The Aβ42 Peptide and IAPP Physically Interact in a Yeast-Based Assay
Abstract
:1. Introduction
2. Results
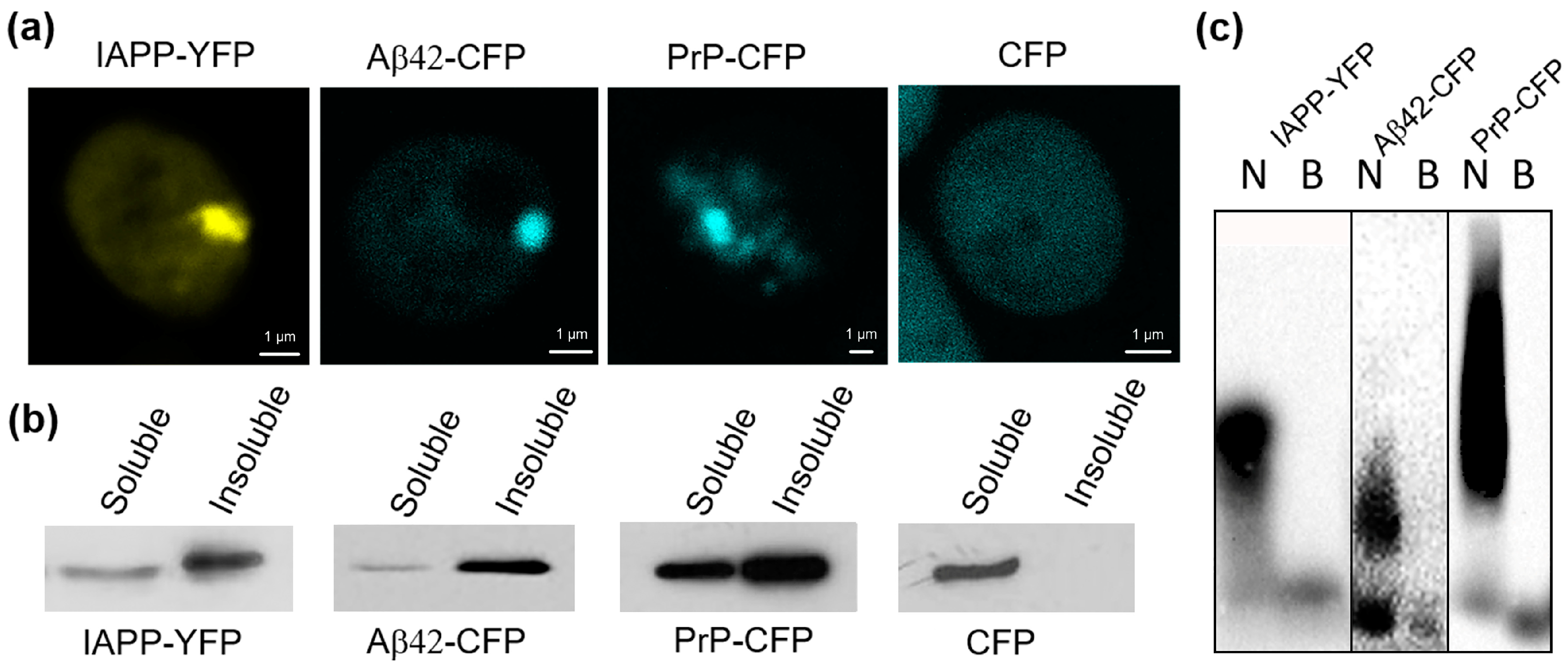
2.1. Human IAPP Fused with YFP Form Amyloid-like Oligomers in Yeast
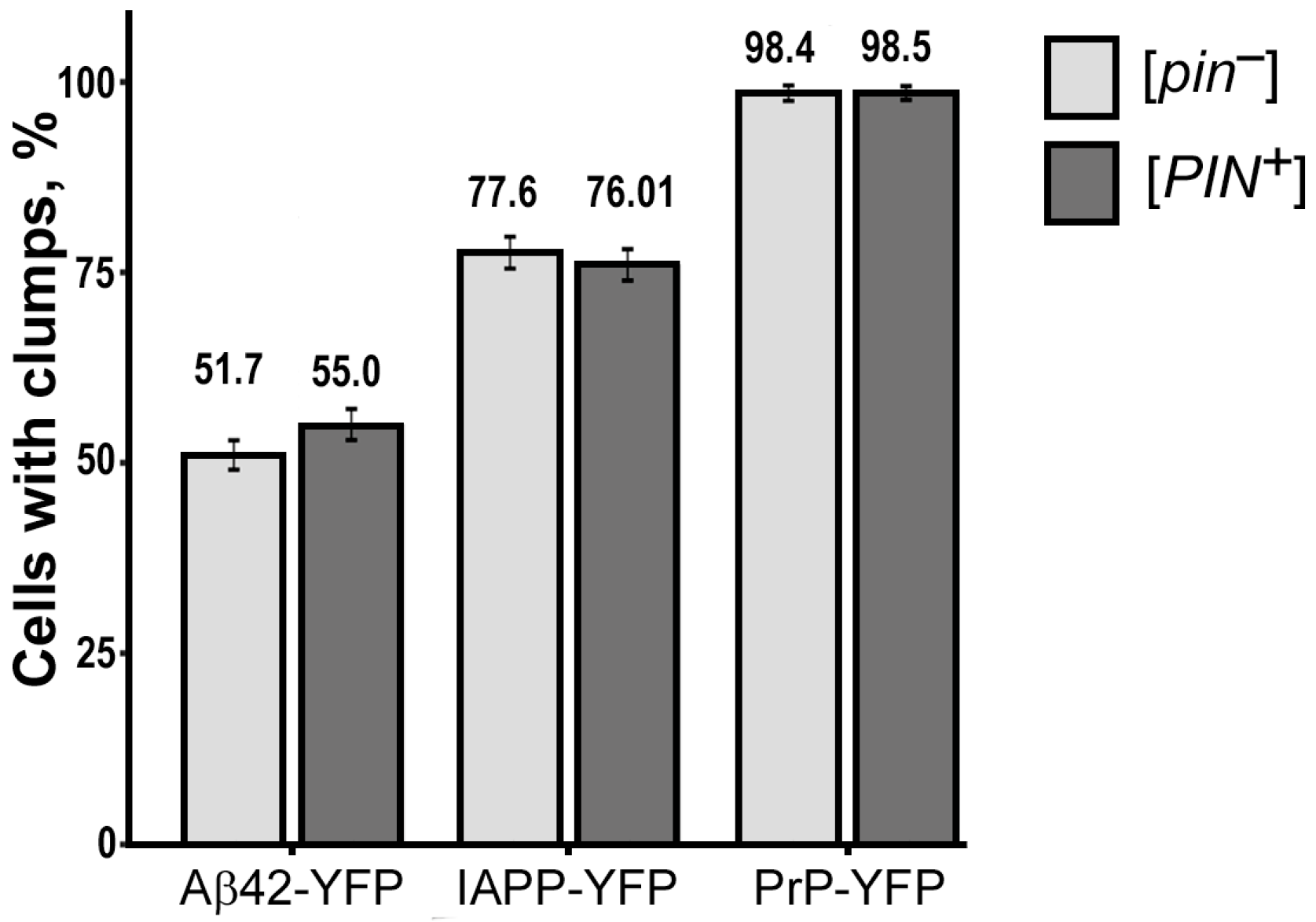
2.2. The [PIN+] Factor Does Not Affect the Aggregation of Heterologous Proteins PrP-CFP, Aβ42-CFP, and IAPP-YFP in the Yeast S. cerevisiae
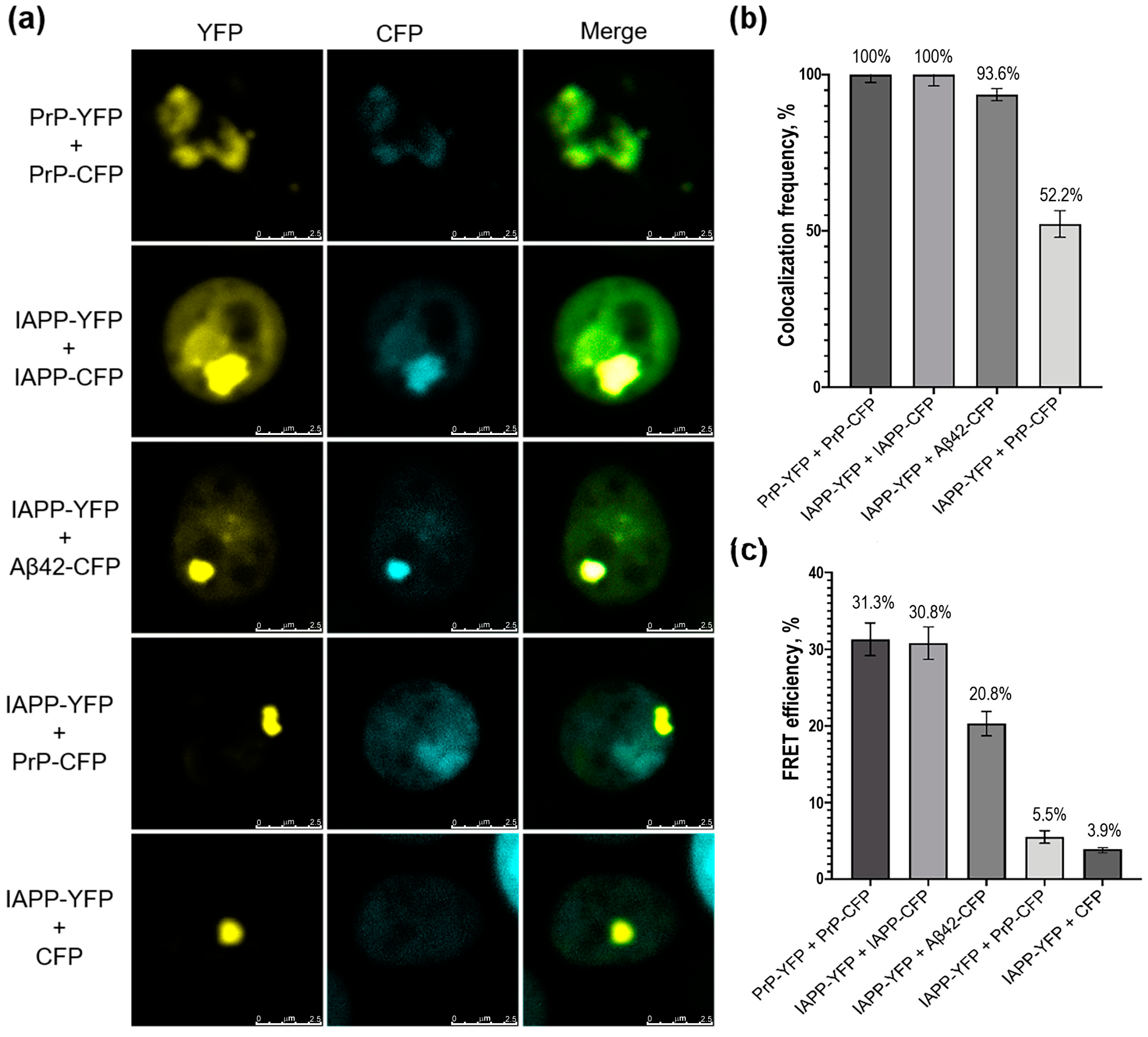
2.3. Human IAPP Colocalizes and Physically Interacts with Aβ42 in Yeast Cells
3. Discussion
4. Materials and Methods
4.1. Plasmids, Strains, Media, and Growth Conditions
4.2. DNA Assays
4.3. Protein Isolation and Analysis
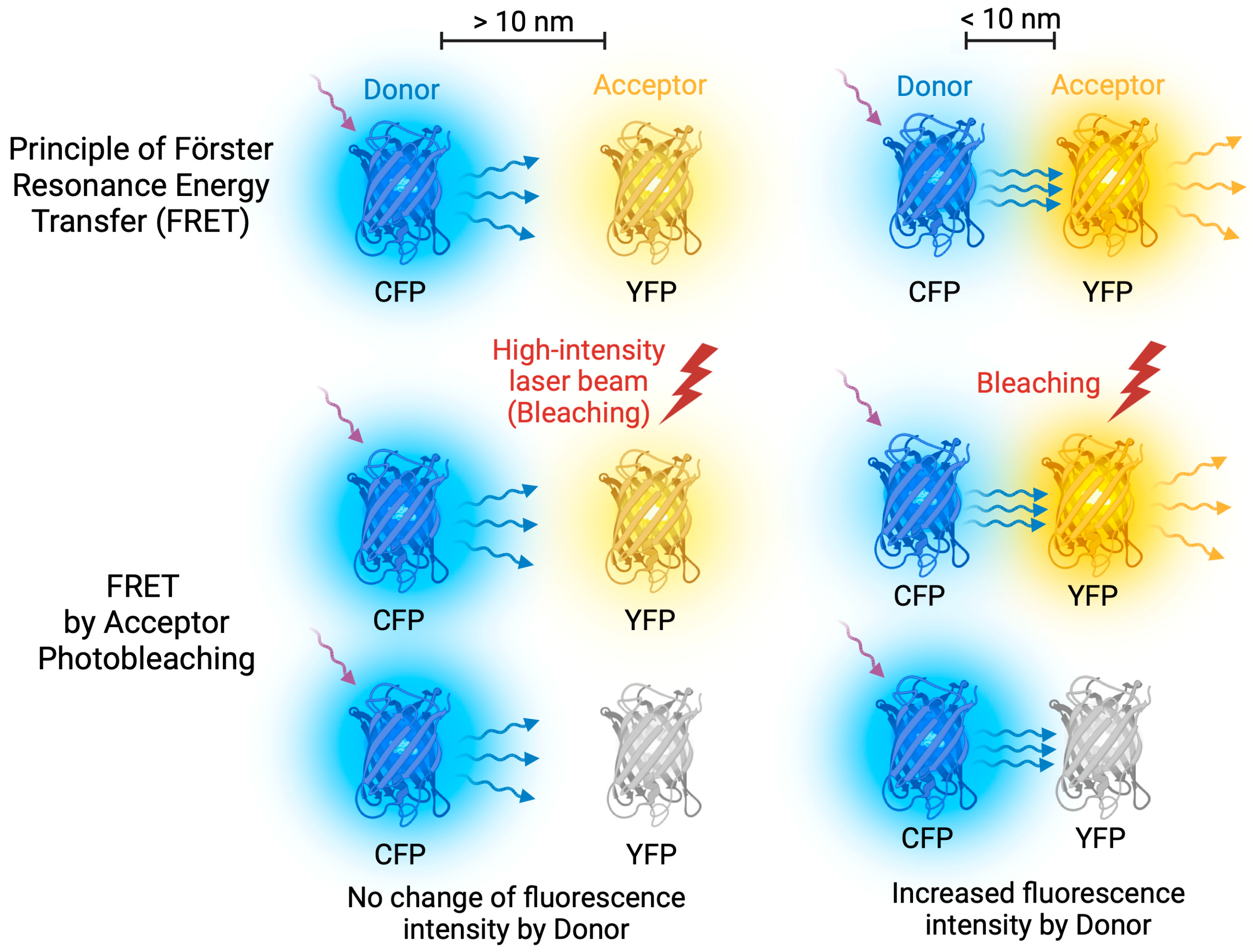
4.4. Fluorescence Microscopy
4.5. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| Aβ | Amyloid beta |
| AB | acceptor bleaching |
| AD | Alzheimer’s disease |
| BiFC | bimolecular fluorescence complementation |
| CFP | cyan fluorescent protein |
| Em | Emission |
| Ex | Excitation |
| FRET | Förster resonance energy transfer |
| HD | Huntington’s disease |
| Htt | Huntingtin |
| Hs | Homo sapiens |
| IAPP | islet amyloid polypeptide |
| OD | optical density |
| PD | Parkinson’s disease |
| PMD | protein misfolding disorders |
| PrP | Prion Protein |
| SDS | sodium dodecyl sulfate |
| SDD-AGE | semi-denaturing detergent agarose electrophoresis |
| T2DM | type 2 diabetes mellitus |
| TSE | transmissible spongiform encephalopathies |
| YFP | yellow fluorescent protein |
| YNB | Yeast nitrogen base |
| YPD | Yeast Extract–Peptone–Dextrose |
Appendix A
| Protein Name | [PIN+] Status | Total Number of Cells | Number of Cells with Fluorescent Glow | Number of Cells with Fluorescent Clumps | % of Cells with Fluorescent Clumps | p-Value |
|---|---|---|---|---|---|---|
| PrP-YFP | [PIN+] | 3939 | 819 | 807 | 98.53 ± 0.42 | 1 |
| [pin−] | 3783 | 739 | 727 | 98.38 ± 0.46 | ||
| IAPP-YFP | [PIN+] | 4031 | 429 | 326 | 75.99 ± 2.06 | 0.874 |
| [pin−] | 3998 | 397 | 308 | 77.58 ± 2.09 | ||
| Aβ42-YFP | [PIN+] | 3039 | 596 | 328 | 55.03 ± 2.04 | 0.534 |
| [pin−] | 3256 | 667 | 345 | 51.72 ±1.93 |
References
- Benson, M.D.; Buxbaum, J.N.; Eisenberg, D.S.; Merlini, G.; Saraiva, M.J.M.; Sekijima, Y.; Sipe, J.D.; Westermark, P. Amyloid nomenclature 2018: Recommendations by the International Society of Amyloidosis (ISA) nomenclature committee. Amyloid 2018, 4, 215–219. [Google Scholar] [CrossRef] [PubMed]
- Buxbaum, J.N.; Dispenzieri, A.; Eisenberg, D.S.; Fändrich, M.; Merlini, G.; Saraiva, M.J.M.; Sekijima, Y.; Westermark, P. Amyloid nomenclature 2022: Update, novel proteins, and recommendations by the International Society of Amyloidosis (ISA) Nomenclature Committee. Amyloid 2022, 29, 213–219. [Google Scholar] [CrossRef]
- Hardy, J.; Allsop, D. Amyloid Deposition as the Central Event in the Aetiology of Alzheimer’s Disease. Trends Pharmacol. Sci. 1991, 12, 383–388. [Google Scholar] [CrossRef] [PubMed]
- Kremer, B.; Goldberg, P.; Andrew, S.E.; Theilmann, J.; Telenius, H.; Zeisler, J.; Squitieri, F.; Lin, B.; Bassett, A.; Almqvist, E.; et al. A Worldwide Study of the Huntington’s Disease Mutation: The Sensitivity and Specificity of Measuring CAG Repeats. N. Engl. J. Med. 1994, 330, 1401–1406. [Google Scholar] [CrossRef] [PubMed]
- Baba, M.; Nakajo, S.; Tu, P.H.; Tomita, T.; Nakaya, K.; Lee, V.M.; Trojanowski, J.Q.; Iwatsubo, T. Aggregation of Alpha-Synuclein in Lewy Bodies of Sporadic Parkinson’s Disease and Dementia with Lewy Bodies. Am. J. Pathol. 1998, 152, 879–884. [Google Scholar] [PubMed]
- Johnson, K.H.; O’Brien, T.D.; Betsholtz, C.; Westermark, P. Islet Amyloid Polypeptide: Mechanisms of Amyloidogenesis in the Pancreatic Islets and Potential Roles in Diabetes Mellitus. Lab. Investig. 1992, 66, 522–535. [Google Scholar]
- Mackenzie, I.R.; Rademakers, R.; Neumann, M. TDP-43 and FUS in amyotrophic lateral sclerosis and frontotemporal dementia. Lancet Neurol. 2010, 10, 995–1007. [Google Scholar] [CrossRef]
- Prusiner, S.B. Novel Proteinaceous Infectious Particles Cause Scrapie. Science 1982, 216, 136–144. [Google Scholar] [CrossRef]
- Giasson, B.I.; Forman, M.S.; Higuchi, M.; Golbe, L.I.; Graves, C.L.; Kotzbauer, P.T.; Trojanowski, J.Q.; Lee, V.M.-Y. Initiation and Synergistic Fibrillization of Tau and Alpha-Synuclein. Science 2003, 300, 636–640. [Google Scholar] [CrossRef]
- Twohig, D.; Nielsen, H.M. α-Synuclein in the Pathophysiology of Alzheimer’s Disease. Mol. Neurodegener. 2019, 14, 23. [Google Scholar] [CrossRef]
- Chang, X.-L.; Tan, M.-S.; Tan, L.; Yu, J.-T. The Role of TDP-43 in Alzheimer’s Disease. Mol. Neurobiol. 2016, 53, 3349–3359. [Google Scholar] [CrossRef] [PubMed]
- Kovács, G.G.; Trabattoni, G.; Hainfellner, J.A.; Ironside, J.W.; Knight, R.S.G.; Budka, H. Mutations of the Prion Protein Gene. J. Neurol. 2002, 249, 1567–1582. [Google Scholar] [CrossRef] [PubMed]
- Morales, R.; Estrada, L.D.; Diaz-Espinoza, R.; Morales-Scheihing, D.; Jara, M.C.; Castilla, J.; Soto, C. Molecular Cross Talk between Misfolded Proteins in Animal Models of Alzheimer’s and Prion Diseases. J. Neurosci. 2010, 30, 4528–4535. [Google Scholar] [CrossRef] [PubMed]
- Jackson, K.; Barisone, G.A.; Diaz, E.; Jin, L.; DeCarli, C.; Despa, F. Amylin Deposition in the Brain: A Second Amyloid in Alzheimer Disease? Ann. Neurol. 2013, 74, 517–526. [Google Scholar] [CrossRef]
- Horvath, I.; Wittung-Stafshede, P. Cross-talk between amyloidogenic proteins in type-2 diabetes and Parkinson’s disease. Proc. Natl. Acad. Sci. USA 2016, 113, 12473–12477. [Google Scholar] [CrossRef]
- Miklossy, J.; Qing, H.; Radenovic, A.; Kis, A.; Vileno, B.; Làszló, F.; Miller, L.; Martins, R.N.; Waeber, G.; Mooser, V.; et al. Beta Amyloid and Hyperphosphorylated Tau Deposits in the Pancreas in Type 2 Diabetes. Neurobiol. Aging 2010, 31, 1503–1515. [Google Scholar] [CrossRef]
- Morales, R.; Green, K.M.; Soto, C. Cross currents in protein misfolding disorders: Interactions and therapy. CNS Neurol. Disord. Drug Targets 2009, 8, 363–371. [Google Scholar] [CrossRef]
- Moreno-Gonzalez, I.; Edwards Iii, G.; Salvadores, N.; Shahnawaz, M.; Diaz-Espinoza, R.; Soto, C. Molecular interaction between type 2 diabetes and Alzheimer’s disease through cross-seeding of protein misfolding. Mol. Psychiatry 2017, 22, 1327–1334. [Google Scholar] [CrossRef]
- Oskarsson, M.E.; Paulsson, J.F.; Schultz, S.W.; Ingelsson, M.; Westermark, P.; Westermark, G.T. In vivo seeding and cross-seeding of localized amyloidosis: A molecular link between type 2 diabetes and Alzheimer disease. Am. J. Pathol. 2015, 185, 834–846. [Google Scholar] [CrossRef]
- Chernova, T.A.; Chernoff, Y.O.; Wilkinson, K.D. Yeast Models for Amyloids and Prions: Environmental Modulation and Drug Discovery. Molecules 2019, 24, 3388. [Google Scholar] [CrossRef]
- Tuite, M.F. Yeast Models of Neurodegenerative Diseases. In Progress in Molecular Biology and Translational Science; Elsevier: Amsterdam, The Netherlands, 2019; Volume 168, pp. 351–379. ISBN 978-0-12-817874-4. [Google Scholar]
- Chernoff, Y.O.; Grizel, A.V.; Rubel, A.A.; Zelinsky, A.A.; Chandramowlishwaran, P.; Chernova, T.A. Application of Yeast to Studying Amyloid and Prion Diseases. In Advances in Genetics; Elsevier: Amsterdam, The Netherlands, 2020; Volume 105, pp. 293–380. ISBN 978-0-12-821685-9. [Google Scholar]
- Rubel, A.A.; Ryzhova, T.A.; Antonets, K.S.; Chernoff, Y.O.; Galkin, A.P. Identification of PrP sequences essential for the interaction between the PrP polymers and Aβ peptide in a yeast-based assay. Prion 2013, 7, 469–476. [Google Scholar] [CrossRef] [PubMed]
- Ciaccioli, G.; Martins, A.; Rodrigues, C.; Vieira, H.; Calado, P. A Powerful Yeast Model to Investigate the Synergistic Interaction of α-Synuclein and Tau in Neurodegeneration. PLoS ONE 2013, 8, e55848. [Google Scholar] [CrossRef] [PubMed]
- Grizel, A.V.; Rubel, A.A.; Chernoff, Y.O. Strain Conformation Controls the Specificity of Cross-Species Prion Transmission in the Yeast Model. Prion 2016, 10, 269–282. [Google Scholar] [CrossRef]
- Sergeeva, A.V.; Sopova, J.V.; Belashova, T.A.; Siniukova, V.A.; Chirinskaite, A.V.; Galkin, A.P.; Zadorsky, S.P. Amyloid Properties of the Yeast Cell Wall Protein Toh1 and Its Interaction with Prion Proteins Rnq1 and Sup35. Prion 2019, 13, 21–32. [Google Scholar] [CrossRef] [PubMed]
- Chernoff, Y.O.; Uptain, S.M.; Lindquist, S.L. Analysis of Prion Factors in Yeast. In Methods in Enzymology; Elsevier: Amsterdam, The Netherlands, 2002; Volume 351, pp. 499–538. ISBN 978-0-12-182254-5. [Google Scholar]
- Bagriantsev, S.N.; Kushnirov, V.V.; Liebman, S.W. Analysis of Amyloid Aggregates Using Agarose Gel Electrophoresis. In Methods in Enzymology; Elsevier: Amsterdam, The Netherlands, 2006; Volume 412, pp. 33–48. ISBN 978-0-12-182817-2. [Google Scholar]
- Prusiner, S.B.; Scott, M.R.; DeArmond, S.J.; Cohen, F.E. Prion Protein Biology. Cell 1998, 93, 337–348. [Google Scholar] [CrossRef] [PubMed]
- Kushnirov, V.V.; Alexandrov, I.M.; Mitkevich, O.V.; Shkundina, I.S.; Ter-Avanesyan, M.D. Purification and analysis of prion and amyloid aggregates. Methods 2006, 39, 50–55. [Google Scholar] [CrossRef] [PubMed]
- Derkatch, I.L.; Chernoff, Y.O.; Kushnirov, V.V.; Inge-Vechtomov, S.G.; Liebman, S.W. Genesis and variability of [PSI+] Prion Factors Saccharomyces cerevisiae. Genetics 1996, 144, 1375–1386. [Google Scholar] [CrossRef]
- Derkatch, I.L.; Bradley, M.E.; Hong, J.Y.; Liebman, S.W. Prions affect the appearance of other prions: The story of [PIN+]. Cell 2001, 106, 171–182. [Google Scholar] [CrossRef]
- Halfmann, R.; Wright, J.J.R.; Alberti, S.; Lindquist, S.; Rexach, M. Prion formation by a yeast GLFG nucleoporin. Prion 2012, 6, 391–399. [Google Scholar] [CrossRef]
- Bradley, M.E.; Edskes, H.K.; Hong, J.Y.; Wickner, R.B.; Liebman, S.W. Interactions among prions and prion “strains” in yeast. Proc. Natl. Acad. Sci. USA 2002, 99, 16392–16399. [Google Scholar] [CrossRef]
- Danilov, L.G.; Sukhanova, X.V.; Rogoza, T.M.; Antonova, E.Y.; Trubitsina, N.P.; Zhouravleva, G.A.; Bondarev, S.A. Identification of New FG-Repeat Nucleoporins with Amyloid Properties. Int. J. Mol. Sci. 2023, 24, 8571. [Google Scholar] [CrossRef] [PubMed]
- Meriin, A.B.; Zhang, X.; He, X.; Newnam, G.P.; Chernoff, Y.O.; Sherman, M.Y. Huntingtin toxicity in yeast model depends on polyglutamine aggregation mediated by a prion-like protein Rnq1. J. Cell Biol. 2002, 157, 997–1004. [Google Scholar] [CrossRef] [PubMed]
- Karpova, T.S.; Baumann, C.T.; He, L.; Wu, X.; Grammer, A.; Lipsky, P.; Hager, G.L.; McNally, J.G. Fluorescence resonance energy transfer from cyan to yellow fluorescent protein detected by acceptor photobleaching using confocal microscopy and a single laser. J. Microsc. 2003, 209, 56–70. [Google Scholar] [CrossRef]
- Cui, Z.; Zhang, K.; Zhang, Z.; Liu, Y.; Zhou, Y.; Wei, H.; Zhang, X.-E. Visualization of the dynamic multimerization of human Cytomegalovirus pp65 in punctuate nuclear foci. Virology 2009, 392, 169–177. [Google Scholar] [CrossRef]
- Ishikawa-Ankerhold, H.C.; Ankerhold, R.; Drummen, G.P.C. Advanced fluorescence microscopy techniques–FRAP, FLIP, FLAP, FRET and FLIM. Molecules 2012, 17, 4047–4132. [Google Scholar] [CrossRef]
- Stynen, B.; Tournu, H.; Tavernier, J.; Van Dijck, P. Diversity in genetic in vivo methods for protein-protein interaction studies: From the yeast two-hybrid system to the mammalian split-luciferase system. Microbiol. Mol. Biol. Rev. 2012, 76, 331–382. [Google Scholar] [CrossRef] [PubMed]
- Arvanitakis, Z.; Wilson, R.S.; Bienias, J.L.; Evans, D.A.; Bennett, D.A. Diabetes Mellitus and Risk of Alzheimer Disease and Decline in Cognitive Function. Arch. Neurol. 2004, 61, 661–666. [Google Scholar] [CrossRef] [PubMed]
- Bharadwaj, P.; Wijesekara, N.; Liyanapathirana, M.; Newsholme, P.; Ittner, L.; Fraser, P.; Verdile, G. The Link between Type 2 Diabetes and Neurodegeneration: Roles for Amyloid-β, Amylin, and Tau Proteins. J. Alzheimer’s Dis. 2017, 59, 421–432. [Google Scholar] [CrossRef]
- Karki, R.; Kodamullil, A.T.; Hofmann-Apitius, M. Comorbidity Analysis between Alzheimer’s Disease and Type 2 Diabetes Mellitus (T2DM) Based on Shared Pathways and the Role of T2DM Drugs. J. Alzheimer’s Dis. 2017, 60, 721–731. [Google Scholar] [CrossRef]
- Biessels, G.J.; Despa, F. Cognitive decline and dementia in diabetes mellitus: Mechanisms and clinical implications. Nat. Rev. Endocrinol. 2018, 14, 591–604. [Google Scholar] [CrossRef]
- Verdile, G.; Fuller, S.J.; Martins, R.N. The role of type 2 diabetes in neurodegeneration. Neurobiol. Dis. 2015, 84, 22–38. [Google Scholar] [CrossRef]
- Salas, I.H.; De Strooper, B. Diabetes and Alzheimer’s Disease: A Link Not as Simple as It Seems. Neurochem. Res. 2019, 44, 1271–1278. [Google Scholar] [CrossRef] [PubMed]
- Silva, M.V.F.; Loures, C.d.M.G.; Alves, L.C.V.; de Souza, L.C.; Borges, K.B.G.; das Graças Carvalho, M. Alzheimer’s Disease: Risk Factors and Potentially Protective Measures. J. Biomed. Sci. 2019, 26, 33. [Google Scholar] [CrossRef]
- Biessels, G.J.; Kappelle, L.J. Utrecht Diabetic Encephalopathy Study Group. Increased risk of Alzheimer’s disease in Type II diabetes: Insulin resistance of the brain or insulin-induced amyloid pathology? Biochem. Soc. Trans. 2005, 33 Pt 5, 1041–1044. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Valbuena, I.; Valenti-Azcarate, R.; Amat-Villegas, I.; Marcilla, I.; Marti-Andres, G.; Caballero, M.-C.; Riverol, M.; Tuñon, M.-T.; Fraser, P.E.; Luquin, M.-R. Mixed pathologies in pancreatic β cells from subjects with neurodegenerative diseases and their interaction with prion protein. Acta Neuropathol. Commun. 2021, 9, 64. [Google Scholar] [CrossRef] [PubMed]
- Young, L.M.; Mahood, R.A.; Saunders, J.C.; Tu, L.-H.; Raleigh, D.P.; Radford, S.E.; Ashcroft, A.E. Insights into the consequences of co-polymerisation in the early stages of IAPP and Aβ peptide assembly from mass spectrometry. Analyst 2015, 140, 6990–6999. [Google Scholar] [CrossRef] [PubMed]
- Bharadwaj, P.; Solomon, T.; Sahoo, B.R.; Ignasiak, K.; Gaskin, S.; Rowles, J. Amylin and beta amyloid proteins interact to form amorphous heterocomplexes with enhanced toxicity in neuronal cells. Sci. Rep. 2020, 10, 10356. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Westermark, G.T. The Amyloid Forming Peptides Islet Amyloid Polypeptide and Amyloid β Interact at the Molecular Level. Int. J. Mol. Sci. 2021, 22, 11153. [Google Scholar] [CrossRef]
- Ueki, S.; Lacroix, B.; Citovsky, V. Protein membrane overlay assay: A protocol to test interaction between soluble and insoluble proteins in vitro. J. Vis. Exp. 2011, 54, 2961. [Google Scholar] [CrossRef]
- Zamyatnin AAJr Solovyev, A.G.; Bozhkov, P.V.; Valkonen, J.P.; Morozov, S.Y.; Savenkov, E.I. Assessment of the integral membrane protein topology in living cells. Plant J. 2006, 46, 145–154. [Google Scholar] [CrossRef]
- Hanahan, D.; Glover, D. DNA Cloning: A Practical Approach; Oxford University Press: Oxford, UK, 1985; Volume 1, pp. 109–135. [Google Scholar]
- Kaiser, P.; Mansourl, H.A.; Greeten, T.; Auer, B.; Schweiger, M.; Schneider, R. The Human Ubiquitin-Conjugating Enzyme UbcH1 Is Involved in the Repair of UV-Damaged, Alkylated and Cross-Linked DNA. FEBS Lett. 1994, 350, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Chandramowlishwaran, P.; Sun, M.; Casey, K.L.; Romanyuk, A.V.; Grizel, A.V.; Sopova, J.V.; Rubel, A.A.; Nussbaum-Krammer, C.; Vorberg, I.M.; Chernoff, Y.O. Mammalian amyloidogenic proteins promote prion nucleation in yeast. J. Biol. Chem. 2018, 293, 3436–3450. [Google Scholar] [CrossRef] [PubMed]
- Rubel, A.A.; Korzhova, V.V.; Saifitdinova, A.F.; Antonets, K.S.; Inge-Vechtomov, S.G.; Galkin, A.P. PrP protein and Amyloid beta peptide interact in yeasts Saccharomyces cerevisiae. Ecol. Genet. 2012, 10, 74–80. [Google Scholar] [CrossRef]
- Green, M.R.; Sambrook, J. Molecular Cloning: A Laboratory Manual; Cold Spring Harbor Laboratory Press: Long Island, NY, USA, 2012; ISBN 9781936113415. [Google Scholar]
- Kachkin, D.V.; Khorolskaya, J.I.; Ivanova, J.S.; Rubel, A.A. An Efficient Method for Isolation of Plasmid DNA for Transfection of Mammalian Cell Cultures. Methods Protoc. 2020, 3, 69. [Google Scholar] [CrossRef]
- Rose, D.; Thomas, W.; Holm, C. Segregation of recombined chromosomes in meiosis I requires DNA topoisomerase II. Cell 1990, 60, 1009–1017. [Google Scholar] [CrossRef]
- Fisher, R.A. The logic of inductive inference. J. R. Stat. Soc. 1935, 98, 39–82. [Google Scholar] [CrossRef]
| Plasmid Name | Promoter/Expression Cassette | Yeast Marker | Source |
|---|---|---|---|
| pmCUP1-Sup35NM-IAPP | PCUP1/Sup35NM-IAPP | URA3 | [57] |
| pGPD-PrP23-YFP (URA3) | PGPD/mmPrP(23-231)-YFP | URA3 | [23] |
| pGPD-PrP23-CFP (LEU2) | PGPD/mmPrP(23-231)-CFP | LEU2 | [23] |
| pGPD-Aβ42-CFP (LEU2) | PGPD/hsAβ(1-42)-CFP | LEU2 | This study |
| pGPD-YFP (URA3) | PGPD/YFP | URA3 | [23] |
| pGPD-Ab-YFP (LEU2) | PGPD/hsAβ(1-40)-YFP | LEU2 | [58] |
| pGPD-CFP (LEU2) | PGPD/CFP | LEU2 | [23] |
| pGPD-IAPP-YFP (URA3) | PGPD/hsIAPP-YFP | URA3 | This study |
| pGPD-IAPP-CFP (LEU2) | PGPD/hsIAPP-CFP | LEU2 | This study |
| Primer Name | Sequence |
|---|---|
| Amy1-F | 5′-GCGGATCCATGGCCACACAAAGATTGGCTAATTTCC-3′ |
| Amy1-R | 5′-TTACCGCGGCACCGCGGTGGCGGCC-3′ |
| Aβ-F | 5′-CGGGATCCAATATGGATGCAGAGTTCC-3′ |
| Aβ-R | 5′-TCCCCGCGGCGCTATGACAACACC-3′ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kachkin, D.V.; Lashkul, V.V.; Gorsheneva, N.A.; Fedotov, S.A.; Rubel, M.S.; Chernoff, Y.O.; Rubel, A.A. The Aβ42 Peptide and IAPP Physically Interact in a Yeast-Based Assay. Int. J. Mol. Sci. 2023, 24, 14122. https://doi.org/10.3390/ijms241814122
Kachkin DV, Lashkul VV, Gorsheneva NA, Fedotov SA, Rubel MS, Chernoff YO, Rubel AA. The Aβ42 Peptide and IAPP Physically Interact in a Yeast-Based Assay. International Journal of Molecular Sciences. 2023; 24(18):14122. https://doi.org/10.3390/ijms241814122
Chicago/Turabian StyleKachkin, Daniel V., Veronika V. Lashkul, Natalia A. Gorsheneva, Sergey A. Fedotov, Maria S. Rubel, Yury O. Chernoff, and Aleksandr A. Rubel. 2023. "The Aβ42 Peptide and IAPP Physically Interact in a Yeast-Based Assay" International Journal of Molecular Sciences 24, no. 18: 14122. https://doi.org/10.3390/ijms241814122






