Ohnishi, T.; Nomura, T.; Watanabe, B.; Ohta, D.; Yokota, T.; Miyagawa, H.; Sakata, K.; Mizutani, M. Tomato cytochrome P450 CYP734A7 functions in brassinosteroid catabolism. Phytochemistry 2006, 67, 1895–1906. [Google Scholar] [CrossRef]
- Sakamoto, T.; Kawabe, A.; Tokida-Segawa, A.; Shimizu, B.; Takatsuto, S.; Shimada, Y.; Fujioka, S.; Mizutani, M. Rice CYP734As function as multisubstrate and multifunctional enzymes in brassinosteroid catabolism. Plant J. 2011, 67, 1–12. [Google Scholar] [CrossRef]
- Fang, S.T.; Liu, X.; Kong, N.N.; Liu, S.J.; Xia, C.H. Two new withanolides from the halophyte Datura stramonium L. Nat. Prod. Res. 2013, 27, 1965–1970. [Google Scholar] [CrossRef]
- Yang, Z.; Zhang, C.; Yang, X.; Liu, K.; Wu, Z.; Zhang, X.; Zheng, W.; Xun, Q.; Liu, C.; Lu, L.; et al. PAG1, a cotton brassinosteroid catabolism gene, modulates fiber elongation. New Phytol. 2014, 203, 437–448. [Google Scholar] [CrossRef]
- Bancos, S.; Nomura, T.; Sato, T.; Molnar, G.; Bishop, G.J.; Koncz, C.; Yokota, T.; Nagy, F.; Szekeres, M. Regulation of transcript levels of the Arabidopsis cytochrome p450 genes involved in brassinosteroid biosynthesis. Plant Physiol. 2002, 130, 504–513. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Bubner, B.; Baldwin, I.T. Use of real-time PCR for determining copy number and zygosity in transgenic plants. Plant Cell Rep. 2004, 23, 263–271. [Google Scholar] [CrossRef]
- Armstrong, R.A.; Eperjesi, F.; Gilmartin, B. The application of analysis of variance (ANOVA) to different experimental designs in optometry. Ophthalmic Physiol. Opt. J. Br. Coll. Ophthalmic Opt. 2002, 22, 248–256. [Google Scholar] [CrossRef] [PubMed]
- Thornton, L.E.; Peng, H.; Neff, M.M. Rice CYP734A cytochrome P450s inactivate brassinosteroids in Arabidopsis. Planta 2011, 234, 1151–1162. [Google Scholar] [CrossRef]
- Yang, Y.; Li, W.; Pang, J.; Jiang, L.; Qu, X.; Pu, X.; Zhang, G.; Luo, Y. Correction to Bifunctional Cytochrome P450 Enzymes Involved in Camptothecin Biosynthesis. ACS Chem. Biol. 2021, 16, 1298. [Google Scholar] [CrossRef]
- Salim, V.; Yu, F.; Altarejos, J.; De Luca, V. Virus-induced gene silencing identifies Catharanthus roseus 7-deoxyloganic acid-7-hydroxylase, a step in iridoid and monoterpene indole alkaloid biosynthesis. Plant J. 2013, 76, 754–765. [Google Scholar] [CrossRef]
- Takemura, T.; Ikezawa, N.; Iwasa, K.; Sato, F. Molecular cloning and characterization of a cytochrome P450 in sanguinarine biosynthesis from Eschscholzia californica cells. Phytochemistry 2013, 91, 100–108. [Google Scholar] [CrossRef]
- Kim, T.W.; Hwang, J.Y.; Kim, Y.S.; Joo, S.H.; Chang, S.C.; Lee, J.S.; Takatsuto, S.; Kim, S.K. Arabidopsis CYP85A2, a cytochrome P450, mediates the Baeyer-Villiger oxidation of castasterone to brassinolide in brassinosteroid biosynthesis. Plant Cell 2005, 17, 2397–2412. [Google Scholar] [CrossRef]
- Joo, S.H.; Kim, T.W.; Son, S.H.; Lee, W.S.; Yokota, T.; Kim, S.K. Biosynthesis of a cholesterol-derived brassinosteroid, 28-norcastasterone, in Arabidopsis thaliana. J. Exp. Bot. 2012, 63, 1823–1833. [Google Scholar] [CrossRef]
- Jin, Y.L.; Tang, R.J.; Wang, H.H.; Jiang, C.M.; Bao, Y.; Yang, Y.; Liang, M.X.; Sun, Z.C.; Kong, F.J.; Li, B.; et al. Overexpression of Populus trichocarpa CYP85A3 promotes growth and biomass production in transgenic trees. Plant Biotechnol. J. 2017, 15, 1309–1321. [Google Scholar] [CrossRef] [PubMed]
- Tohge, T.; Fernie, A.R. Co-Regulation of Clustered and Neo-Functionalized Genes in Plant-Specialized Metabolism. Plants 2020, 9, 622. [Google Scholar] [CrossRef] [PubMed]
- Yan, B.; Zhang, Y.; Li, J.; Fang, J.; Liu, T.; Dong, L. Transcriptome profiling to identify cytochrome P450 genes involved in penoxsulam resistance in Echinochloa glabrescens. Pestic. Biochem. Physiol. 2019, 158, 112–120. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Hou, L.; Li, S.; Li, J. The Functional Characterization of DzCYP72A12-4 Related to Diosgenin Biosynthesis and Drought Adaptability in Dioscorea zingiberensis. Int. J. Mol. Sci. 2023, 24, 8430. [Google Scholar] [CrossRef]
- Geisler, K.; Jensen, N.B.; Yuen, M.M.; Madilao, L.; Bohlmann, J. Modularity of Conifer Diterpene Resin Acid Biosynthesis: P450 Enzymes of Different CYP720B Clades Use Alternative Substrates and Converge on the Same Products. Plant Physiol. 2016, 171, 152–164. [Google Scholar] [CrossRef]
- Hamberger, B.; Bohlmann, J. Cytochrome P450 mono-oxygenases in conifer genomes: Discovery of members of the terpenoid oxygenase superfamily in spruce and pine. Biochem. Soc. Trans. 2006, 34, 1209–1214. [Google Scholar] [CrossRef]
- Zhang, Y.N.; Ding, X.Y.; Luan, Q.F.; Jiang, J.M.; Diao, S. Identification and Tissue-Specific Expression Analysis of CYP720B Subfamily Genes in Slash Pine and Loblolly Pine. Forests 2022, 13, 283. [Google Scholar] [CrossRef]
- Gan, Q.Q.; Luan, M.B.; Hu, M.L.; Liu, Z.S.; Zhang, Z.Q. Functional study of CYP90A1 and ALDH3F1 gene obtained by transcriptome sequencing analysis of Brassica napus seedlings treated with brassinolide. Front. Plant Sci. 2022, 13, 1040511. [Google Scholar] [CrossRef]
- Ohnishi, T.; Szatmari, A.M.; Watanabe, B.; Fujita, S.; Bancos, S.; Koncz, C.; Lafos, M.; Shibata, K.; Yokota, T.; Sakata, K.; et al. C-23 hydroxylation by Arabidopsis CYP90C1 and CYP90D1 reveals a novel shortcut in brassinosteroid biosynthesis. Plant Cell 2006, 18, 3275–3288. [Google Scholar] [CrossRef]
- Sakamoto, T.; Ohnishi, T.; Fujioka, S.; Watanabe, B.; Mizutani, M. Rice CYP90D2 and CYP90D3 catalyze C-23 hydroxylation of brassinosteroids in vitro. Plant Physiol. Bioch. 2012, 58, 220–226. [Google Scholar] [CrossRef]
- Han, J.Y.; Hwang, H.S.; Choi, S.W.; Kim, H.J.; Choi, Y.E. Cytochrome P450 CYP716A53v2 catalyzes the formation of protopanaxatriol from protopanaxadiol during ginsenoside biosynthesis in Panax ginseng. Plant Cell Physiol. 2012, 53, 1535–1545. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.C.; Lu, Y.; Liu, Y.; Zhou, J.W.; Zhang, Y.F.; Gao, H.Y.; Li, D.; Gao, W. Identification of a cytochrome P450 from Tripterygium hypoglaucum (Levl.) Hutch that catalyzes polpunonic acid formation in celastrol biosynthesis. Chin. J. Nat. Med. 2022, 20, 691–700. [Google Scholar] [CrossRef] [PubMed]
- Morikawa, T.; Saga, H.; Hashizume, H.; Ohta, D. CYP710A genes encoding sterol C22-desaturase in Physcomitrella patens as molecular evidence for the evolutionary conservation of a sterol biosynthetic pathway in plants. Planta 2009, 229, 1311–1322. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, A.; Hamberger, B. P450s controlling metabolic bifurcations in plant terpene specialized metabolism. Phytochem. Rev. 2018, 17, 81–111. [Google Scholar] [CrossRef] [PubMed]
- Wei, K.F.; Chen, H.Q. Global identification, structural analysis and expression characterization of cytochrome P450 monooxygenase superfamily in rice. BMC Genom. 2018, 19, 35. [Google Scholar] [CrossRef]
- Schmelz, E.A.; Huffaker, A.; Sims, J.W.; Christensen, S.A.; Lu, X.; Okada, K.; Peters, R.J. Biosynthesis, elicitation and roles of monocot terpenoid phytoalexins. Plant J. 2014, 79, 659–678. [Google Scholar] [CrossRef]
- Yamada, Y.; Motomura, Y.; Sato, F. CjbHLH1 homologs regulate sanguinarine biosynthesis in Eschscholzia californica cells. Plant Cell Physiol. 2015, 56, 1019–1030. [Google Scholar] [CrossRef]
- Yamada, Y.; Shimada, T.; Motomura, Y.; Sato, F. Modulation of benzylisoquinoline alkaloid biosynthesis by heterologous expression of CjWRKY1 in Eschscholzia californica cells. PLoS ONE 2017, 12, 186953. [Google Scholar] [CrossRef]
- Vasav, A.P.; Godbole, R.C.; Darshetkar, A.M.; Pable, A.A.; Barvkar, V.T. Functional genomics-enabled characterization of CYP81B140 and CYP81B141 from Plumbago zeylanica L. substantiates their involvement in plumbagin biosynthesis. Planta 2022, 256, 102. [Google Scholar] [CrossRef]
- Liu, C.J.; Huhman, D.; Sumner, L.W.; Dixon, R.A. Regiospecific hydroxylation of isoflavones by cytochrome P450 81E enzymes from Medicago truncatula. Plant J. 2003, 36, 471–484. [Google Scholar] [CrossRef]
- Akashi, T.; Aoki, T.; Ayabe, S. CYP81E1, a cytochrome P450 cDNA of licorice (Glycyrrhiza echinata L.), encodes isoflavone 2′-hydroxylase. Biochem. Biophys. Res. Commun. 1998, 251, 67–70. [Google Scholar] [CrossRef] [PubMed]
- Overkamp, S.; Hein, F.; Barz, W. Cloning and characterization of eight cytochrome P450 cDNAs from chickpea (Cicer arietinum L.) cell suspension cultures. Plant Sci. 2000, 155, 101–108. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hua, C.; Xu, Z.; Tang, N.; Xu, Y.; Zhang, Y.; Li, C. Identification of P450 Candidates Associated with the Biosynthesis of Physalin-Class Compounds in Physalis angulata. Int. J. Mol. Sci. 2023, 24, 14077. https://doi.org/10.3390/ijms241814077
Hua C, Xu Z, Tang N, Xu Y, Zhang Y, Li C. Identification of P450 Candidates Associated with the Biosynthesis of Physalin-Class Compounds in Physalis angulata. International Journal of Molecular Sciences. 2023; 24(18):14077. https://doi.org/10.3390/ijms241814077
Chicago/Turabian StyleHua, Congkun, Zhengqin Xu, Nan Tang, Yehan Xu, Yansheng Zhang, and Changfu Li. 2023. "Identification of P450 Candidates Associated with the Biosynthesis of Physalin-Class Compounds in Physalis angulata" International Journal of Molecular Sciences 24, no. 18: 14077. https://doi.org/10.3390/ijms241814077
APA StyleHua, C., Xu, Z., Tang, N., Xu, Y., Zhang, Y., & Li, C. (2023). Identification of P450 Candidates Associated with the Biosynthesis of Physalin-Class Compounds in Physalis angulata. International Journal of Molecular Sciences, 24(18), 14077. https://doi.org/10.3390/ijms241814077





