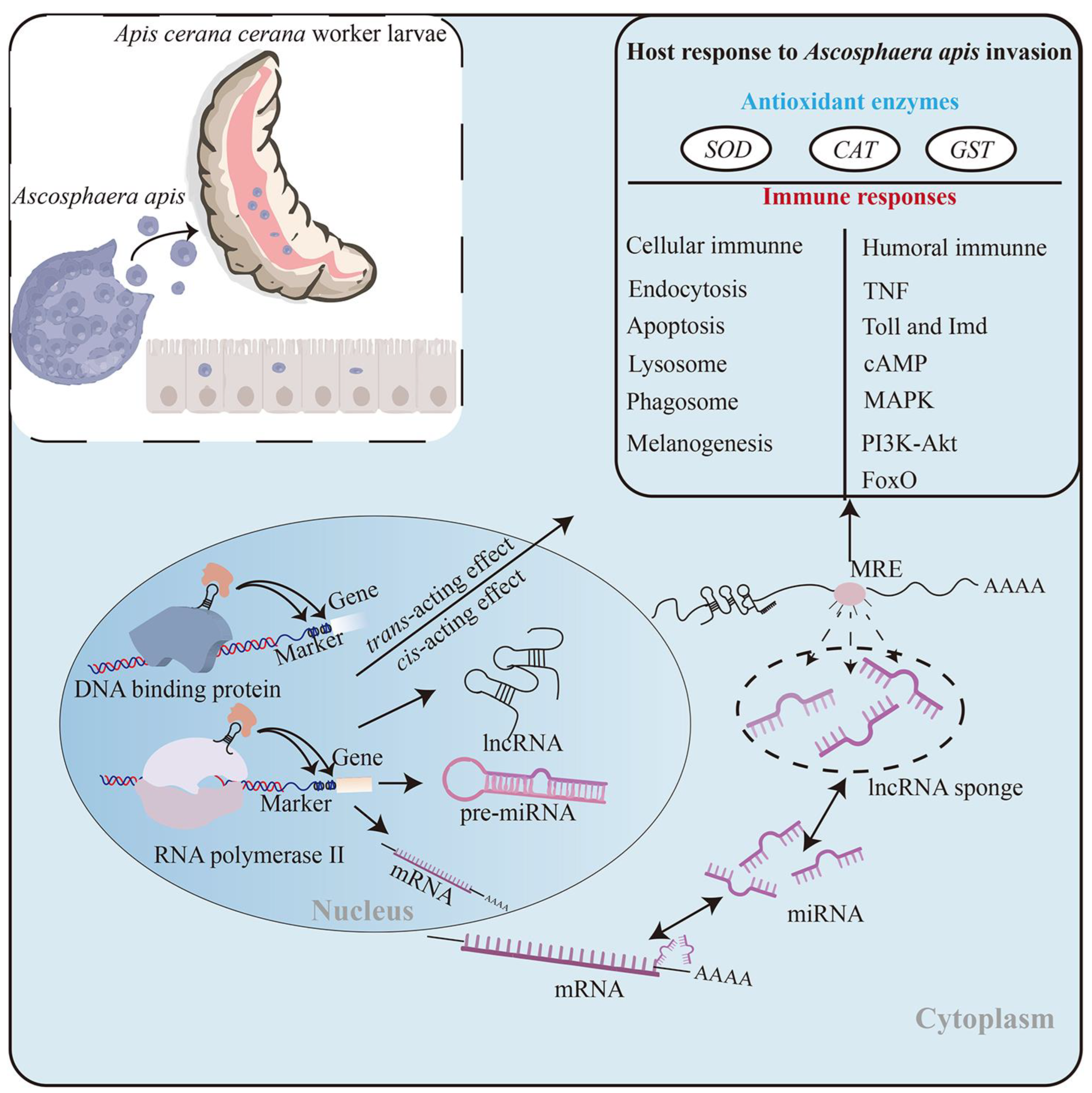
Regulatory Roles of Long Non-Coding RNAs Relevant to Antioxidant Enzymes and Immune Responses of Apis cerana Larvae Following Ascosphaera apis Invasion
Abstract
:1. Introduction
2. Results
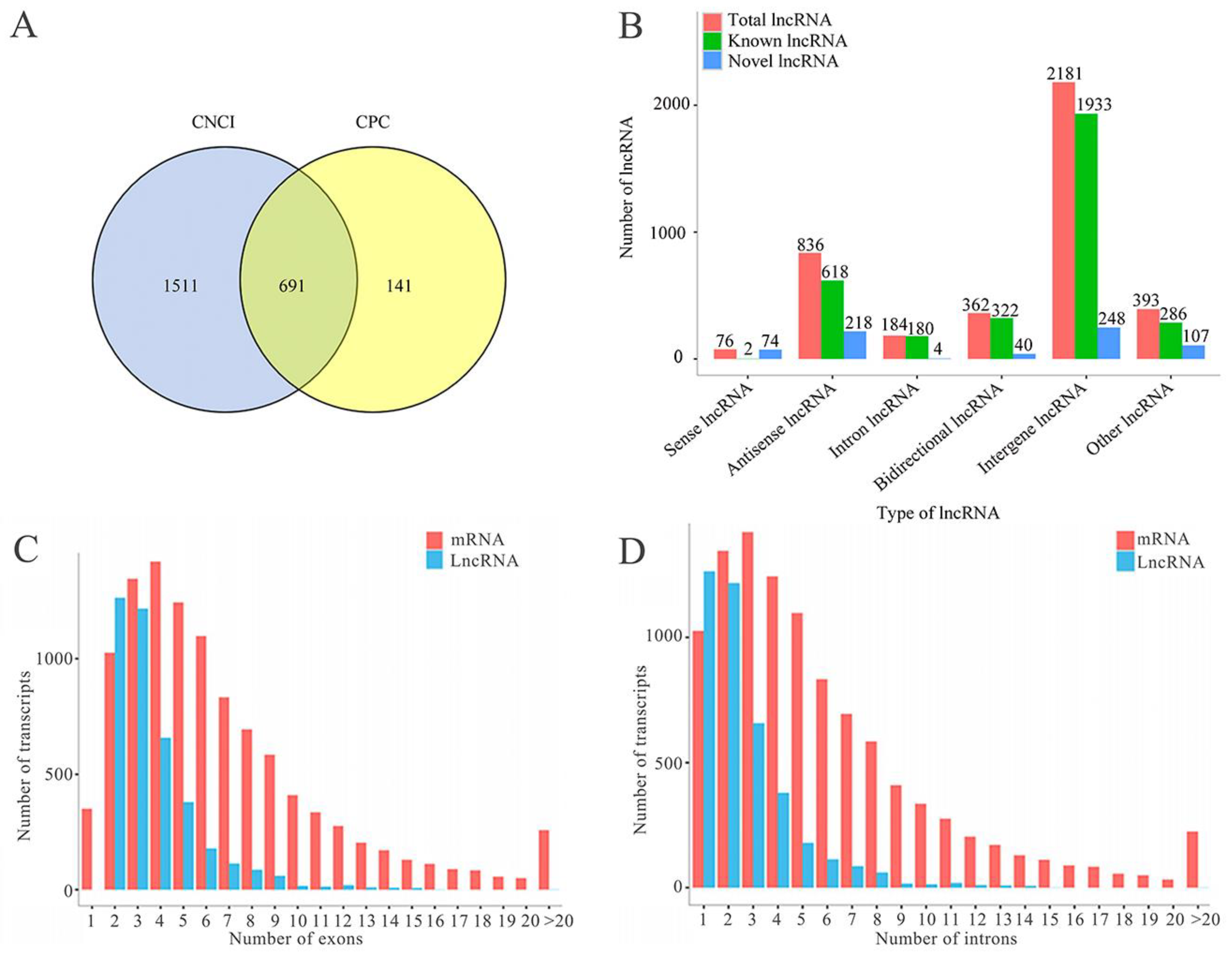
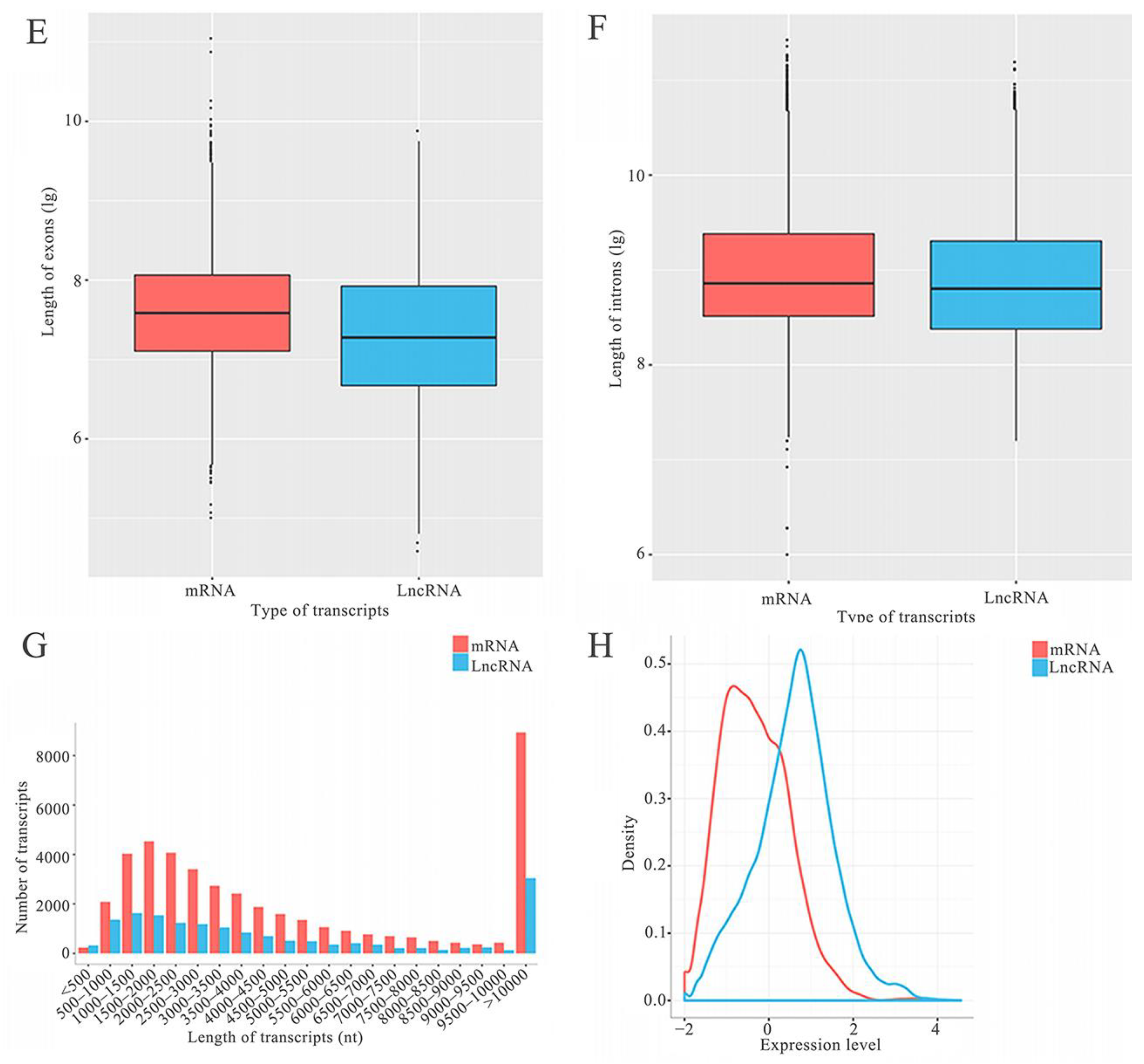
2.1. Number, Type, and Property of lncRNAs in A. c. cerana Larval Guts
2.2. Analysis of DElncRNAs as Putative miRNA Precursors
2.3. Differential Expression Profile of lncRNAs in Larval Guts Following A. apis Infection
2.4. Cis-Acting Regulatory Manner of Host DElncRNAs
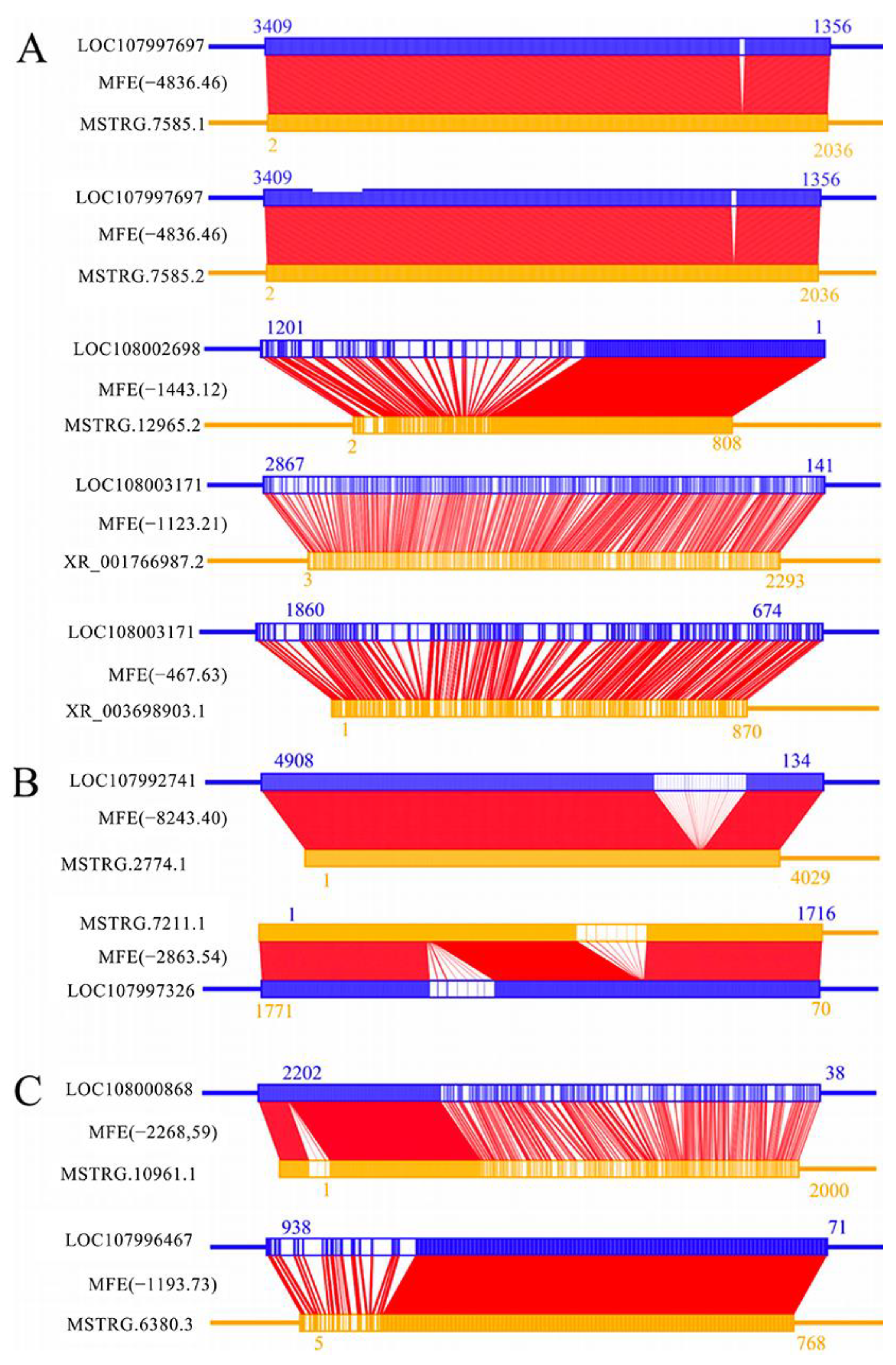
2.5. Investigation of Antisense DElncRNAs
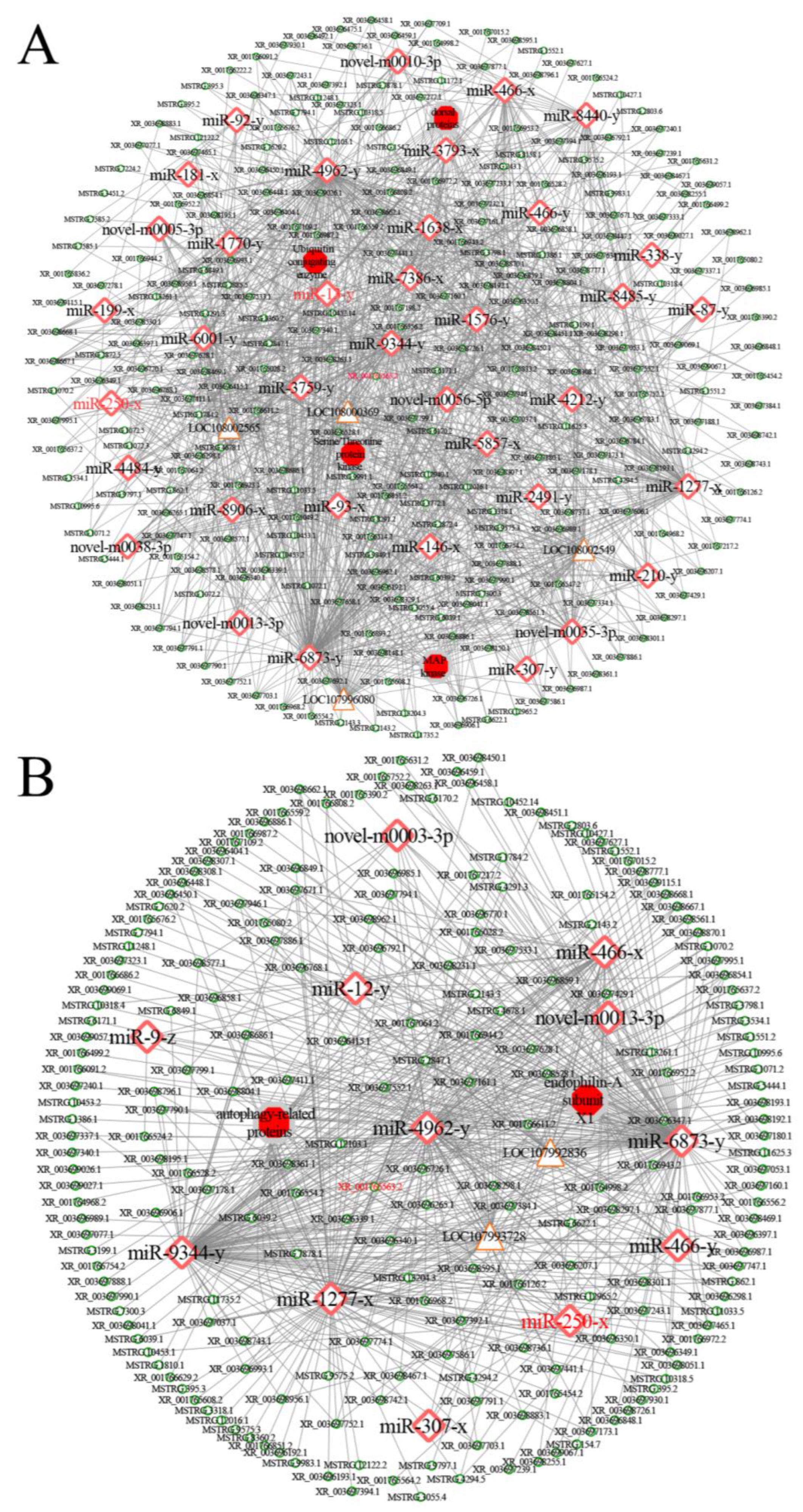
2.6. Analysis of DElncRNA-DEmiRNA Regulatory Networks
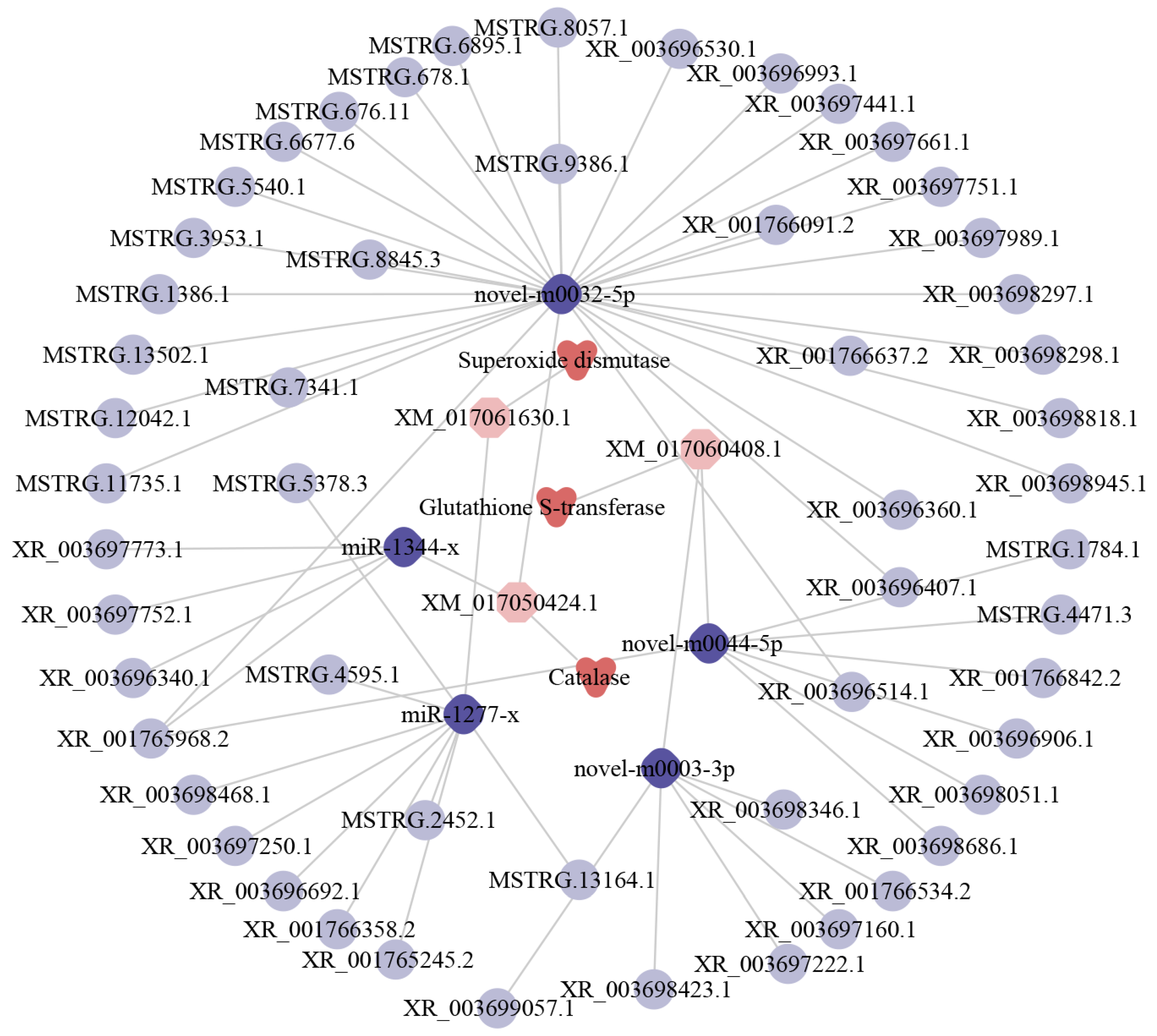
2.7. Antioxidant Enzyme-Associated ceRNA Sub-Network of Host DElncRNAs
2.8. Immune Response-Associated ceRNA Sub-Network of Host DElncRNAs
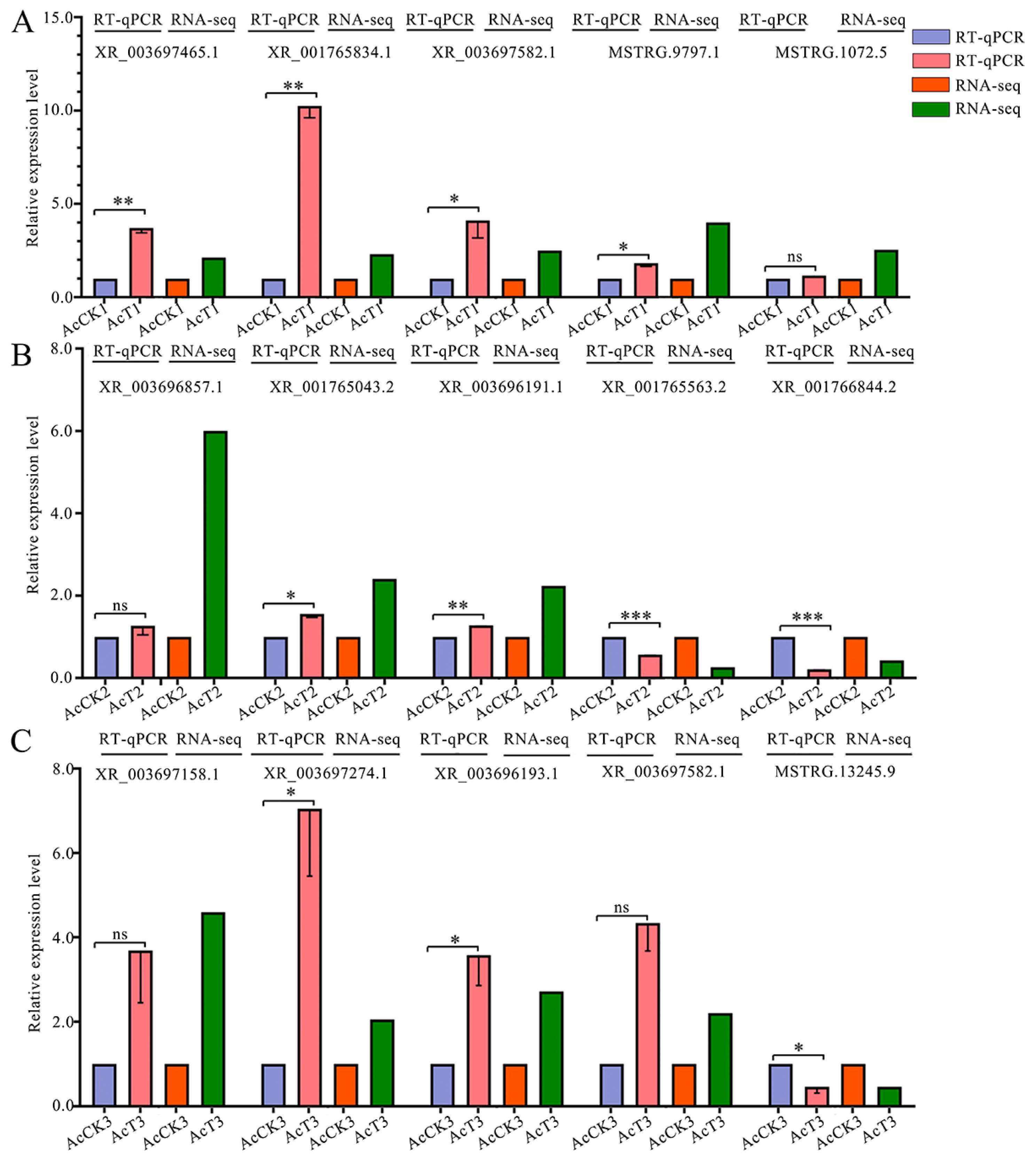
2.9. RT-qPCR Detection of DElncRNAs
3. Discussion
3.1. A. c. cerana lncRNAs Shared Analogous Structural Property with Those in Other Animals, Plants, and Microorganisms
3.2. A. apis Invasion Gave Rise to the Alteration of Overall Expression Pattern of lncRNAs in the Larval Guts
3.3. A. c. cerana lncRNAs May Be a Source for the Generation of Abundant miRNAs
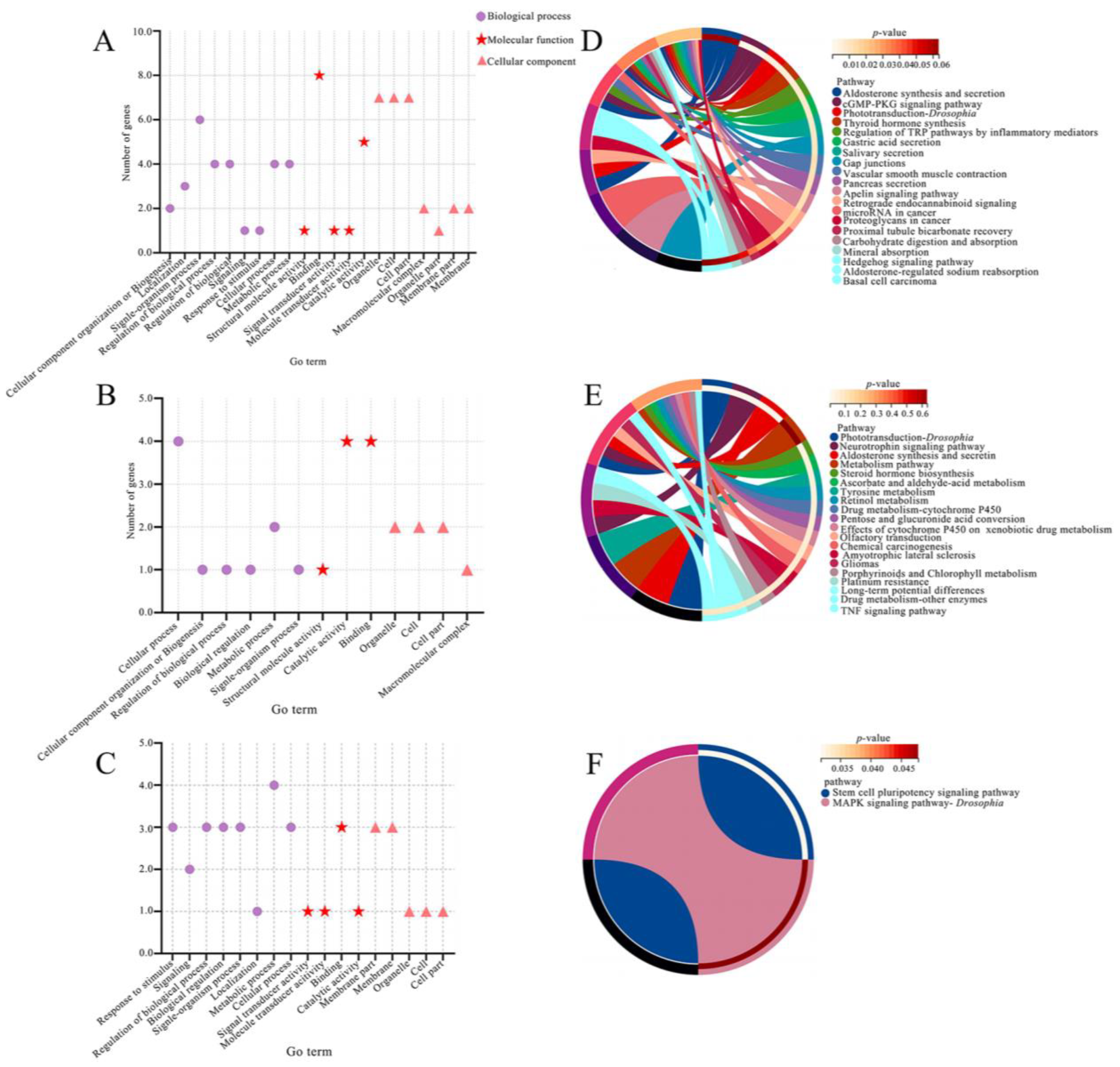
3.4. DElncRNAs Potentially Modulated Host Response to A. apis Invasion by Regulating the Transcription of Up- and Down-Stream Genes
3.5. Antisense DElncRNAs May Participate in the Response of Larvae to A. apis Invasion by Interacting with Corresponding Sense-Strand mRNAs
3.6. DElncRNAs Putatively Participated in the Regulation of Larval A. apis—Response through Modulating the Expression of Target Genes via ceRNA Networks
4. Materials and Methods
4.1. Fungi and Bee Larvae
4.2. Transcriptome Data Source
4.3. Quality Control and Genomic Mapping
4.4. Identification of lncRNAs
4.5. Analysis of lncRNAs as Potential miRNA Precursors
4.6. Differential Analysis of lncRNAs
4.7. Investigation of the Cis-Acting Effect of DElncRNAs
4.8. Investigation of the Trans-Acting Effect of DElncRNAs
4.9. Construction and Analysis of ceRNA Networks of DElncRNAs
4.10. Investigation of Antioxidant Enzyme- and Immune Response-Associated DElncRNAs and the Corresponding Regulatory Network
4.11. RT-qPCR Validation of DElnRNAs
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Li, T.; Wang, S.; Wu, R.; Zhou, X.; Zhu, D.; Zhang, Y. Identification of long non-protein coding RNAs in chicken skeletal muscle using next generation sequencing. Genomics 2012, 99, 292–298. [Google Scholar] [CrossRef] [PubMed]
- Fatica, A.; Bozzoni, I. Long non-coding RNAs: New players in cell differentiation and development. Nat. Rev. Genet. 2014, 15, 7–21. [Google Scholar] [CrossRef] [PubMed]
- Delás, M.J.; Hannon, G.J. lncRNAs in development and disease: From functions to mechanisms. Open Biol. 2017, 7, 170121. [Google Scholar] [CrossRef] [PubMed]
- Ponting, C.P.; Oliver, P.L.; Reik, W. Evolution and functions of long noncoding RNAs. Cell 2009, 136, 629–641. [Google Scholar] [CrossRef]
- Huang, J.Z.; Chen, M.; Chen, D.; Gao, X.C.; Zhu, S.; Huang, H.; Hu, M.; Zhu, H.; Yan, G.R. A Peptide encoded by a putative lncRNA HOXB-AS3 suppresses colon cancer growth. Mol. Cell 2017, 68, 171–184.e6. [Google Scholar] [CrossRef]
- Yu, X.J.; Jin, K.; Jin, J.; Zuo, Q.S.; Zhao, R.F.; Li, B.C. Encoding small petide by lncRNA-CEPT8 in chicken primordial germ cells. J. Yangzhou Univ. (Agric. Life Sci. Ed.) 2019, 40, 83–88. (In Chinese) [Google Scholar]
- Natarajan, P.; Shrinivas, K.; Chakraborty, A.K. A model for cis-regulation of transcriptional condensates and gene expression by proximal lncRNAs. Biophys. J. 2023, 122, 2757–2772. [Google Scholar] [CrossRef]
- Li, R.; Xu, H.; Gao, X. The ceRNA network regulates epithelial-mesenchymal transition in colorectal cancer. Heliyon 2023, 9, e14143. [Google Scholar] [CrossRef] [PubMed]
- Jadaliha, M.; Gholamalamdari, O.; Tang, W.; Zhang, Y.; Petracovici, A.; Hao, Q.; Tariq, A.; Kim, T.G.; Holton, S.E.; Singh, D.K.; et al. A natural antisense lncRNA controls breast cancer progression by promoting tumor suppressor gene mRNA stability. PLoS Genet. 2018, 14, e1007802. [Google Scholar] [CrossRef]
- Lim, Y.H.; Yoon, G.; Ryu, Y.; Jeong, D.; Song, J.; Kim, Y.S.; Ahn, Y.; Kook, H.; Kim, Y. Human lncRNA SUGCT-AS1 regulates the proinflammatory response of macrophage. Int. J. Mol. Sci. 2023, 24, 13315. [Google Scholar] [CrossRef]
- Luo, H.; Zhu, G.; Zha, J.; Yan, B.; Guo, Y.; Xu, J.; Lai, Q.; Xu, B.; Yang, F.; Li, W. Activation of hottip lncRNA perturbs HSC function leading to AML like disease in mice. Blood 2018, 132, 3877. [Google Scholar] [CrossRef]
- Dou, X.; Yang, W.; Ding, Q.; Han, Q.; Qian, Q.; Du, Z.; Fan, Y.; Wang, C.; Li, S. Comprehensive analysis of the expression profiles of hepatic lncRNAs in the mouse model of alcoholic liver disease. Front. Pharmacol. 2021, 12, 709287. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Li, J.; Lian, B.; Gu, H.; Li, Y.; Qi, Y. Global identification of Arabidopsis lncRNAs reveals the regulation of MAF4 by a natural antisense RNA. Nat. Commun. 2018, 9, 5056. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Cheng, T.; Liu, C.; Liu, D.; Zhang, Q.; Long, R.; Zhao, P.; Xia, Q. Systematic identification and characterization of long non-coding RNAs in the silkworm, Bombyx mori. PLoS ONE 2016, 11, e0147147. [Google Scholar] [CrossRef]
- Xu, X.; Wang, K.; Zha, X. An antisense lncRNA functions in alternative splicing of Bmdsx in the silkworm, Bombyx mori. Biochem. Biophys. Res. 2019, 516, 639–644. [Google Scholar] [CrossRef] [PubMed]
- Jayakodi, M.; Jung, J.W.; Park, D.; Ahn, Y.J.; Lee, S.C.; Shin, S.Y.; Shin, C.; Yang, T.J.; Kwon, H.W. Genome-wide characterization of long intergenic non-coding RNAs (lincRNAs) provides new insight into viral diseases in honey bees Apis cerana and Apis mellifera. BMC Genom. 2015, 16, 680. [Google Scholar] [CrossRef]
- Aronstein, K.A.; Murray, K.D. Chalkbrood disease in honey bees. J. Invertebr. Pathol. 2009, 103 (Suppl. 1), S20–S29. [Google Scholar] [CrossRef]
- Chen, D.; Guo, R.; Xu, X.; Xiong, C.; Liang, Q.; Zheng, Y.; Luo, Q.; Zhang, Z.; Huang, Z.; Kumar, D.; et al. Uncovering the immune responses of Apis mellifera ligustica larval gut to Ascosphaera apis infection utilizing transcriptome sequencing. Gene 2017, 621, 40–50. [Google Scholar] [CrossRef]
- Du, Y.; Tong, X.Y.; Zhou, D.D.; Chen, D.F.; Xiong, C.L.; Zheng, Y.Z.; Xu, G.J.; Wang, H.P.; Chen, H.Z.; Guo, Y.L.; et al. MicroRNA responses in the larve gut of Apis cerana cerana to Ascosphaera apis stress. Acta Microbiol. Sin. 2019, 59, 1747–1764. (In Chinese) [Google Scholar]
- Guo, R.; Chen, D.; Diao, Q.; Xiong, C.; Zheng, Y.; Hou, C. Transcriptomic investigation of immune responses of the Apis cerana cerana larval gut infected by Ascosphaera apis. J. Invertebr. Pathol. 2019, 166, 107210. [Google Scholar] [CrossRef]
- Chen, D.; Guo, R.; Xiong, C.; Zheng, Y.; Hou, C.; Fu, Z. Morphological and molecular identification of chalkbrood disease pathogen Ascosphaera apis in Apis cerana cerana. J. Apic. Res. 2018, 57, 516–521. [Google Scholar] [CrossRef]
- Wang, X. The progress of study on the physiology and histopatology of insect idgut. J. Zhongkai Agrotech. Coll. 2000, 13, 58–68. (In Chinese) [Google Scholar]
- Li, Y.; Zhang, C.; Qin, L.; Li, D.; Zhou, G.; Dang, D.; Chen, S.; Sun, T.; Zhang, R.; Wu, W. Characterization of critical functions of long non-coding RNAs and mRNAs in rhabdomyosarcoma cells and mouse skeletal muscle infected by Enterovirus 71 using RNA-Seq. Viruses 2018, 10, 556. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Xu, W.; Gao, X.; Li, W.; Qi, S.; Guo, D.; Ajayi, O.E.; Ding, S.W.; Wu, Q. lncRNA sensing of a viral suppressor of RNAi activates non-canonical innate immune signaling in Drosophila. Cell Host Microbe 2020, 27, 115–128.e8. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.H.; Dong, Y.; Wu, W.; Yi, D.S.; Wang, M.; Wang, H.T.; Xu, Q.F. Comprehensive identification and characterization of long non-coding RNAs associated with Rice black-streaked dwarf virus infection in Laodelphax striatellus (Fallén) midgut. Front. Physiol. 2020, 11, 1011. [Google Scholar] [CrossRef]
- Guo, R.; Zhang, K.; Gao, X.; Jing, X.; Guo, S.; Zang, H.; Song, Y.; Li, K.; Zou, Y.; Chen, M.; et al. CircRNAs and their regulatory networks associated with an-tioxidant enzymes and immune responses of Asian honey bee larvae to fungal infection. bioRxiv 2023, 7, 549790. [Google Scholar]
- Wang, Z.; Wang, S.; Fan, X.; Zhang, K.; Zhang, J.; Zhao, H.; Gao, X.; Zhang, Y.; Guo, S.; Zhou, D.; et al. Systematic characterization and regulatory role of lncRNAs in Asian honey bees responding to microsporidian infestation. Int. J. Mol. Sci. 2023, 24, 5886. [Google Scholar] [CrossRef]
- Djebali, S.; Davis, C.A.; Merkel, A.; Dobin, A.; Lassmann, T.; Mortazavi, A.; Tanzer, A.; Lagarde, J.; Lin, W.; Schlesinger, F.; et al. Landscape of transcription in human cells. Nature 2012, 489, 101–108. [Google Scholar] [CrossRef]
- Zhu, B.; Xu, M.; Shi, H.; Gao, X.; Liang, P. Genome-wide identification of lncRNAs associated with chlorantraniliprole resistance in diamondback moth Plutella xylostella (L.). BMC Genom. 2017, 18, 380. [Google Scholar] [CrossRef]
- Zhao, J.; Ajadi, A.A.; Wang, Y.; Tong, X.; Wang, H.; Tang, L.; Li, Z.; Shu, Y.; Liu, X.; Li, S.; et al. Genome-wide identification of lncRNAs during rice seed development. Genes 2020, 11, 243. [Google Scholar] [CrossRef]
- Guo, R.; Chen, D.; Xiong, C.; Hou, C.; Zheng, Y.; Fu, Z.; Liang, Q.; Diao, Q.; Zhang, L.; Wang, H.; et al. First identification of long non-coding RNAs in fungal parasite Nosema ceranae. Apidologie 2018, 49, 660–670. [Google Scholar] [CrossRef]
- Shang, Y.; Feng, Y.; Ren, L.; Zhang, X.; Yang, F.; Zhang, C.; Guo, Y. Genome-wide analysis of long noncoding RNAs and their association in regulating the metamorphosis of the Sarcophaga peregrina (Diptera: Sarcophagidae). PLoS Negl. Trop. Dis. 2023, 17, e0011411. [Google Scholar] [CrossRef]
- Belavilas-Trovas, A.; Tastsoglou, S.; Dong, S.; Kefi, M.; Tavadia, M.; Mathiopoulos, K.D.; Dimopoulos, G. Long non-coding RNAs regulate Aedes aegypti vector competence for Zika virus and reproduction. PLoS Pathog. 2023, 19, e1011440. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.X.; Andoh, V.; Chen, L. Multi-omics study and ncRNA regulation of anti-BmNPV in silkworms, Bombyx mori: An update. Front Microbiol. 2023, 14, 1123448. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Zhao, X.; Liu, Q.; Li, C.; Graves-Deal, R.; Cao, Z.; Singh, B.; Franklin, J.L.; Wang, J.; Hu, H.; et al. lncRNA MIR100HG-derived miR-100 and miR-125b mediate cetuximab resistance via Wnt/β-catenin signaling. Nat. Med. 2017, 23, 1331–1341. [Google Scholar] [CrossRef] [PubMed]
- Zafar, J.; Huang, J.; Xu, X.; Jin, F. Analysis of long non-coding RNA-mediated regulatory networks of Plutella xylostella in response to Metarhizium anisopliae infection. Insects 2022, 13, 916. [Google Scholar] [CrossRef]
- Ling, Y.; Xu, L.; Zhu, L.; Sui, M.; Zheng, Q.; Li, W.; Liu, Y.; Fang, F.; Zhang, X. Identification and analysis of differentially expressed long non-coding RNAs between multiparous and uniparous goat (Capra hircus) ovaries. PLoS ONE 2017, 12, e0183163. [Google Scholar] [CrossRef]
- Zhou, X.; Cui, J.; Cui, H.; Jiang, N.; Hou, X.; Liu, S.; Gao, P.; Luan, Y.; Meng, J.; Luan, F. Identification of lncRNAs and their regulatory relationships with target genes and corresponding miRNAs in melon response to powdery mildew fungi. Gene 2020, 735, 144403. [Google Scholar] [CrossRef]
- Shilo, B.Z. The regulation and functions of MAPK pathways in Drosophila. Methods 2014, 68, 151–159. [Google Scholar] [CrossRef]
- Towarnicki, S.G.; Youngson, N.A.; Corley, S.M.; St John, J.C.; Melvin, R.G.; Turner, N.; Morris, M.J.; Ballard, J.W.O. Ancestral dietary change alters the development of Drosophila larvae through MAPK signalling. Fly 2022, 16, 299–311. [Google Scholar] [CrossRef]
- Frappaolo, A.; Karimpour-Ghahnavieh, A.; Cesare, G.; Sechi, S.; Fraschini, R.; Vaccari, T.; Giansanti, M.G. GOLPH3 protein controls organ growth by interacting with TOR signaling proteins in Drosophila. Cell Death Dis. 2022, 13, 1003. [Google Scholar] [CrossRef] [PubMed]
- McMenamin, A.J.; Daughenbaugh, K.F.; Parekh, F.; Pizzorno, M.C.; Flenniken, M.L. Honey bee and bumble bee antiviral defense. Viruses 2018, 10, 395. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Wu, C.; Ding, X.; Li, W.; Xue, L. Toll signaling promotes JNK-dependent apoptosis in Drosophila. Cell Div. 2020, 15, 7. [Google Scholar] [CrossRef]
- Guo, Z.; Kang, S.; Sun, D.; Gong, L.; Zhou, J.; Qin, J.; Guo, L.; Zhu, L.; Bai, Y.; Ye, F.; et al. MAPK-dependent hormonal signaling plasticity contributes to overcoming Bacillus thuringiensis toxin action in an insect host. Nat. Commun. 2020, 11, 3003. [Google Scholar] [CrossRef]
- Broughton, S.J.; Piper, M.D.; Ikeya, T.; Bass, T.M.; Jacobson, J.; Driege, Y.; Martinez, P.; Hafen, E.; Withers, D.J.; Leevers, S.J. Longer lifespan, altered metabolism, and stress resistance in Drosophila from ablation of cells making insulin-like ligands. Proc. Natl. Acad. Sci. USA 2005, 102, 3105–3110. [Google Scholar] [CrossRef] [PubMed]
- Schwenke, R.A.; Lazzaro, B.P.; Wolfner, M.F. Reproduction-immunity trade-offs in insects. Annu. Rev. Entomol. 2016, 61, 239–256. [Google Scholar] [CrossRef] [PubMed]
- McCormack, S.; Yadav, S.; Shokal, U.; Kenney, E.; Cooper, D.; Eleftherianos, I. The insulin receptor substrate Chico regulates antibacterial immune function in Drosophila. Immun. Ageing 2016, 13, 15. [Google Scholar] [CrossRef]
- Wang, C.; Feng, T.; Wan, Q.; Kong, Y.; Yuan, L. miR-124 controls Drosophila behavior and is required for neural development. Int. J. Dev. Neurosci. 2014, 38, 105–112. [Google Scholar] [CrossRef]
- Yuan, L.; Ren, X.; Zheng, Y.; Qian, J.; Xu, L.; Sun, M. MiR-315 is required for neural development and represses the expression of dFMR1 in Drosophila melanogaster. Biochem. Biophys. Res. Commun. 2020, 525, 469–476. [Google Scholar] [CrossRef]
- Sen, R.; Ghosal, S.; Das, S.; Balti, S.; Chakrabarti, J. Competing endogenous RNA: The key to posttranscriptional regulation. Sci. World J. 2014, 2014, 896206. [Google Scholar] [CrossRef]
- Zhang, S.; Wang, N.; Ma, Q.; Fan, F.; Ma, X. LncRNA TUG1 acts as a competing endogenous RNA to mediate CTGF expression by sponging miR-133b in myocardial fibrosis after myocardial infarction. Cell Biol. Int. 2021, 45, 2534–2543. [Google Scholar] [CrossRef] [PubMed]
- Jiang, N.; Cui, J.; Shi, Y.; Yang, G.; Zhou, X.; Hou, X.; Meng, J.; Luan, Y. Tomato lncRNA23468 functions as a competing endogenous RNA to modulate NBS-LRR genes by decoying miR482b in the tomato-Phytophthora infestans interaction. Hortic Res. 2019, 6, 28. [Google Scholar] [CrossRef] [PubMed]
- Birben, E.; Sahiner, U.M.; Sackesen, C.; Erzurum, S.; Kalayci, O. Oxidative stress and antioxidant defense. World Allergy Organ. J. 2012, 5, 9–19. [Google Scholar] [CrossRef] [PubMed]
- Braun, B.C.; Halaski, N.; Painer, J.; Krause, E.; Jewgenow, K. The antioxidative enzyme SOD2 is important for physiological persistence of corpora lutea in lynxes. Sci Rep. 2020, 10, 3681. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Hou, M.; Qiu, Y.; Zhao, B.; Nie, H.; Su, S. Changes in antioxidant enzymes activity and metabolomic profiles in the guts of honey bee (Apis mellifera) larvae infected with Ascosphaera apis. Insects 2020, 11, 419. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Tower, J. FLP recombinase-mediated induction of Cu/Zn-superoxide dismutase transgene expression can extend the life span of adult Drosophila melanogaster flies. Mol. Cell. Biol. 1999, 19, 216–228. [Google Scholar] [CrossRef]
- Li, H.M.; Buczkowski, G.; Mittapalli, O.; Xie, J.; Wu, J.; Westerman, R.; Schemerhorn, B.J.; Murdock, L.L.; Pittendrigh, B.R. Transcriptomic profiles of Drosophila melanogaster third instar larval midgut and responses to oxidative stress. Insect Mol. Biol. 2008, 17, 325–339. [Google Scholar] [CrossRef]
- Abbas, R.; Larisch, S. Killing by degradation: Regulation of apoptosis by the ubiquitin-proteasome-system. Cells 2021, 10, 3465. [Google Scholar] [CrossRef]
- Mevissen, T.E.T.; Komander, D. Mechanisms of deubiquitinase specificity and regulation. Annu. Rev. Biochem. 2006, 86, 159–192. [Google Scholar] [CrossRef]
- McBride, W.H.; Iwamoto, K.S.; Syljuasen, R.; Pervan, M.; Pajonk, F. The role of the ubiquitin/proteasome system in cellular responses to radiation. Oncogene 2003, 22, 5755–5773. [Google Scholar] [CrossRef]
- Choi, B.; Lim, C.; Lee, H.; Lee, J.; Kim, J.; Chung, C.; Cho, K. Neuroprotective effects of linear ubiquitin E3 ligase against aging-induced DNA damage and amyloid β neurotoxicity in the brain of Drosophila melanogaster. Biochem. Biophys. Res. Commun. 2022, 637, 196–202. [Google Scholar] [CrossRef]
- Kanoh, H.; Tong, L.L.; Kuraishi, T.; Suda, Y.; Momiuchi, Y.; Shishido, F.; Kurata, S. Genome-wide RNAi screening implicates the E3 ubiquitin ligase Sherpa in mediating innate immune signaling by Toll in Drosophila adults. Sci. Signal. 2015, 8, ra107. [Google Scholar] [CrossRef]
- Chen, D.; Du, Y.; Chen, H.; Fan, Y.; Fan, X.; Zhu, Z.; Wang, J.; Xiong, C.; Zheng, Y.; Hou, C.; et al. Comparative identification of microRNAs in Apis cerana cerana workers’ midguts in responseto Nosema ceranae invasion. Insects 2019, 10, 258. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Luo, H.; Bu, D.; Zhao, G.; Yu, K.; Zhang, C.; Liu, Y.; Chen, R.; Zhao, Y. Utilizing sequence intrinsic composition to classify protein-coding and long non-coding transcripts. Nucleic Acids Res. 2013, 41, e166. [Google Scholar] [CrossRef] [PubMed]
- Kong, L.; Zhang, Y.; Ye, Z.Q.; Liu, X.Q.; Zhao, S.Q.; Wei, L.; Gao, G. CPC: Assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res. 2007, 35, W345–W349. [Google Scholar] [CrossRef]
- Wu, Y.; Wei, B.; Liu, H.; Li, T.; Rayner, S. MiRPara: A SVM-based software tool for prediction of most probable microRNA coding regions in genome scale sequences. BMC Bioinform. 2011, 12, 107. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. EdgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Stewart, G.; Sage, A.; Enfield, K.; Marshall, E.A.; Cohn, D.E.; Lam, W.L. Deregulation of a cis-acting lncRNA in non-small cell lung cancer may control HMGA1 expression. Front. Genet. 2021, 11, 615378. [Google Scholar] [CrossRef]
- Chen, N.; Guo, D.; Xu, Q.; Yang, M.; Wang, D.; Peng, M.; Ding, Y.; Wang, S.; Zhou, J. Long non-coding RNA FEZF1-AS1 facilitates cell proliferation and migration in colorectal carcinoma. Oncotarget 2016, 7, 11271–11283. [Google Scholar] [CrossRef]
- Tafer, H.; Hofacker, I. RNAplex: A fast tool for RNA-RNA interaction search. Bioinformatics 2008, 24, 2657–2663. [Google Scholar] [CrossRef]
- Wang, L.X.; Wan, C.; Dong, Z.B.; Wang, B.H.; Liu, H.Y.; Li, Y. Integrative analysis of long noncoding RNA (lncRNA), microRNA (miRNA) and mRNA expression and construction of a competing endogenous RNA (ceRNA) network in metastatic melanoma. Med. Sci. Monit. 2019, 25, 2896–2907. [Google Scholar] [CrossRef]
- Sadeghi, M.; Bahrami, A.; Hasankhani, A.; Kioumarsi, H.; Nouralizadeh, R.; Abdulkareem, S.A.; Ghafouri, F.; Barkema, H.W. lncRNA-miRNA-mRNA ceRNA network involved in sheep prolificacy: An integrated approach. Genes 2022, 13, 1295. [Google Scholar] [CrossRef] [PubMed]
- Krüger, J.; Rehmsmeier, M. RNAhybrid: microRNA target prediction easy, fast and flexible. Nucleic Acids Res. 2006, 34, W451–W454. [Google Scholar] [CrossRef] [PubMed]
- Kropinski, A.M.; Clokie, M.R. Methods in molecular biology. Introduction. Methods Mol. Biol. 2009, 502, xiii–xxii. [Google Scholar]
- Allen, E.; Xie, Z.; Gustafson, A.M.; Carrington, J.C. MicroRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 2005, 121, 207–221. [Google Scholar] [CrossRef] [PubMed]
- Smoot, M.E.; Ono, K.; Ruscheinski, J.; Wang, P.L.; Ideker, T. Cytoscape 2.8: New features for data integration and network visualization. Bioinformatics 2011, 27, 431–432. [Google Scholar] [CrossRef] [PubMed]
- Guo, R.; Geng, S.; Zheng, Y.; Fu, Z.; Wang, H.; Du, Y.; Tong, X.; Zhao, H.; Chen, D. Differential expression analysis of long non-coding RNAs during the developmental process of Apis mellifera ligustica worker’s midgut. J. China Agric. Univ. 2018, 51, 3600–3613. (In Chinese) [Google Scholar]
- Wang, Y.; Duan, R.; Zhang, J. Differentiating collagens based on mitochondrion 12SrRNA gene. Food Chem. 2017, 234, 139–143. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]


| Pathway | AcCK1 vs. AcT1 | AcCK2 vs. AcT2 | AcCK3 vs. AcT3 | |
|---|---|---|---|---|
| Cellular immune pathways | Apoptosis | 1 | 1 | 0 |
| Apoptosis-fly | 0 | 1 | ||
| Phagosome | 1 | 1 | 0 | |
| Fc-γ-Receptor-Mediated Phagocytosis | 1 | 0 | 0 | |
| Autophagy-animal | 1 | 0 | 0 | |
| Melanogenesis | 0 | 1 | 0 | |
| Necroptosis | 0 | 1 | 0 | |
| Humoral immune pathways | cAMP signaling pathway | 1 | 1 | 0 |
| MAPK signaling pathway | 0 | 1 | 0 | |
| Toll and Imd signaling pathway | 0 | 1 | 0 | |
| TNF signaling pathway | 0 | 1 | 0 | |
| MAPK signaling pathway-fly | 0 | 1 | 1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, R.; Wang, S.; Guo, S.; Fan, X.; Zang, H.; Gao, X.; Jing, X.; Liu, Z.; Na, Z.; Zou, P.; et al. Regulatory Roles of Long Non-Coding RNAs Relevant to Antioxidant Enzymes and Immune Responses of Apis cerana Larvae Following Ascosphaera apis Invasion. Int. J. Mol. Sci. 2023, 24, 14175. https://doi.org/10.3390/ijms241814175
Guo R, Wang S, Guo S, Fan X, Zang H, Gao X, Jing X, Liu Z, Na Z, Zou P, et al. Regulatory Roles of Long Non-Coding RNAs Relevant to Antioxidant Enzymes and Immune Responses of Apis cerana Larvae Following Ascosphaera apis Invasion. International Journal of Molecular Sciences. 2023; 24(18):14175. https://doi.org/10.3390/ijms241814175
Chicago/Turabian StyleGuo, Rui, Siyi Wang, Sijia Guo, Xiaoxue Fan, He Zang, Xuze Gao, Xin Jing, Zhitan Liu, Zhihao Na, Peiyuan Zou, and et al. 2023. "Regulatory Roles of Long Non-Coding RNAs Relevant to Antioxidant Enzymes and Immune Responses of Apis cerana Larvae Following Ascosphaera apis Invasion" International Journal of Molecular Sciences 24, no. 18: 14175. https://doi.org/10.3390/ijms241814175
APA StyleGuo, R., Wang, S., Guo, S., Fan, X., Zang, H., Gao, X., Jing, X., Liu, Z., Na, Z., Zou, P., & Chen, D. (2023). Regulatory Roles of Long Non-Coding RNAs Relevant to Antioxidant Enzymes and Immune Responses of Apis cerana Larvae Following Ascosphaera apis Invasion. International Journal of Molecular Sciences, 24(18), 14175. https://doi.org/10.3390/ijms241814175






