Short Survey on the Protein Modifications in Plasma during SARS-CoV-2 Infection
Abstract
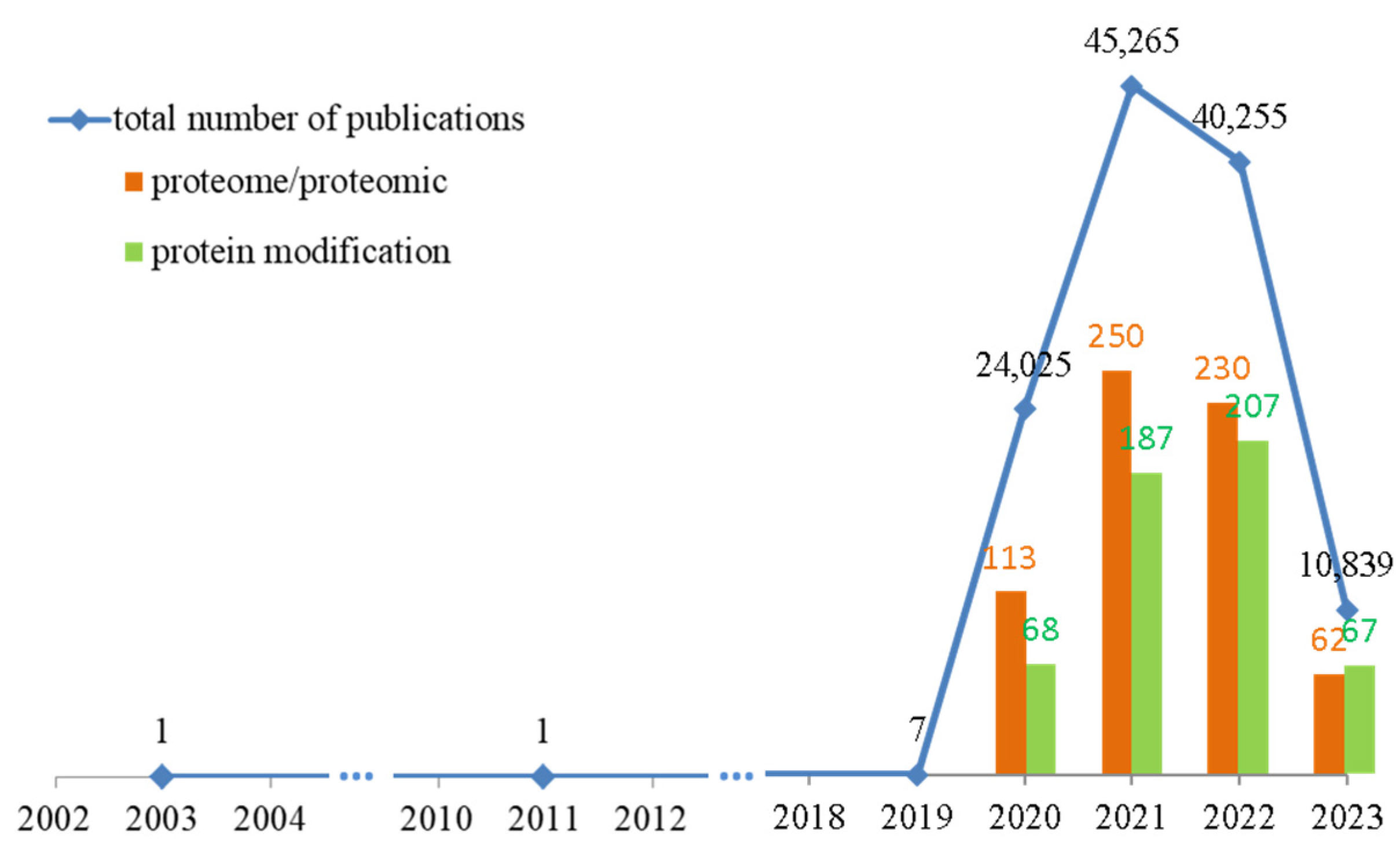
:1. Introduction
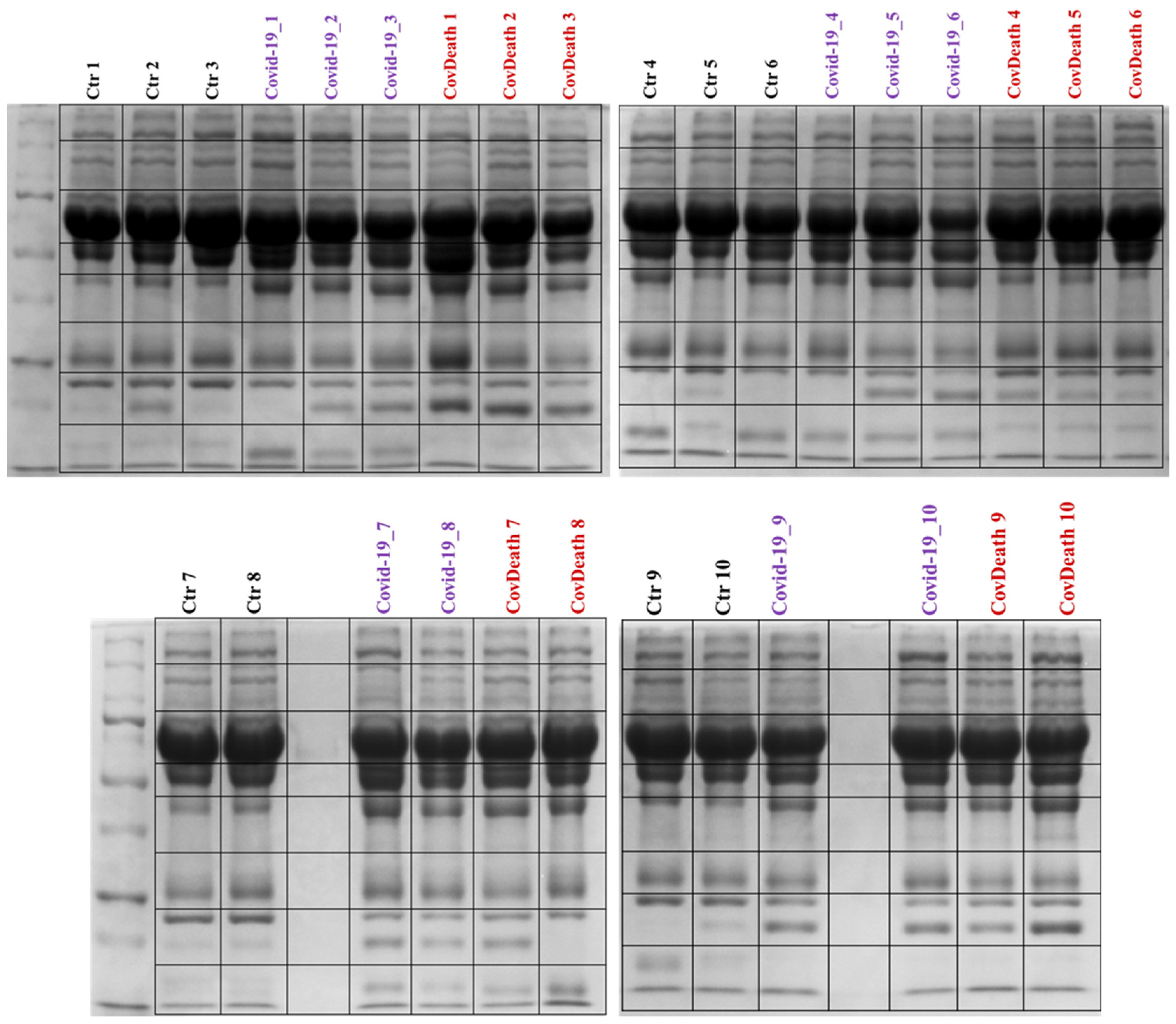
2. Results
3. Discussion
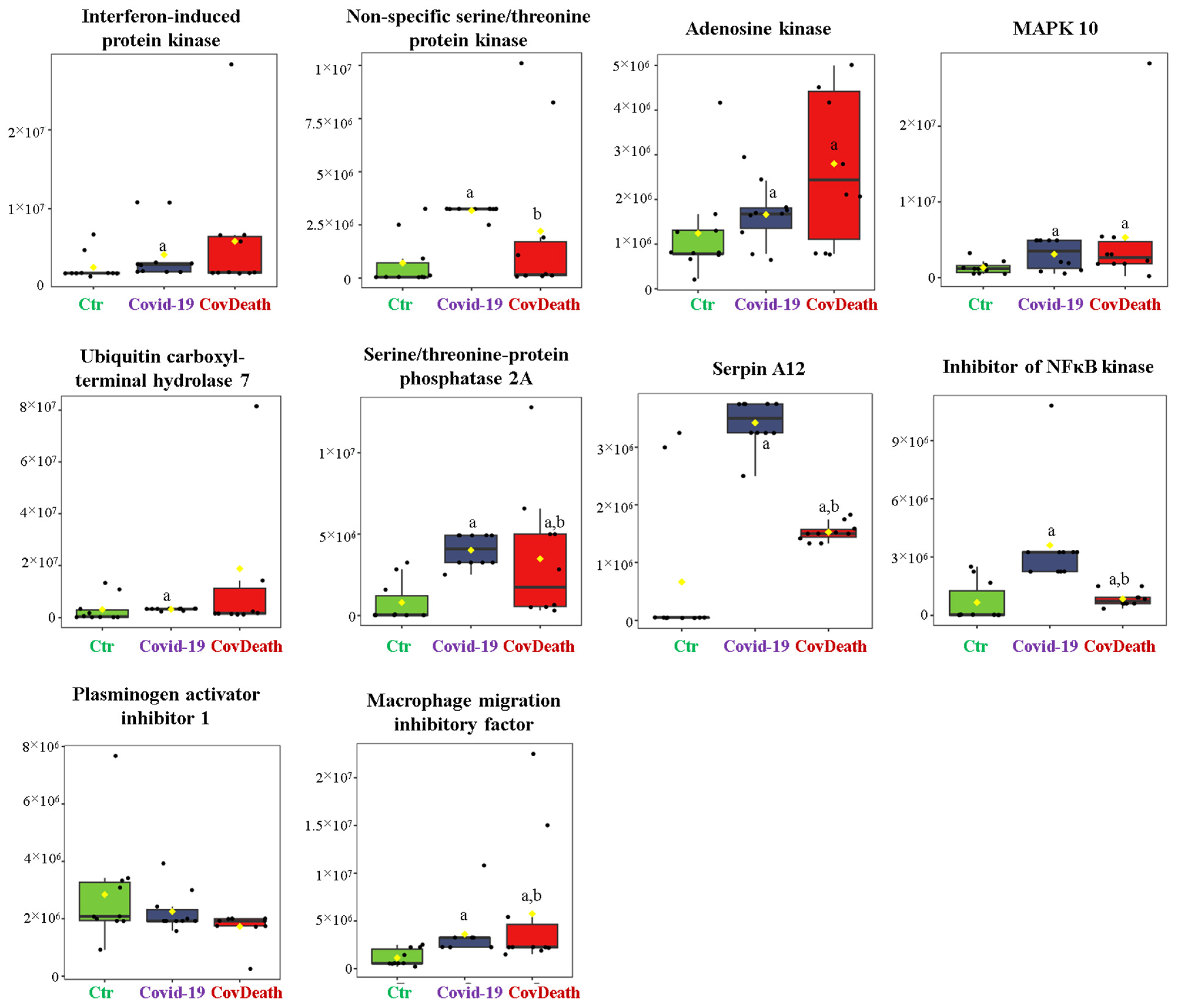
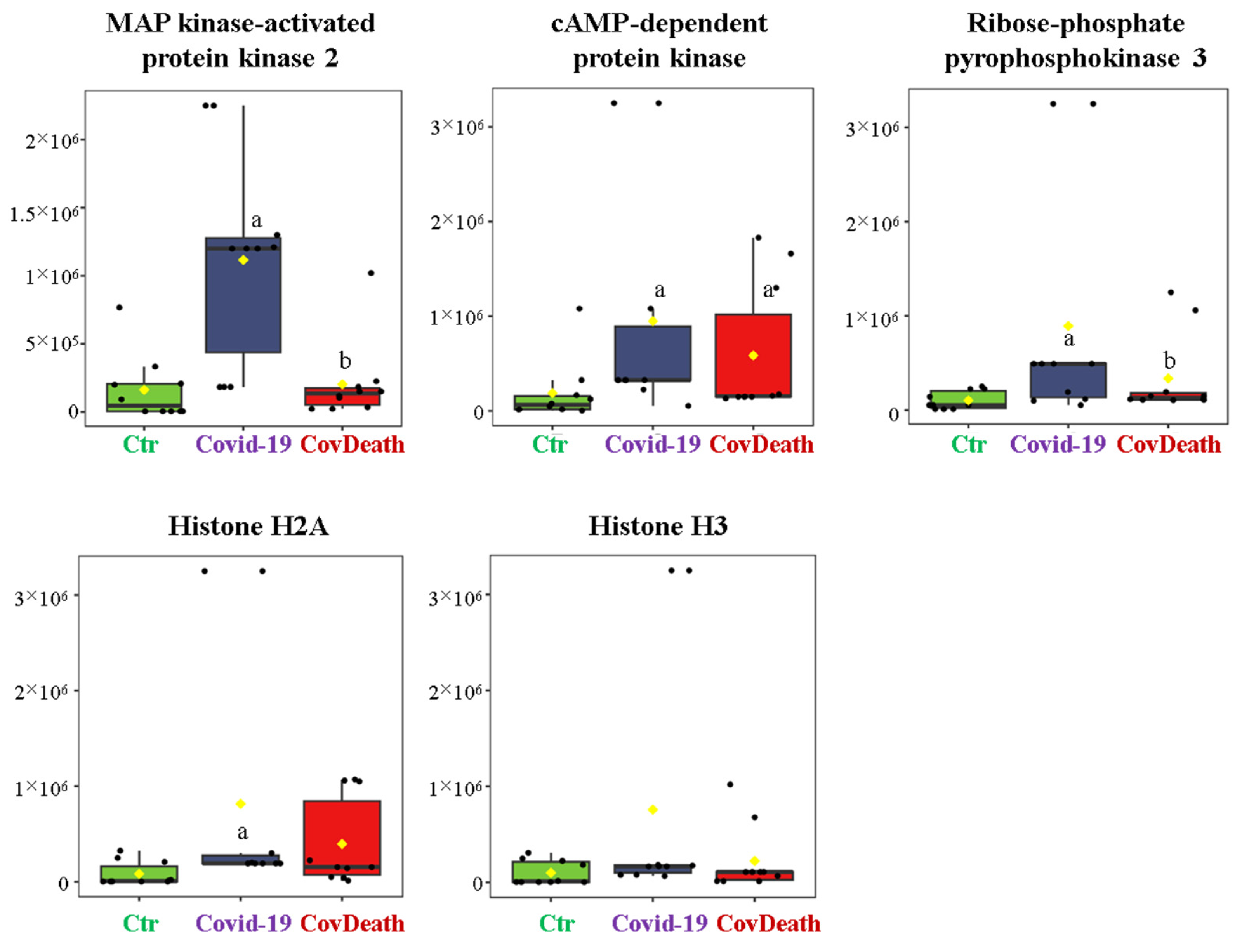
3.1. COVID-19-Induced Changes in Protein Profile
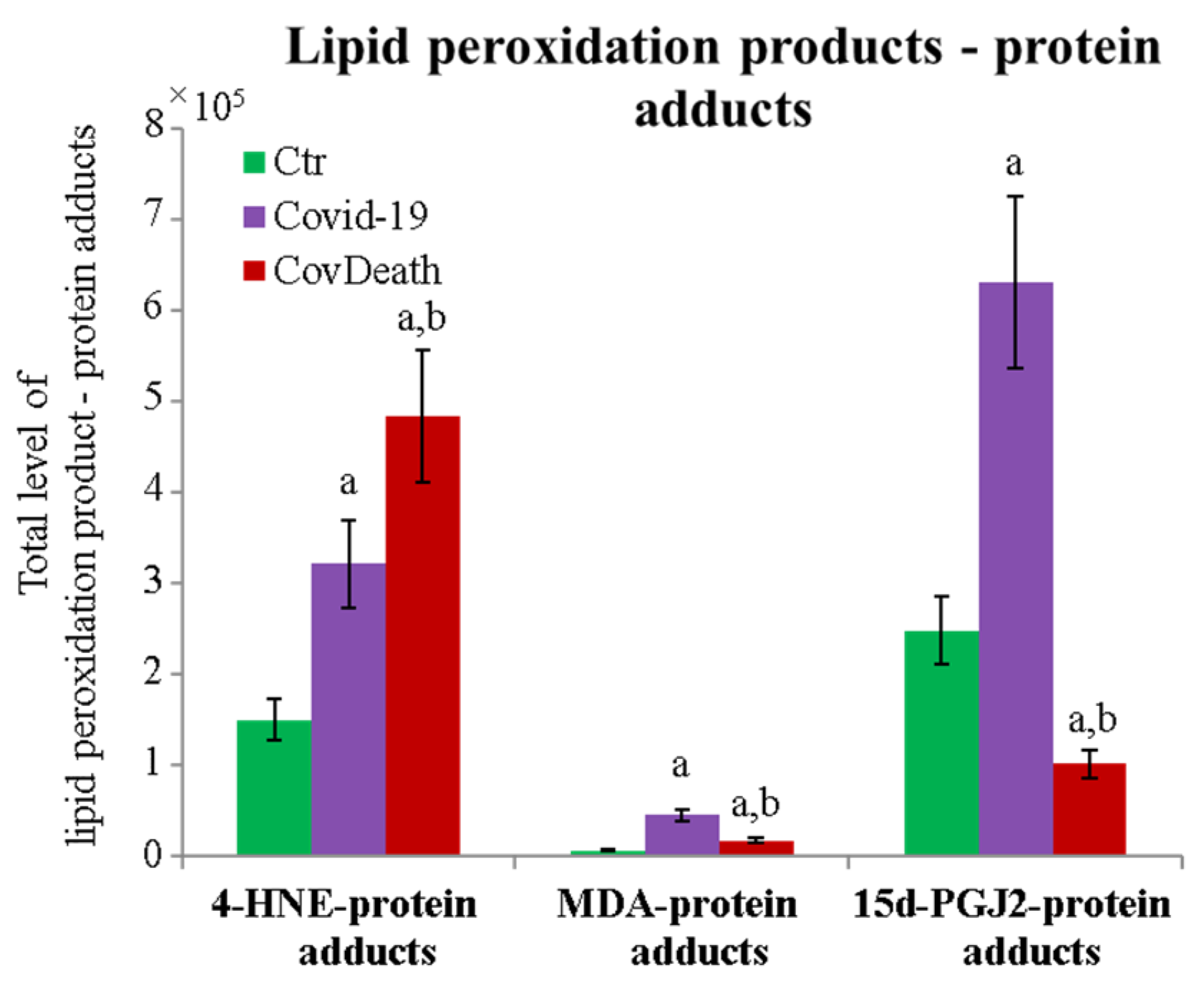
3.2. Protein Modifications by Reactive Aldehydes in COVID-19 Patients
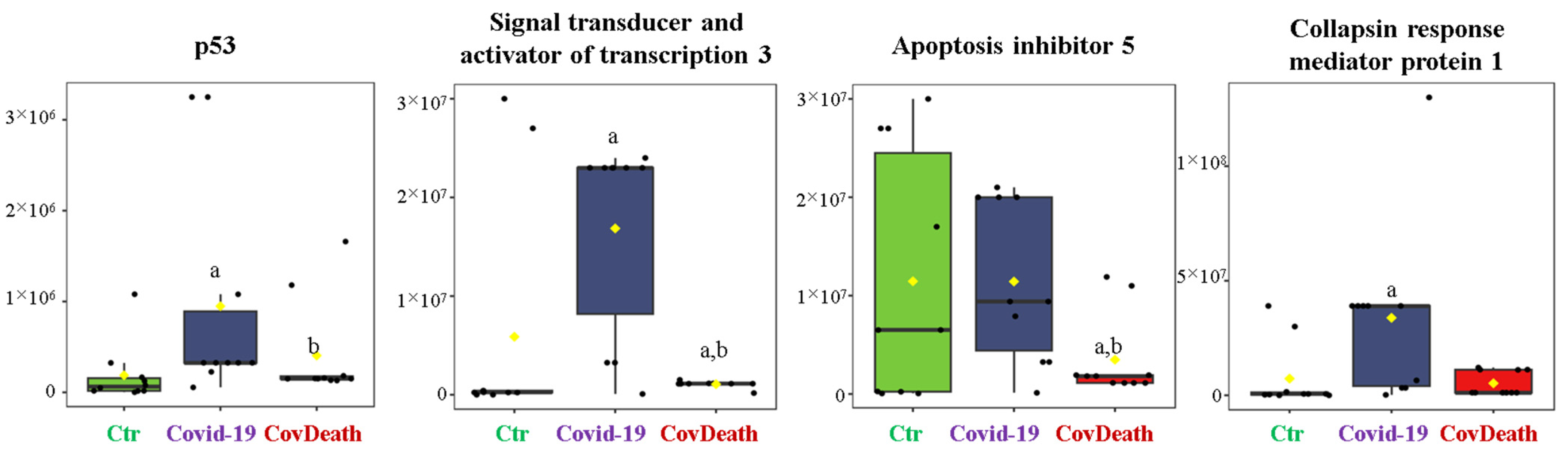
3.3. Formation of Prostaglandin–Protein Adducts in Plasma of COVID-19 Patients
3.4. Limitations
4. Materials and Methods
4.1. Samples Collection
4.2. Protein Separation and Digestion
4.3. Proteomic Analysis and Protein Identification
4.4. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- WHO. Coronavirus (COVID-19) Dashboard. Available online: https://covid19.who.int (accessed on 26 June 2023).
- Evolution of CT Manifestations in a Patient Recovered from 2019 Novel Coronavirus (2019-nCoV) Pneumonia in Wuhan, China. Radiology. Available online: https://pubs.rsna.org/doi/full/10.1148/radiol.2020200269 (accessed on 26 June 2023).
- Web of Science Core Collection. Available online: https://www-1webofscience-1com-1bcnm7wly0ab5.han.umb.edu.pl/wos/woscc/basic-search (accessed on 26 June 2023).
- Žarković, N.; Jastrząb, A.; Jarocka-Karpowicz, I.; Orehovec, B.; Baršić, B.; Tarle, M.; Kmet, M.; Lukšić, I.; Łuczaj, W.; Skrzydlewska, E. The Impact of Severe COVID-19 on Plasma Antioxidants. Molecules 2022, 27, 5323. [Google Scholar] [CrossRef] [PubMed]
- Žarković, N.; Łuczaj, W.; Jarocka-Karpowicz, I.; Orehovec, B.; Baršić, B.; Tarle, M.; Kmet, M.; Lukšić, I.; Biernacki, M.; Skrzydlewska, E. Diversified Effects of COVID-19 as a Consequence of the Differential Metabolism of Phospholipids and Lipid Peroxidation Evaluated in the Plasma of Survivors and Deceased Patients upon Admission to the Hospital. Int. J. Mol. Sci. 2022, 23, 11810. [Google Scholar] [CrossRef] [PubMed]
- Žarković, N.; Orehovec, B.; Baršić, B.; Tarle, M.; Kmet, M.; Lukšić, I.; Tatzber, F.; Wonisch, W.; Skrzydlewska, E.; Łuczaj, W. Lipidomics Revealed Plasma Phospholipid Profile Differences between Deceased and Recovered COVID-19 Patients. Biomolecules 2022, 12, 1488. [Google Scholar] [CrossRef] [PubMed]
- Žarković, N.; Orehovec, B.; Milković, L.; Baršić, B.; Tatzber, F.; Wonisch, W.; Tarle, M.; Kmet, M.; Mataić, A.; Jakovčević, A.; et al. Preliminary Findings on the Association of the Lipid Peroxidation Product 4-Hydroxynonenal with the Lethal Outcome of Aggressive COVID-19. Antioxidants 2021, 10, 1341. [Google Scholar] [CrossRef] [PubMed]
- Negre-Salvayre, A.; Auge, N.; Ayala, V.; Basaga, H.; Boada, J.; Brenke, R.; Chapple, S.; Cohen, G.; Feher, J.; Grune, T.; et al. Pathological Aspects of Lipid Peroxidation. Free Radic. Res. 2010, 44, 1125–1171. [Google Scholar] [CrossRef] [PubMed]
- Yadav, U.C.S.; Ramana, K.V. Regulation of NF-kB-Induced Inflammatory Signaling by Lipid Peroxidation-Derived Aldehydes. Oxidative Med. Cell. Longev. 2013, 2013, e690545. [Google Scholar] [CrossRef] [PubMed]
- Urbiola-Salvador, V.; Lima de Souza, S.; Grešner, P.; Qureshi, T.; Chen, Z. Plasma Proteomics Unveil Novel Immune Signatures and Biomarkers upon SARS-CoV-2 Infection. Int. J. Mol. Sci. 2023, 24, 6276. [Google Scholar] [CrossRef]
- Zarkovic, N.; Jakovcevic, A.; Mataic, A.; Jaganjac, M.; Vukovic, T.; Waeg, G.; Zarkovic, K. Post-Mortem Findings of Inflammatory Cells and the Association of 4-Hydroxynonenal with Systemic Vascular and Oxidative Stress in Lethal COVID-19. Cells 2022, 11, 444. [Google Scholar] [CrossRef]
- Gęgotek, A.; Skrzydlewska, E. Biological Effect of Protein Modifications by Lipid Peroxidation Products. Chem. Phys. Lipids 2019, 221, 46–52. [Google Scholar] [CrossRef]
- Muralidharan, A.; Bauer, C.; Katafiasz, D.M.; Pham, D.; Oyewole, O.O.; Morwitzer, M.J.; Roy, E.; Bailey, K.L.; Reid, S.P.; Wyatt, T.A. Malondialdehyde Acetaldehyde Adduction of Surfactant Protein D Attenuates SARS-CoV-2 Spike Protein Binding and Virus Neutralization. Alcohol 2023, 47, 95–103. [Google Scholar] [CrossRef]
- Yang, L.; Liu, S.; Liu, J.; Zhang, Z.; Wan, X.; Huang, B.; Chen, Y.; Zhang, Y. COVID-19: Immunopathogenesis and Immunotherapeutics. Signal Transduct. Target. Ther. 2020, 5, 128. [Google Scholar] [CrossRef] [PubMed]
- Apostolo, D.; D’Onghia, D.; Tonello, S.; Minisini, R.; Baricich, A.; Gramaglia, C.; Patrucco, F.; Zeppegno, P.; Acquaviva, A.; Balbo, P.E.; et al. Decreased Gas6 and sAxl Plasma Levels Are Associated with Hair Loss in COVID-19 Survivors. Int. J. Mol. Sci. 2023, 24, 6257. [Google Scholar] [CrossRef] [PubMed]
- Yuki, K.; Fujiogi, M.; Koutsogiannaki, S. COVID-19 Pathophysiology: A Review. Clin. Immunol. 2020, 215, 108427. [Google Scholar] [CrossRef] [PubMed]
- Bhargavan, B.; Kanmogne, G.D. SARS-CoV-2 Spike Proteins and Cell–Cell Communication Induce P-Selectin and Markers of Endothelial Injury, NETosis, and Inflammation in Human Lung Microvascular Endothelial Cells and Neutrophils: Implications for the Pathogenesis of COVID-19 Coagulopathy. Int. J. Mol. Sci. 2023, 24, 12585. [Google Scholar] [CrossRef]
- Bergantini, L.; d’Alessandro, M.; Gangi, S.; Bianchi, F.; Cameli, P.; Perea, B.; Meocci, M.; Fabbri, G.; Marrucci, S.; Ederbali, M.; et al. Predictive Role of Cytokine and Adipokine Panel in Hospitalized COVID-19 Patients: Evaluation of Disease Severity, Survival and Lung Sequelae. Int. J. Mol. Sci. 2023, 24, 12994. [Google Scholar] [CrossRef]
- Salton, F.; Confalonieri, P.; Campisciano, G.; Cifaldi, R.; Rizzardi, C.; Generali, D.; Pozzan, R.; Tavano, S.; Bozzi, C.; Lapadula, G.; et al. Cytokine Profiles as Potential Prognostic and Therapeutic Markers in SARS-CoV-2-Induced ARDS. J. Clin. Med. 2022, 11, 2951. [Google Scholar] [CrossRef]
- Arthur, L.; Esaulova, E.; Mogilenko, D.A.; Tsurinov, P.; Burdess, S.; Laha, A.; Presti, R.; Goetz, B.; Watson, M.A.; Goss, C.W.; et al. Cellular and Plasma Proteomic Determinants of COVID-19 and Non-COVID-19 Pulmonary Diseases Relative to Healthy Aging. Nat. Aging 2021, 1, 535–549. [Google Scholar] [CrossRef]
- Wang, L.; Western, D.; Timsina, J.; Repaci, C.; Song, W.-M.; Norton, J.; Kohlfeld, P.; Budde, J.; Climer, S.; Butt, O.H.; et al. Plasma Proteomics of SARS-CoV-2 Infection and Severity Reveals Impact on Alzheimer and Coronary Disease Pathways. medRxiv 2022. [Google Scholar] [CrossRef]
- Li, H.; Li, X.; Wu, Q.; Wang, X.; Qin, Z.; Wang, Y.; He, Y.; Wu, Q.; Li, L.; Chen, H. Plasma Proteomic and Metabolomic Characterization of COVID-19 Survivors 6 Months after Discharge. Cell Death Dis. 2022, 13, 235. [Google Scholar] [CrossRef]
- Shu, T.; Ning, W.; Wu, D.; Xu, J.; Han, Q.; Huang, M.; Zou, X.; Yang, Q.; Yuan, Y.; Bie, Y.; et al. Plasma Proteomics Identify Biomarkers and Pathogenesis of COVID-19. Immunity 2020, 53, 1108–1122.e5. [Google Scholar] [CrossRef]
- Chen, D.; Zheng, Q.; Sun, L.; Ji, M.; Li, Y.; Deng, H.; Zhang, H. ORF3a of SARS-CoV-2 Promotes Lysosomal Exocytosis-Mediated Viral Egress. Dev. Cell 2021, 56, 3250–3263.e5. [Google Scholar] [CrossRef] [PubMed]
- Sciaudone, A.; Corkrey, H.; Humphries, F.; Koupenova, M. Platelets and SARS-CoV-2 During COVID-19: Immunity, Thrombosis, and Beyond. Circ. Res. 2023, 132, 1272–1289. [Google Scholar] [CrossRef] [PubMed]
- Fitzgerald, J.R.; Foster, T.J.; Cox, D. The Interaction of Bacterial Pathogens with Platelets. Nat. Rev. Microbiol. 2006, 4, 445–457. [Google Scholar] [CrossRef] [PubMed]
- Cox, D.; Kerrigan, S.W.; Watson, S.P. Platelets and the Innate Immune System: Mechanisms of Bacterial-induced Platelet Activation. J. Thromb. Haemost. 2011, 9, 1097–1107. [Google Scholar] [CrossRef]
- Fard, M.B.; Fard, S.B.; Ramazi, S.; Atashi, A.; Eslamifar, Z. Thrombosis in COVID-19 Infection: Role of Platelet Activation-Mediated Immunity. Thromb. J. 2021, 19, 59. [Google Scholar] [CrossRef]
- Zaid, Y.; Puhm, F.; Allaeys, I.; Naya, A.; Oudghiri, M.; Khalki, L.; Limami, Y.; Zaid, N.; Sadki, K.; Ben El Haj, R.; et al. Platelets Can Associate With SARS-CoV-2 RNA and Are Hyperactivated in COVID-19. Circ. Res. 2020, 127, 1404–1418. [Google Scholar] [CrossRef]
- Jevtic, S.D.; Nazy, I. The COVID Complex: A Review of Platelet Activation and Immune Complexes in COVID-19. Front. Immunol. 2022, 13, 807934. [Google Scholar] [CrossRef]
- Liu, L.; Meng, T.; Zheng, X.; Liu, Y.; Hao, R.; Yan, Y.; Chen, S.; You, H.; Xing, J.; Dong, Y. Transgelin 2 Promotes Paclitaxel Resistance, Migration, and Invasion of Breast Cancer by Directly Interacting with PTEN and Activating PI3K/Akt/GSK-3β Pathway. Mol. Cancer Ther. 2019, 18, 2457–2468. [Google Scholar] [CrossRef]
- Yin, L.-M.; Xu, Y.-D.; Peng, L.-L.; Duan, T.-T.; Liu, J.-Y.; Xu, Z.; Wang, W.-Q.; Guan, N.; Han, X.-J.; Li, H.-Y.; et al. Transgelin-2 as a Therapeutic Target for Asthmatic Pulmonary Resistance. Sci. Transl. Med. 2018, 10, eaam8604. [Google Scholar] [CrossRef]
- Moin, A.S.M.; Al-Qaissi, A.; Sathyapalan, T.; Atkin, S.L.; Butler, A.E. Mapping of Type 2 Diabetes Proteins to COVID-19 Biomarkers: A Proteomic Analysis. Metab. Open 2020, 9, 100074. [Google Scholar] [CrossRef]
- Woziwodzka, K.; Małyszko, J.; Koc-Żórawska, E.; Żórawski, M.; Dumnicka, P.; Jurczyszyn, A.; Batko, K.; Mazur, P.; Banaszkiewicz, M.; Krzanowski, M.; et al. Transgelin-2 in Multiple Myeloma: A New Marker of Renal Impairment? Molecules 2022, 27, 79. [Google Scholar] [CrossRef]
- Yin, L.-M.; Ulloa, L.; Yang, Y.-Q. Transgelin-2: Biochemical and Clinical Implications in Cancer and Asthma. Trends Biochem. Sci. 2019, 44, 885–896. [Google Scholar] [CrossRef] [PubMed]
- Sayre, L.M.; Moreira, P.I.; Smith, M.A.; Perry, G. Metal Ions and Oxidative Protein Modification in Neurological Disease. Ann. Ist. Super. Sanità 2005, 41, 143–164. [Google Scholar] [PubMed]
- Beal, M.F. Oxidatively Modified Proteins in Aging and Disease. Free Radic. Biol. Med. 2002, 32, 797–803. [Google Scholar] [CrossRef] [PubMed]
- Vedula, P.; Tang, H.-Y.; Speicher, D.W.; Kashina, A.; UPenn COVID Processing Unit. Protein Posttranslational Signatures Identified in COVID-19 Patient Plasma. Front. Cell Dev. Biol. 2022, 10, 807149. [Google Scholar] [CrossRef]
- Ayala, A.; Muñoz, M.F.; Argüelles, S. Lipid Peroxidation: Production, Metabolism, and Signaling Mechanisms of Malondialdehyde and 4-Hydroxy-2-Nonenal. Oxidative Med. Cell. Longev. 2014, 2014, 360438. [Google Scholar] [CrossRef]
- Jacobs, W.; Lammens, M.; Kerckhofs, A.; Voets, E.; Van San, E.; Van Coillie, S.; Peleman, C.; Mergeay, M.; Sirimsi, S.; Matheeussen, V.; et al. Fatal Lymphocytic Cardiac Damage in Coronavirus Disease 2019 (COVID-19): Autopsy Reveals a Ferroptosis Signature. ESC Heart Fail. 2020, 7, 3772–3781. [Google Scholar] [CrossRef]
- Chen, H.A.; Davids, J.; Zheng, D.; Bryant, M.; Bot, I.; J.C. van Berckel, T.; Biessen, E.; Pepine, C.; Ryman, K.; Progulski-Fox, A.; et al. The Serpin Solution; Targeting Thrombotic and Thrombolytic Serine Proteases in Inflammation. Cardiovasc. Haematol. Disord.—Drug Targets 2013, 13, 99–110. [Google Scholar] [CrossRef]
- Janciauskiene, S. Conformational Properties of Serine Proteinase Inhibitors (Serpins) Confer Multiple Pathophysiological Roles. Biochim. Biophys. Acta (BBA)—Mol. Basis Dis. 2001, 1535, 221–235. [Google Scholar] [CrossRef]
- Łuczaj, W.; Gęgotek, A.; Skrzydlewska, E. Antioxidants and HNE in Redox Homeostasis. Free Radic. Biol. Med. 2017, 111, 87–101. [Google Scholar] [CrossRef]
- Grieb, G.; Merk, M.; Bernhagen, J.; Bucala, R. Macrophage Migration Inhibitory Factor (MIF): A Promising Biomarker. Drug News Perspect. 2010, 23, 257–264. [Google Scholar] [CrossRef] [PubMed]
- Shin, J.J.; Fan, W.; Par-Young, J.; Piecychna, M.; Leng, L.; Israni-Winger, K.; Qing, H.; Gu, J.; Zhao, H.; Schulz, W.L.; et al. MIF Is a Common Genetic Determinant of COVID-19 Symptomatic Infection and Severity. QJM Int. J. Med. 2023, 116, 205–212. [Google Scholar] [CrossRef] [PubMed]
- Hemmat, N.; Asadzadeh, Z.; Ahangar, N.K.; Alemohammad, H.; Najafzadeh, B.; Derakhshani, A.; Baghbanzadeh, A.; Baghi, H.B.; Javadrashid, D.; Najafi, S.; et al. The Roles of Signaling Pathways in SARS-CoV-2 Infection; Lessons Learned from SARS-CoV and MERS-CoV. Arch. Virol. 2021, 166, 675–696. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Tang, J.; Cao, J.; Liu, F.; Fu, M.; Xue, B.; Zhou, A.; Chen, S.; Liu, J.; Zhou, Y.; et al. SARS-CoV-2 Infection Activates CREB/CBP in Cellular Cyclic AMP-Dependent Pathways. J. Med. Virol. 2023, 95, e28383. [Google Scholar] [CrossRef]
- Ekaney, M.L.; Otto, G.P.; Sossdorf, M.; Sponholz, C.; Boehringer, M.; Loesche, W.; Rittirsch, D.; Wilharm, A.; Kurzai, O.; Bauer, M.; et al. Impact of Plasma Histones in Human Sepsis and Their Contribution to Cellular Injury and Inflammation. Crit. Care 2014, 18, 543. [Google Scholar] [CrossRef]
- de Vries, F.; Huckriede, J.; Wichapong, K.; Reutelingsperger, C.; Nicolaes, G.A.F. The Role of Extracellular Histones in COVID-19. J. Intern. Med. 2023, 293, 275–292. [Google Scholar] [CrossRef]
- Kreuz, S.; Fischle, W. Oxidative Stress Signaling to Chromatin in Health and Disease. Epigenomics 2016, 8, 843–862. [Google Scholar] [CrossRef]
- Ricciotti, E.; FitzGerald, G.A. Prostaglandins and Inflammation. Arterioscler. Thromb. Vasc. Biol. 2011, 31, 986–1000. [Google Scholar] [CrossRef]
- Figueiredo-Pereira, M.E.; Corwin, C.; Babich, J. Prostaglandin J2: A Potential Target for Halting Inflammation-Induced Neurodegeneration. Ann. N. Y. Acad. Sci. 2016, 1363, 125–137. [Google Scholar] [CrossRef]
- Shahzad, S.; Willcox, M. Immuno-Pathogenesis of nCOVID-19 and a Possible Host-Directed Therapy Including Anti-Inflammatory and Anti-Viral Prostaglandin (PG J2) for Effective Treatment and Reduction in the Death Toll. Med. Hypotheses 2020, 143, 110080. [Google Scholar] [CrossRef]
- Aggarwal, B.B.; Kunnumakkara, A.B.; Harikumar, K.B.; Gupta, S.R.; Tharakan, S.T.; Koca, C.; Dey, S.; Sung, B. Signal Transducer and Activator of Transcription-3, Inflammation, and Cancer. Ann. N. Y. Acad. Sci. 2009, 1171, 59–76. [Google Scholar] [CrossRef] [PubMed]
- Jin, S.; Levine, A.J. The P53 Functional Circuit. J. Cell Sci. 2001, 114, 4139–4140. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.-J.; Cho, N.-C.; Han, B.; Kim, K.; Hahn, Y.-I.; Kim, K.P.; Suh, Y.G.; Choi, B.Y.; Na, H.-K.; Surh, Y.-J. 15-Deoxy-Δ12,14-Prostaglandin J2 Binds and Inactivates STAT3 via Covalent Modification of Cysteine 259 in H-Ras-Transformed Human Breast Epithelial Cells. FEBS Lett. 2021, 595, 604–622. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.-H.; Kim, E.-H.; Na, H.-K.; Sun, Y.; Surh, Y.-J. 15-Deoxy-Δ12,14-Prostaglandin J2 Stabilizes, but Functionally Inactivates P53 by Binding to the Cysteine 277 Residue. Oncogene 2010, 29, 2560–2576. [Google Scholar] [CrossRef]
- Jafarzadeh, A.; Nemati, M.; Jafarzadeh, S. Contribution of STAT3 to the Pathogenesis of COVID-19. Microb. Pathog. 2021, 154, 104836. [Google Scholar] [CrossRef]
- Milani, D.; Caruso, L.; Zauli, E.; Al Owaifeer, A.M.; Secchiero, P.; Zauli, G.; Gemmati, D.; Tisato, V. P53/NF-kB Balance in SARS-CoV-2 Infection: From OMICs, Genomics and Pharmacogenomics Insights to Tailored Therapeutic Perspectives (COVIDomics). Front. Pharmacol. 2022, 13, 871583. [Google Scholar] [CrossRef]
- Levin, Y. The Role of Statistical Power Analysis in Quantitative Proteomics. Proteomics 2011, 11, 2565–2567. [Google Scholar] [CrossRef]
- Bradford, M.M. A Rapid and Sensitive Method for the Quantitation of Microgram Quantities of Protein Utilizing the Principle of Protein-Dye Binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Medzihradszky, K.F. In-Solution Digestion of Proteins for Mass Spectrometry. Methods Enzymol. 2005, 405, 50–65. [Google Scholar] [CrossRef]
- Gęgotek, A.; Domingues, P.; Wroński, A.; Wójcik, P.; Skrzydlewska, E. Proteomic Plasma Profile of Psoriatic Patients. J. Pharm. Biomed. Anal. 2018, 155, 185–193. [Google Scholar] [CrossRef]
- Müller, T.; Winter, D. Systematic Evaluation of Protein Reduction and Alkylation Reveals Massive Unspecific Side Effects by Iodine-Containing Reagents. Mol. Cell. Proteom. 2017, 16, 1173–1187. [Google Scholar] [CrossRef] [PubMed]
- Domingues, M.R.; Fedorova, M.; Domingues, P. Mass Spectrometry Detection of Protein Modification by Cross-Reaction with Lipid Peroxidation Products. React. Oxyg. Species Lipid Peroxidation Protein Oxid. 2015, 3, 61–86. [Google Scholar]
- Pang, Z.; Zhou, G.; Ewald, J.; Chang, L.; Hacariz, O.; Basu, N.; Xia, J. Using MetaboAnalyst 5.0 for LC–HRMS Spectra Processing, Multi-Omics Integration and Covariate Adjustment of Global Metabolomics Data. Nat. Protoc. 2022, 17, 1735–1761. [Google Scholar] [CrossRef] [PubMed]
- Välikangas, T.; Suomi, T.; Elo, L.L. A Systematic Evaluation of Normalization Methods in Quantitative Label-Free Proteomics. Brief. Bioinform. 2018, 19, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Hackstadt, A.J.; Hess, A.M. Filtering for Increased Power for Microarray Data Analysis. BMC Bioinform. 2009, 10, 11. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING V11: Protein-Protein Association Networks with Increased Coverage, Supporting Functional Discovery in Genome-Wide Experimental Datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gęgotek, A.; Zarkovic, N.; Orehovec, B.; Jaganjac, M.; Sunjic, S.B.; Skrzydlewska, E. Short Survey on the Protein Modifications in Plasma during SARS-CoV-2 Infection. Int. J. Mol. Sci. 2023, 24, 14109. https://doi.org/10.3390/ijms241814109
Gęgotek A, Zarkovic N, Orehovec B, Jaganjac M, Sunjic SB, Skrzydlewska E. Short Survey on the Protein Modifications in Plasma during SARS-CoV-2 Infection. International Journal of Molecular Sciences. 2023; 24(18):14109. https://doi.org/10.3390/ijms241814109
Chicago/Turabian StyleGęgotek, Agnieszka, Neven Zarkovic, Biserka Orehovec, Morana Jaganjac, Suzana Borovic Sunjic, and Elżbieta Skrzydlewska. 2023. "Short Survey on the Protein Modifications in Plasma during SARS-CoV-2 Infection" International Journal of Molecular Sciences 24, no. 18: 14109. https://doi.org/10.3390/ijms241814109
APA StyleGęgotek, A., Zarkovic, N., Orehovec, B., Jaganjac, M., Sunjic, S. B., & Skrzydlewska, E. (2023). Short Survey on the Protein Modifications in Plasma during SARS-CoV-2 Infection. International Journal of Molecular Sciences, 24(18), 14109. https://doi.org/10.3390/ijms241814109






