Uncovering the Novel Role of NR1D1 in Regulating BNIP3-Mediated Mitophagy in Ulcerative Colitis
Abstract
:1. Introduction
2. Results
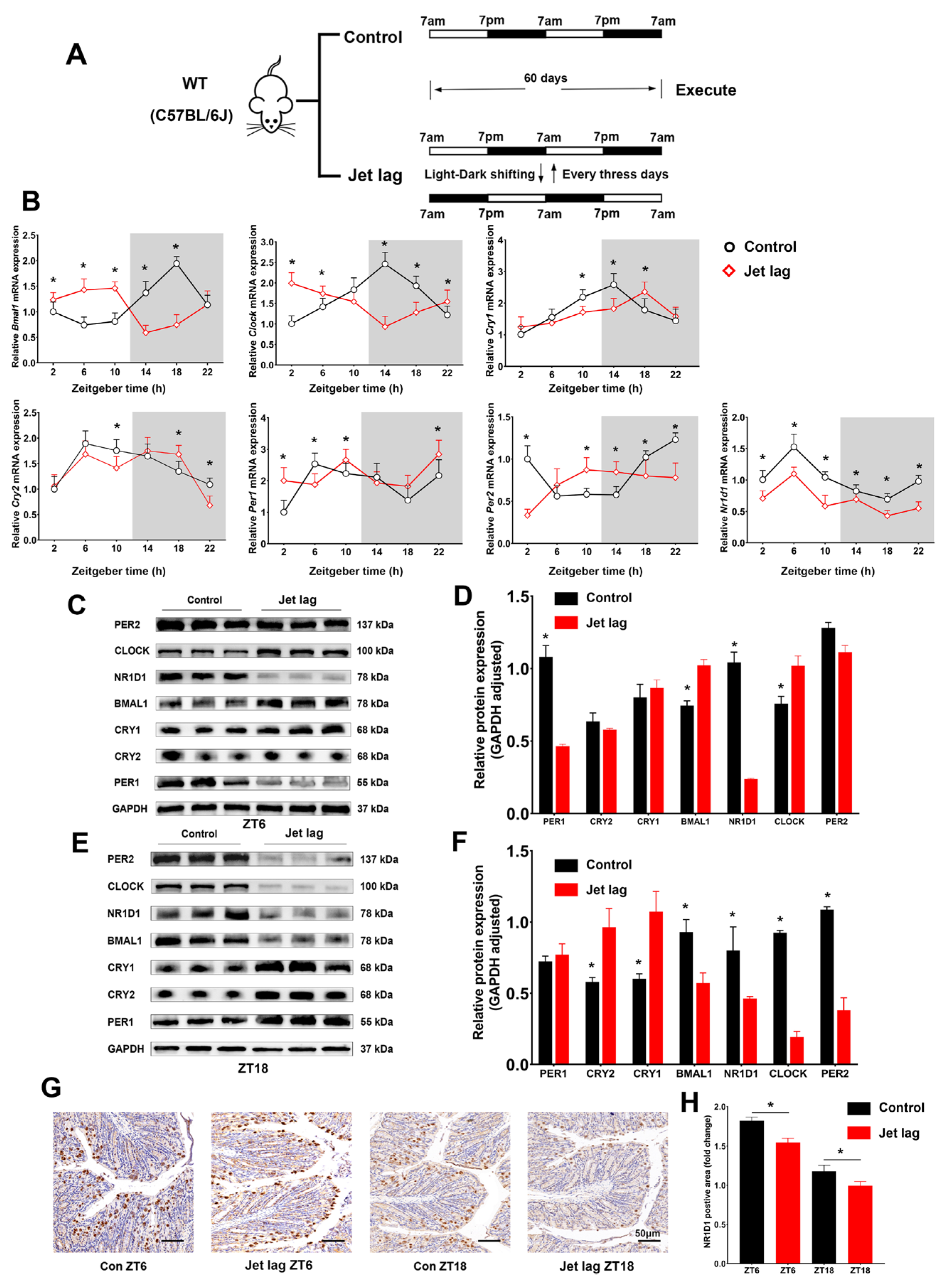
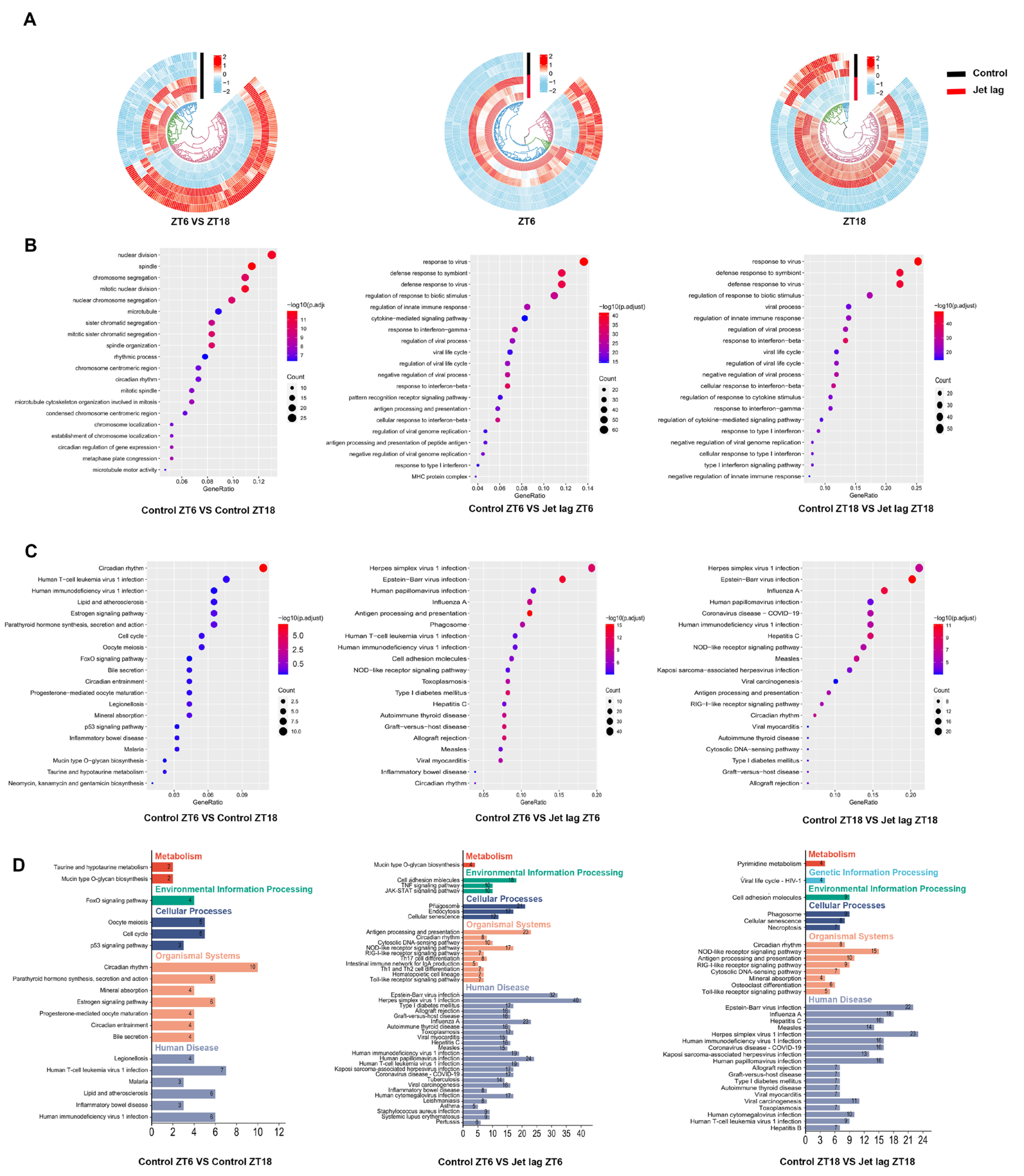
2.1. Circadian Clock Gene Expression Patterns in IECs Altered by Jet Lag and Ulcerative Colitis
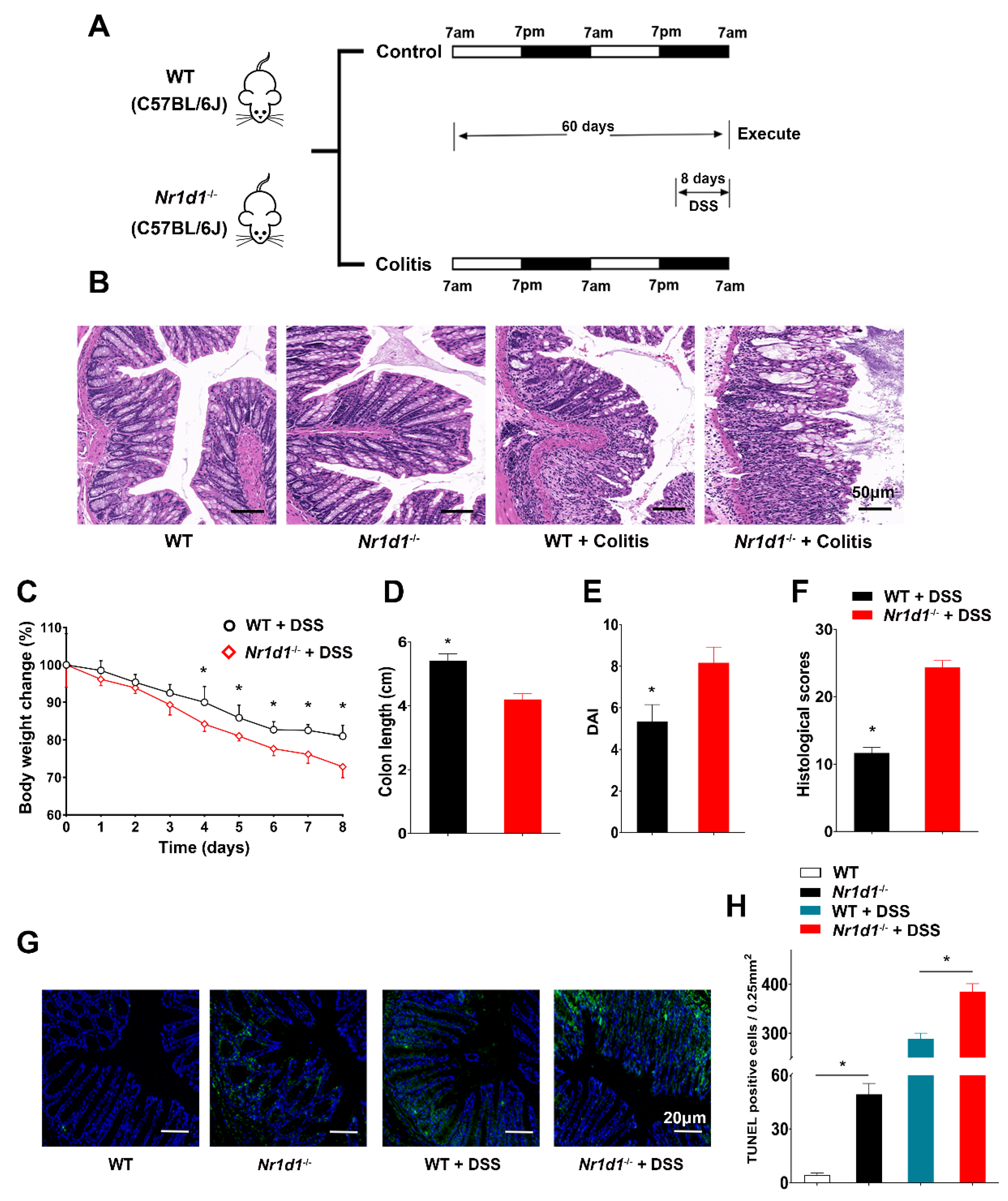
2.2. Downregulation of NR1D1 Impairs Immune Homeostasis in IECs and Exacerbates DSS-Induced Colitis in Mice
2.3. NR1D1 Has Been Identified as a Transcription Factor That Regulates the Expression of BNIP3
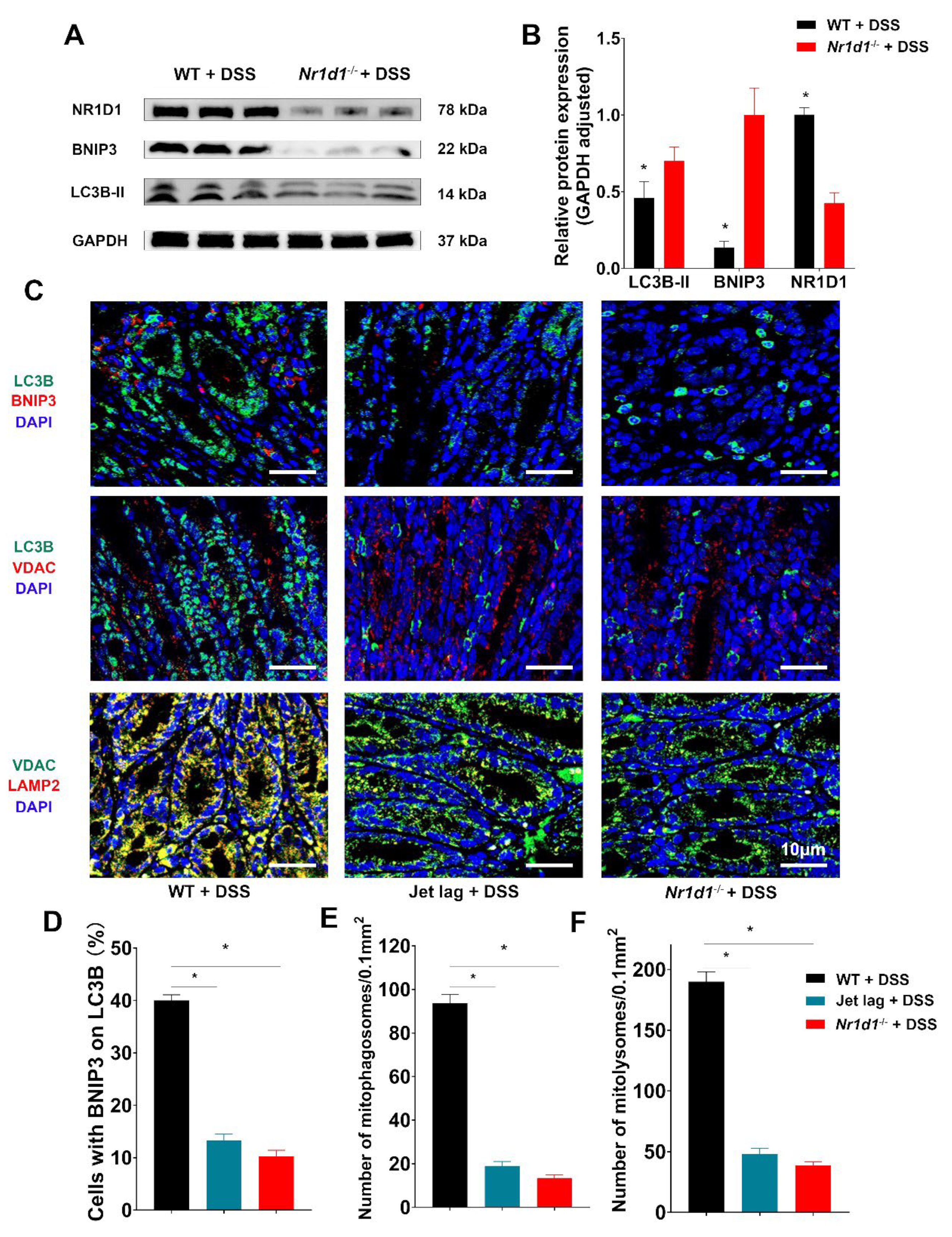
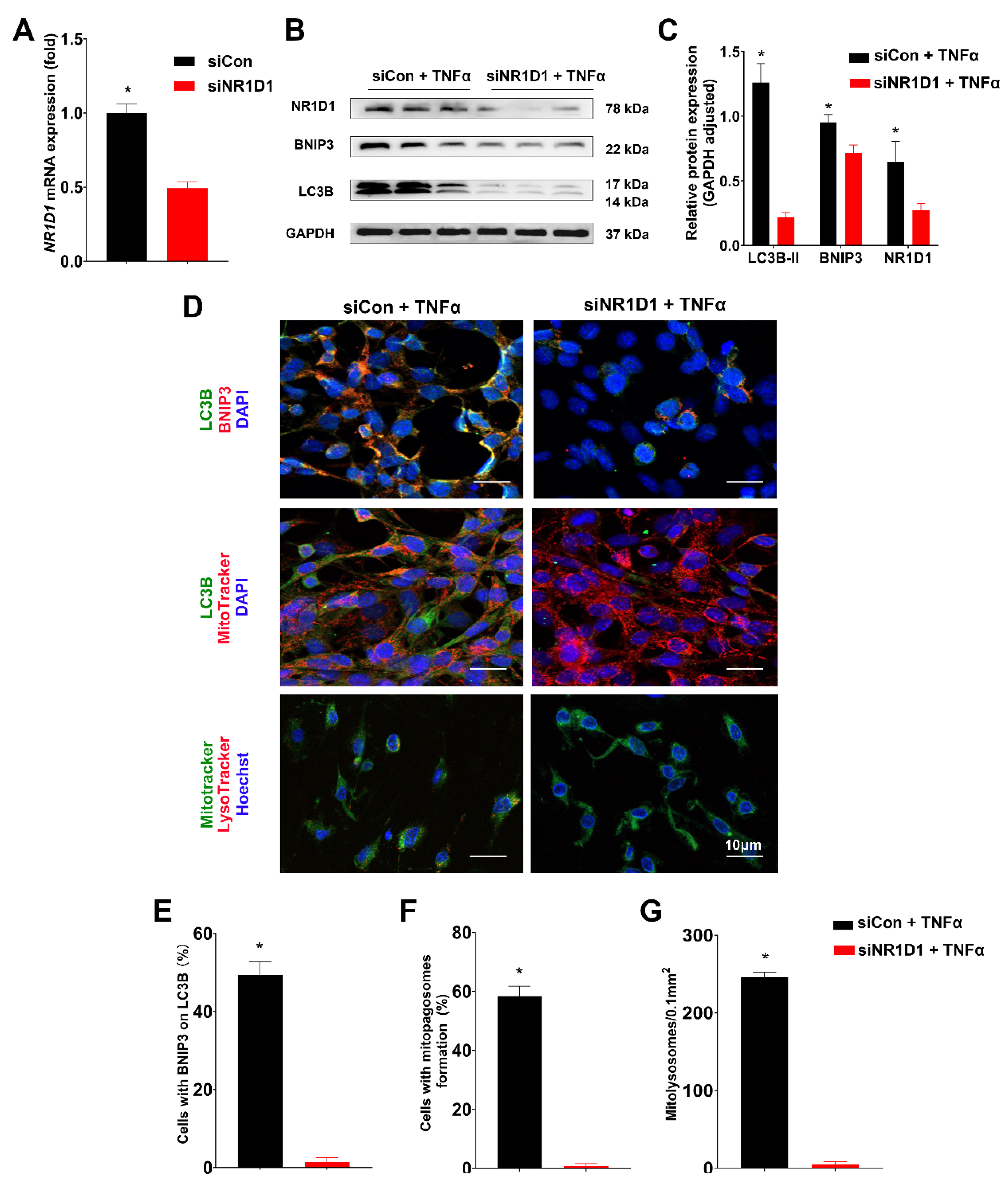
2.4. Nr1d1 KO Leads to Impaired Mitophagy
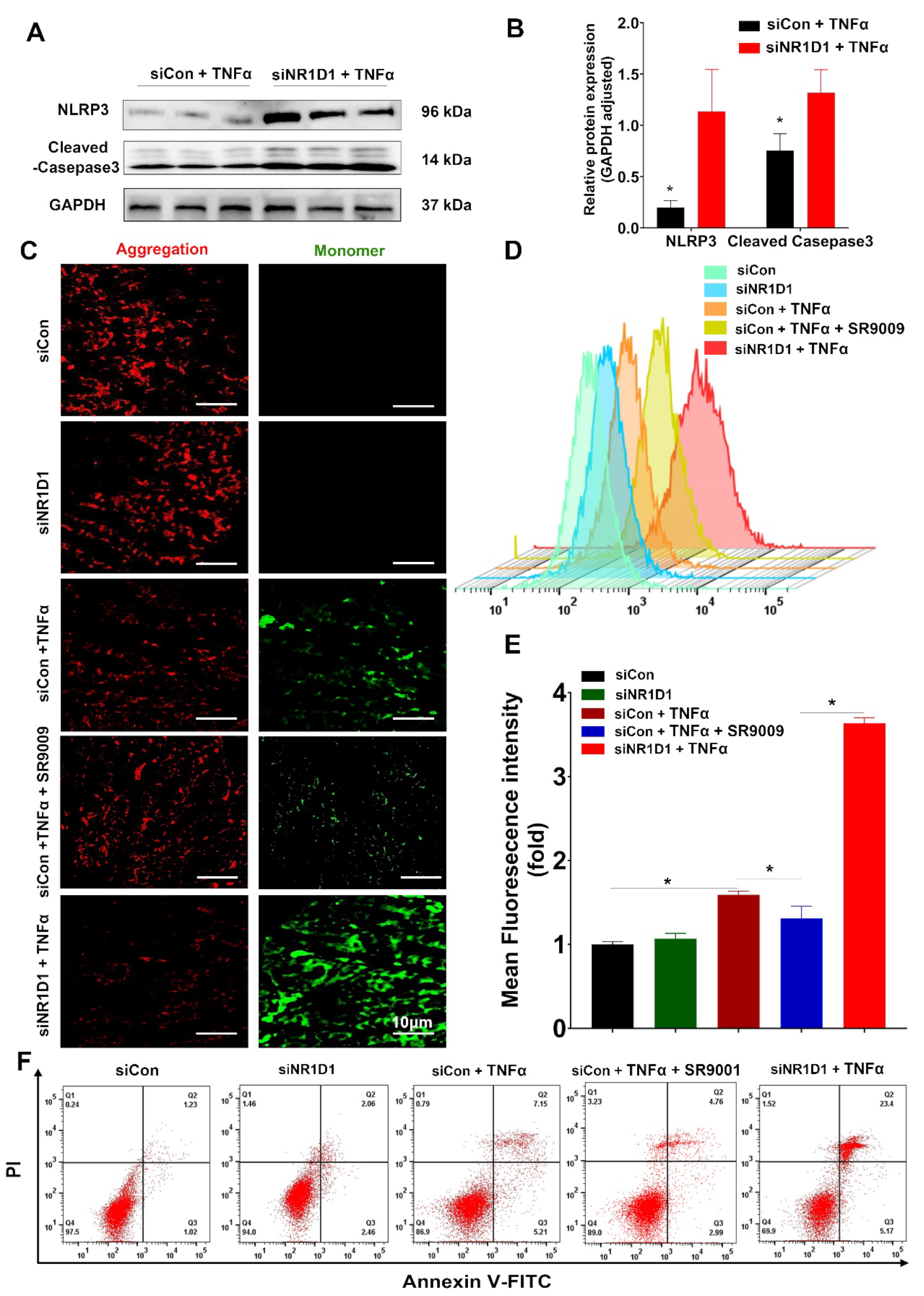
2.5. NR1D1 Modulation by SR9009 Mitigates TNFα-Induced Mitochondrial Injury and Cell Apoptosis
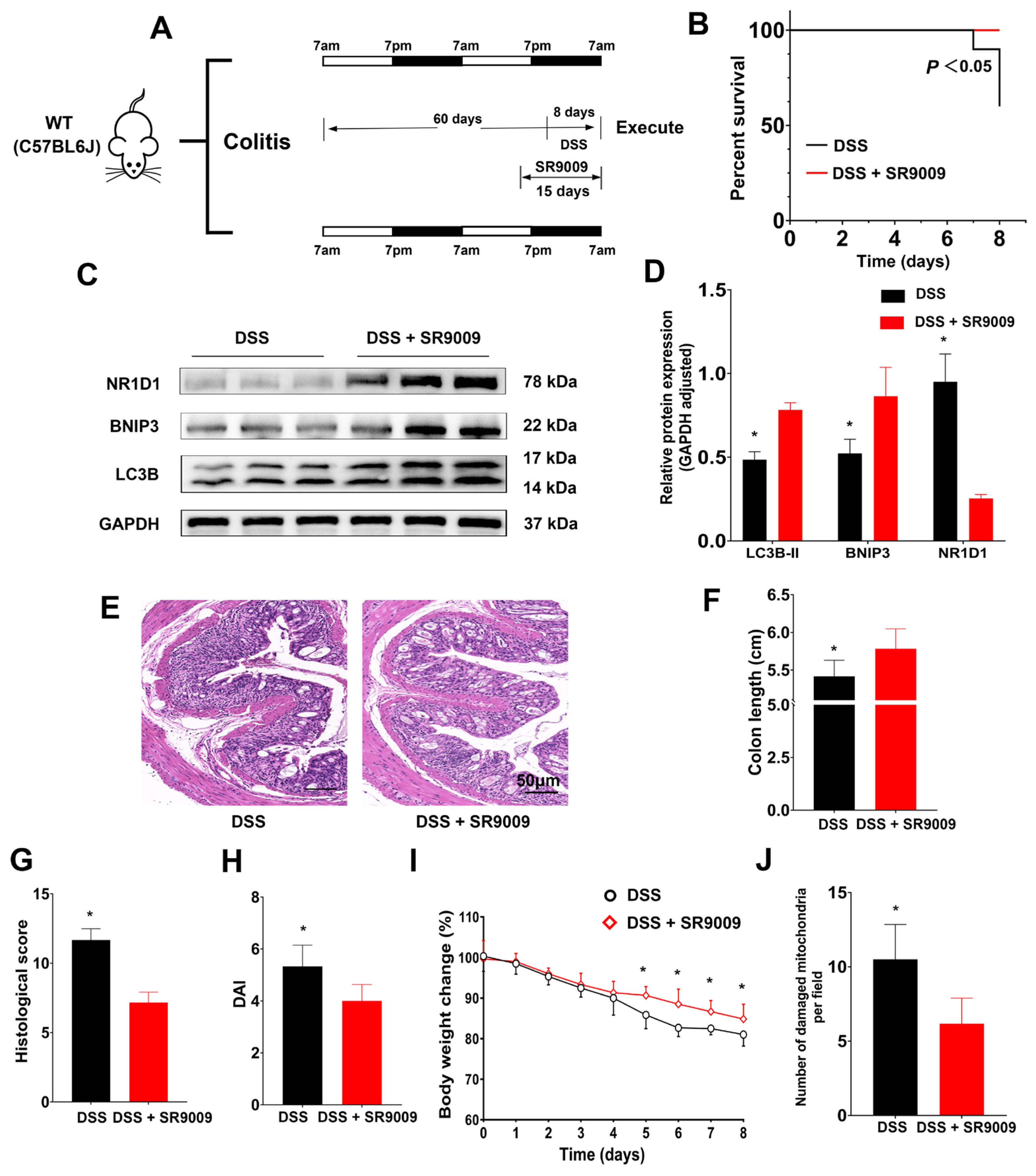
2.6. Amelioration of DSS-Induced Colitis in Mice by the NR1D1 Agonist SR9009
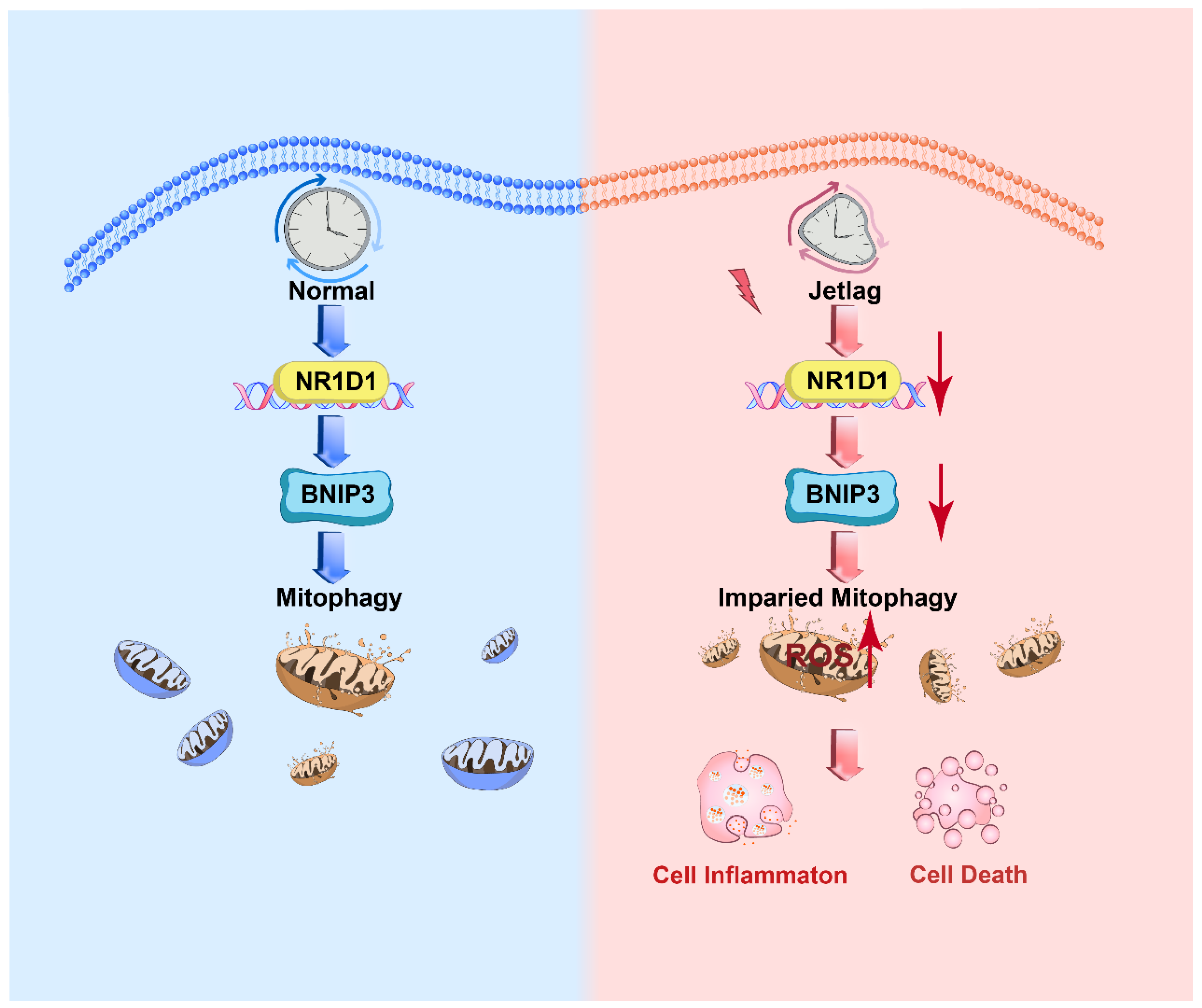
3. Discussion
4. Materials and Methods
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Torres, J.; Mehandru, S.; Colombel, J.-F.; Peyrin-Biroulet, L. Crohn’s Disease. Lancet 2017, 389, 1741–1755. [Google Scholar] [CrossRef]
- Kobayashi, T.; Siegmund, B.; Le Berre, C.; Wei, S.C.; Ferrante, M.; Shen, B.; Bernstein, C.N.; Danese, S.; Peyrin-Biroulet, L.; Hibi, T. Ulcerative Colitis. Nat. Rev. Dis. Primers 2020, 6, 74. [Google Scholar] [CrossRef] [PubMed]
- Rieder, F.; Karrasch, T.; Ben-Horin, S.; Schirbel, A.; Ehehalt, R.; Wehkamp, J.; de Haar, C.; Velin, D.; Latella, G.; Scaldaferri, F.; et al. Results of the 2nd Scientific Workshop of the ECCO (III): Basic Mechanisms of Intestinal Healing. J. Crohns Colitis 2012, 6, 373–385. [Google Scholar] [CrossRef] [PubMed]
- Villablanca, E.J.; Selin, K.; Hedin, C.R.H. Mechanisms of Mucosal Healing: Treating Inflammatory Bowel Disease without Immunosuppression? Nature reviews. Gastroenterol. Hepatol. 2022, 19, 493–507. [Google Scholar] [CrossRef] [PubMed]
- Vancamelbeke, M.; Vermeire, S. The Intestinal Barrier: A Fundamental Role in Health and Disease. Expert Rev. Gastroenterol. Hepatol. 2017, 11, 821–834. [Google Scholar] [CrossRef]
- Peterson, L.W.; Artis, D. Intestinal Epithelial Cells: Regulators of Barrier Function and Immune Homeostasis. Nat. Rev. Immunol. 2014, 14, 141–153. [Google Scholar] [CrossRef]
- Allaire, J.M.; Crowley, S.M.; Law, H.T.; Chang, S.-Y.; Ko, H.-J.; Vallance, B.A. The Intestinal Epithelium: Central Coordinator of Mucosal Immunity. Trends Immunol. 2018, 39, 677–696. [Google Scholar] [CrossRef]
- Nowarski, R.; Jackson, R.; Flavell, R.A. The Stromal Intervention: Regulation of Immunity and Inflammation at the Epithelial-Mesenchymal Barrier. Cell 2017, 168, 362–375. [Google Scholar] [CrossRef]
- Martini, E.; Krug, S.M.; Siegmund, B.; Neurath, M.F.; Becker, C. Mend Your Fences: The Epithelial Barrier and Its Relationship With Mucosal Immunity in Inflammatory Bowel Disease. Cell. Mol. Gastroenterol. Hepatol. 2017, 4, 33–46. [Google Scholar] [CrossRef]
- Rath, E.; Moschetta, A.; Haller, D. Mitochondrial Function—Gatekeeper of Intestinal Epithelial Cell Homeostasis. Nat. Rev. Gastroenterol. Hepatol. 2018, 15, 497–516. [Google Scholar] [CrossRef]
- Orrenius, S.; Zhivotovsky, B.; Nicotera, P. Regulation of Cell Death: The Calcium-Apoptosis Link. Nat. Rev. Mol. Cell Biol. 2003, 4, 552–565. [Google Scholar] [CrossRef] [PubMed]
- Bock, F.J.; Tait, S.W.G. Mitochondria as Multifaceted Regulators of Cell Death. Nat. Rev. Mol. Cell Biol. 2020, 21, 85–100. [Google Scholar] [CrossRef] [PubMed]
- West, A.P.; Brodsky, I.E.; Rahner, C.; Woo, D.K.; Erdjument-Bromage, H.; Tempst, P.; Walsh, M.C.; Choi, Y.; Shadel, G.S.; Ghosh, S. TLR Signalling Augments Macrophage Bactericidal Activity through Mitochondrial ROS. Nature 2011, 472, 476–480. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.-L.; Thiyagarajan, V.; Shen, P.-C.; Mathew, D.C.; Lin, K.-Y.; Liao, J.-W.; Hseu, Y.-C. Anti-EMT Properties of CoQ0 Attributed to PI3K/AKT/NFKB/MMP-9 Signaling Pathway through ROS-Mediated Apoptosis. J. Exp. Clin. Cancer Res. 2019, 38, 186. [Google Scholar] [CrossRef]
- Sosnovski, K.E.; Braun, T.; Amir, A.; Moshel, D.; BenShoshan, M.; VanDussen, K.L.; Levhar, N.; Abbas-Egbariya, H.; Beider, K.; Ben-Yishay, R.; et al. GATA6-AS1 Regulates Intestinal Epithelial Mitochondrial Functions, and Its Reduced Expression Is Linked to Intestinal Inflammation and Less Favorable Disease Course in Ulcerative Colitis (UC). J. Crohns Colitis 2023, 17, 960–971. [Google Scholar] [CrossRef]
- Patankar, J.V.; Becker, C. Cell Death in the Gut Epithelium and Implications for Chronic Inflammation. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 543–556. [Google Scholar] [CrossRef]
- Moehlman, A.T.; Youle, R.J. Mitochondrial Quality Control and Restraining Innate Immunity. Annu. Rev. Cell Dev. Biol. 2020, 36, 265–289. [Google Scholar] [CrossRef]
- Harrington, J.S.; Ryter, S.W.; Plataki, M.; Price, D.R.; Choi, A.M.K. Mitochondria in Health, Disease, and Ageing. Physiol. Rev. 2023, 103, 2349–2422. [Google Scholar] [CrossRef]
- Li, Y.; Zheng, W.; Lu, Y.; Zheng, Y.; Pan, L.; Wu, X.; Yuan, Y.; Shen, Z.; Ma, S.; Zhang, X.; et al. BNIP3L/NIX-Mediated Mitophagy: Molecular Mechanisms and Implications for Human Disease. Cell Death Dis. 2021, 13, 14. [Google Scholar] [CrossRef]
- Liu, B.; Gulati, A.S.; Cantillana, V.; Henry, S.C.; Schmidt, E.A.; Daniell, X.; Grossniklaus, E.; Schoenborn, A.A.; Sartor, R.B.; Taylor, G.A. Irgm1-Deficient Mice Exhibit Paneth Cell Abnormalities and Increased Susceptibility to Acute Intestinal Inflammation. Am. J. Physiol. Gastrointest. Liver Physiol. 2013, 305, G573–G584. [Google Scholar] [CrossRef]
- Lassen, K.G.; Kuballa, P.; Conway, K.L.; Patel, K.K.; Becker, C.E.; Peloquin, J.M.; Villablanca, E.J.; Norman, J.M.; Liu, T.-C.; Heath, R.J.; et al. Atg16L1 T300A Variant Decreases Selective Autophagy Resulting in Altered Cytokine Signaling and Decreased Antibacterial Defense. Proc. Natl. Acad. Sci. USA 2014, 111, 7741–7746. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Shen, J.; Ran, Z. Emerging Views of Mitophagy in Immunity and Autoimmune Diseases. Autophagy 2020, 16, 3–17. [Google Scholar] [CrossRef] [PubMed]
- Bass, J.; Takahashi, J.S. Circadian Integration of Metabolism and Energetics. Science 2010, 330, 1349–1354. [Google Scholar] [CrossRef]
- Panda, S. Circadian Physiology of Metabolism. Science 2016, 354, 1008–1015. [Google Scholar] [CrossRef]
- Allada, R.; Bass, J. Circadian Mechanisms in Medicine. N. Engl. J. Med. 2021, 384, 550–561. [Google Scholar] [CrossRef] [PubMed]
- Woldt, E.; Sebti, Y.; Solt, L.A.; Duhem, C.; Lancel, S.; Eeckhoute, J.; Hesselink, M.K.C.; Paquet, C.; Delhaye, S.; Shin, Y.; et al. Rev-Erb-α Modulates Skeletal Muscle Oxidative Capacity by Regulating Mitochondrial Biogenesis and Autophagy. Nat. Med. 2013, 19, 1039–1046. [Google Scholar] [CrossRef]
- Sulli, G.; Rommel, A.; Wang, X.; Kolar, M.J.; Puca, F.; Saghatelian, A.; Plikus, M.V.; Verma, I.M.; Panda, S. Pharmacological Activation of REV-ERBs Is Lethal in Cancer and Oncogene-Induced Senescence. Nature 2018, 553, 351–355. [Google Scholar] [CrossRef]
- Sun, L.-Y.; Lyu, Y.-Y.; Zhang, H.-Y.; Shen, Z.; Lin, G.-Q.; Geng, N.; Wang, Y.-L.; Huang, L.; Feng, Z.-H.; Guo, X.; et al. Nuclear Receptor NR1D1 Regulates Abdominal Aortic Aneurysm Development by Targeting the Mitochondrial Tricarboxylic Acid Cycle Enzyme Aconitase-2. Circulation 2022, 146, 1591–1609. [Google Scholar] [CrossRef]
- Teng, F.; Goc, J.; Zhou, L.; Chu, C.; Shah, M.A.; Eberl, G.; Sonnenberg, G.F. A Circadian Clock Is Essential for Homeostasis of Group 3 Innate Lymphoid Cells in the Gut. Sci. Immunol. 2019, 4, eaax1215. [Google Scholar] [CrossRef]
- Oh, S.K.; Kim, D.; Kim, K.; Boo, K.; Yu, Y.S.; Kim, I.S.; Jeon, Y.; Im, S.-K.; Lee, S.-H.; Lee, J.M.; et al. RORα Is Crucial for Attenuated Inflammatory Response to Maintain Intestinal Homeostasis. Proc. Natl. Acad. Sci. USA 2019, 116, 21140–21149. [Google Scholar] [CrossRef]
- Parekh, P.J.; Oldfield Iv, E.C.; Challapallisri, V.; Ware, J.C.; Johnson, D.A. Sleep Disorders and Inflammatory Disease Activity: Chicken or the Egg? Am. J. Gastroenterol. 2015, 110, 484–488. [Google Scholar] [CrossRef] [PubMed]
- Chassaing, B.; Aitken, J.D.; Malleshappa, M.; Vijay-Kumar, M. Dextran Sulfate Sodium (DSS)-Induced Colitis in Mice. Curr. Protoc. Immunol. 2014, 104, 15.25.1–15.25.14. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Cen, L.; Zhang, X.; Tang, C.; Chen, Y.; Zhang, Y.; Yu, M.; Lu, C.; Li, M.; Li, S.; et al. MPST Deficiency Promotes Intestinal Epithelial Cell Apoptosis and Aggravates Inflammatory Bowel Disease via AKT. Redox Biol. 2022, 56, 102469. [Google Scholar] [CrossRef]
- Bayrer, J.R.; Wang, H.; Nattiv, R.; Suzawa, M.; Escusa, H.S.; Fletterick, R.J.; Klein, O.D.; Moore, D.D.; Ingraham, H.A. LRH-1 Mitigates Intestinal Inflammatory Disease by Maintaining Epithelial Homeostasis and Cell Survival. Nat. Commun. 2018, 9, 4055. [Google Scholar] [CrossRef] [PubMed]
- Palikaras, K.; Lionaki, E.; Tavernarakis, N. Mechanisms of Mitophagy in Cellular Homeostasis, Physiology and Pathology. Nat. Cell Biol. 2018, 20, 1013–1022. [Google Scholar] [CrossRef]
- Ordás, I.; Eckmann, L.; Talamini, M.; Baumgart, D.C.; Sandborn, W.J. Ulcerative Colitis. Lancet 2012, 380, 1606–1619. [Google Scholar] [CrossRef]
- Meyer, N.; Harvey, A.G.; Lockley, S.W.; Dijk, D.-J. Circadian Rhythms and Disorders of the Timing of Sleep. Lancet 2022, 400, 1061–1078. [Google Scholar] [CrossRef]
- Schmitt, K.; Grimm, A.; Dallmann, R.; Oettinghaus, B.; Restelli, L.M.; Witzig, M.; Ishihara, N.; Mihara, K.; Ripperger, J.A.; Albrecht, U.; et al. Circadian Control of DRP1 Activity Regulates Mitochondrial Dynamics and Bioenergetics. Cell Metab. 2018, 27, 657–666.e5. [Google Scholar] [CrossRef]
- Boulinguiez, A.; Duhem, C.; Mayeuf-Louchart, A.; Pourcet, B.; Sebti, Y.; Kondratska, K.; Montel, V.; Delhaye, S.; Thorel, Q.; Beauchamp, J.; et al. NR1D1 Controls Skeletal Muscle Calcium Homeostasis through Myoregulin Repression. JCI Insight 2022, 7, e153584. [Google Scholar] [CrossRef]
- Weintraub, Y.; Cohen, S.; Chapnik, N.; Ben-Tov, A.; Yerushalmy-Feler, A.; Dotan, I.; Tauman, R.; Froy, O. Clock Gene Disruption Is an Initial Manifestation of Inflammatory Bowel Diseases. Clin. Gastroenterol. Hepatol. 2020, 18, 115–122.e1. [Google Scholar] [CrossRef]
- Liu, X.; Yu, R.; Zhu, L.; Hou, X.; Zou, K. Bidirectional Regulation of Circadian Disturbance and Inflammation in Inflammatory Bowel Disease. Inflamm. Bowel Dis. 2017, 23, 1741–1751. [Google Scholar] [CrossRef]
- Taleb, Z.; Carmona-Alcocer, V.; Stokes, K.; Haireek, M.; Wang, H.; Collins, S.M.; Khan, W.I.; Karpowicz, P. BMAL1 Regulates the Daily Timing of Colitis. Front. Cell. Infect. MicroBiol. 2022, 12, 773413. [Google Scholar] [CrossRef]
- Zhou, Z.; Lin, Y.; Gao, L.; Yang, Z.; Wang, S.; Wu, B. Circadian Pharmacological Effects of Berberine on Chronic Colitis in Mice: Role of the Clock Component Rev-Erbα. Biochem. Pharmacol. 2020, 172, 113773. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.-D.; Zhao, R.-F.; Zheng, G.; Ling, F.-M.; Li, J.-R.; Xu, M.-Y.; Guo, D.; Zhang, Q.-L.; Li, S.; Zhu, L.-R. The Association between Disruption of the Circadian Rhythm and Aggravation of Colitis in Mice. Gastroenterol. Rep. 2022, 10, goac028. [Google Scholar] [CrossRef] [PubMed]
- Nagata, S. Apoptosis and Clearance of Apoptotic Cells. Annu. Rev. Immunol. 2018, 36, 489–517. [Google Scholar] [CrossRef] [PubMed]
- Kojetin, D.J.; Burris, T.P. REV-ERB and ROR Nuclear Receptors as Drug Targets. Nat. Rev. Drug Discov. 2014, 13, 197–216. [Google Scholar] [CrossRef] [PubMed]
- Negoro, H.; Kanematsu, A.; Doi, M.; Suadicani, S.O.; Matsuo, M.; Imamura, M.; Okinami, T.; Nishikawa, N.; Oura, T.; Matsui, S.; et al. Involvement of Urinary Bladder Connexin43 and the Circadian Clock in Coordination of Diurnal Micturition Rhythm. Nat. Commun. 2012, 3, 809. [Google Scholar] [CrossRef]
- Chandra, V.; Bhagyaraj, E.; Nanduri, R.; Ahuja, N.; Gupta, P. NR1D1 Ameliorates Mycobacterium Tuberculosis Clearance through Regulation of Autophagy. Autophagy 2015, 11, 1987–1997. [Google Scholar] [CrossRef]
- Negoro, H.; Okinami, T.; Kanematsu, A.; Imamura, M.; Tabata, Y.; Ogawa, O. Role of Rev-Erbα Domains for Transactivation of the Connexin43 Promoter with Sp1. FEBS Lett. 2013, 587, 98–103. [Google Scholar] [CrossRef]
- Kuo, W.-T.; Zuo, L.; Odenwald, M.A.; Madha, S.; Singh, G.; Gurniak, C.B.; Abraham, C.; Turner, J.R. The Tight Junction Protein ZO-1 Is Dispensable for Barrier Function but Critical for Effective Mucosal Repair. Gastroenterology 2021, 161, 1924–1939. [Google Scholar] [CrossRef]
- Kuwada, T.; Shiokawa, M.; Kodama, Y.; Ota, S.; Kakiuchi, N.; Nannya, Y.; Yamazaki, H.; Yoshida, H.; Nakamura, T.; Matsumoto, S.; et al. Identification of an Anti-Integrin Avβ6 Autoantibody in Patients With Ulcerative Colitis. Gastroenterology 2021, 160, 2383–2394.e21. [Google Scholar] [CrossRef]
- Pourcet, B.; Zecchin, M.; Ferri, L.; Beauchamp, J.; Sitaula, S.; Billon, C.; Delhaye, S.; Vanhoutte, J.; Mayeuf-Louchart, A.; Thorel, Q.; et al. Nuclear Receptor Subfamily 1 Group D Member 1 Regulates Circadian Activity of NLRP3 Inflammasome to Reduce the Severity of Fulminant Hepatitis in Mice. Gastroenterology 2018, 154, 1449–1464.e20. [Google Scholar] [CrossRef]
- Yue, J.; He, J.; Wei, Y.; Shen, K.; Wu, K.; Yang, X.; Liu, S.; Zhang, C.; Yang, H. Decreased Expression of Rev-Erbα in the Epileptic Foci of Temporal Lobe Epilepsy and Activation of Rev-Erbα Have Anti-Inflammatory and Neuroprotective Effects in the Pilocarpine Model. J. Neuroinflammation 2020, 17, 43. [Google Scholar] [CrossRef]
- Ma, H.; Zhong, W.; Jiang, Y.; Fontaine, C.; Li, S.; Fu, J.; Olkkonen, V.M.; Staels, B.; Yan, D. Increased Atherosclerotic Lesions in LDL Receptor Deficient Mice with Hematopoietic Nuclear Receptor Rev-Erbα Knock- Down. J. Am. Heart Assoc. 2013, 2, e000235. [Google Scholar] [CrossRef]
- Sitaula, S.; Billon, C.; Kamenecka, T.M.; Solt, L.A.; Burris, T.P. Suppression of Atherosclerosis by Synthetic REV-ERB Agonist. Biochem. Biophys. Res. Commun. 2015, 460, 566–571. [Google Scholar] [CrossRef]
- Papagiannakopoulos, T.; Bauer, M.R.; Davidson, S.M.; Heimann, M.; Subbaraj, L.; Bhutkar, A.; Bartlebaugh, J.; Vander Heiden, M.G.; Jacks, T. Circadian Rhythm Disruption Promotes Lung Tumorigenesis. Cell Metab. 2016, 24, 324–331. [Google Scholar] [CrossRef] [PubMed]
- Gulbransen, B.D.; Bashashati, M.; Hirota, S.A.; Gui, X.; Roberts, J.A.; MacDonald, J.A.; Muruve, D.A.; McKay, D.M.; Beck, P.L.; Mawe, G.M.; et al. Activation of Neuronal P2X7 Receptor-Pannexin-1 Mediates Death of Enteric Neurons during Colitis. Nat. Med. 2012, 18, 600–604. [Google Scholar] [CrossRef] [PubMed]
- Van der Sluis, M.; De Koning, B.A.E.; De Bruijn, A.C.J.M.; Velcich, A.; Meijerink, J.P.P.; Van Goudoever, J.B.; Büller, H.A.; Dekker, J.; Van Seuningen, I.; Renes, I.B.; et al. Muc2-Deficient Mice Spontaneously Develop Colitis, Indicating That MUC2 Is Critical for Colonic Protection. Gastroenterology 2006, 131, 117–129. [Google Scholar] [CrossRef]
- Gracz, A.D.; Puthoff, B.J.; Magness, S.T. Identification, Isolation, and Culture of Intestinal Epithelial Stem Cells from Murine Intestine. Methods Mol. Biol. 2012, 879, 89–107. [Google Scholar] [CrossRef] [PubMed]


| Gene | Sequence |
|---|---|
| Mouse Nr1d1 | F:ACTTCCCACCATCACCTACTG |
| R:GGGGAGCTATCATCACTGAGA | |
| Mouse Clock | F:ATGGTGTTTACCGTAAGCTGTAG |
| R:CTCGCGTTACCAGGAAGCAT | |
| Mouse Bmal1 | F:ACAGTCAGATTGAAAAGAGGCG |
| R:GCCATCCTTAGCACGGTGAG | |
| Mouse Cry1 | F:CACTGGTTCCGAAAGGGACTC |
| R:CTGAAGCAAAAATCGCCACCT | |
| Mouse Cry2 | F:CACTGGTTCCGCAAAGGACTA |
| R:CCACGGGTCGAGGATGTAGA | |
| Mouse Per1 | F:GAATTGGAGCATATCACATCCGA |
| R:CCCGAAACACATCCCGTTTG | |
| Mouse Per2 | F:CTCCAGCGGAAACGAGAACTG |
| R:TTGGCAGACTGCTCACTACTG | |
| Mouse Gapdh | F:AGGTCGGTGTGAACGGATTTG |
| R:GGGGTCGTTGATGGCAACA | |
| Human NR1D1 | F:TGGACTCCAACAACAACACAG |
| R:GATGGTGGGAAGTAGGTGGG | |
| Human GAPDH | F:GGAGCGAGATCCCTCCAAAAT |
| R:GGCTGTTGTCATACTTCTCATGG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, Y.; Li, J.; Li, S.; Cheng, Y.; Fu, X.; Li, J.; Zhu, L. Uncovering the Novel Role of NR1D1 in Regulating BNIP3-Mediated Mitophagy in Ulcerative Colitis. Int. J. Mol. Sci. 2023, 24, 14222. https://doi.org/10.3390/ijms241814222
Chen Y, Li J, Li S, Cheng Y, Fu X, Li J, Zhu L. Uncovering the Novel Role of NR1D1 in Regulating BNIP3-Mediated Mitophagy in Ulcerative Colitis. International Journal of Molecular Sciences. 2023; 24(18):14222. https://doi.org/10.3390/ijms241814222
Chicago/Turabian StyleChen, Yidong, Junrong Li, Shuang Li, Yiyu Cheng, Xiaoyu Fu, Jiamin Li, and Liangru Zhu. 2023. "Uncovering the Novel Role of NR1D1 in Regulating BNIP3-Mediated Mitophagy in Ulcerative Colitis" International Journal of Molecular Sciences 24, no. 18: 14222. https://doi.org/10.3390/ijms241814222
APA StyleChen, Y., Li, J., Li, S., Cheng, Y., Fu, X., Li, J., & Zhu, L. (2023). Uncovering the Novel Role of NR1D1 in Regulating BNIP3-Mediated Mitophagy in Ulcerative Colitis. International Journal of Molecular Sciences, 24(18), 14222. https://doi.org/10.3390/ijms241814222






