A Distinct Microglial Cell Population Expressing Both CD86 and CD206 Constitutes a Dominant Type and Executes Phagocytosis in Two Mouse Models of Retinal Degeneration
Abstract
1. Introduction
2. Results
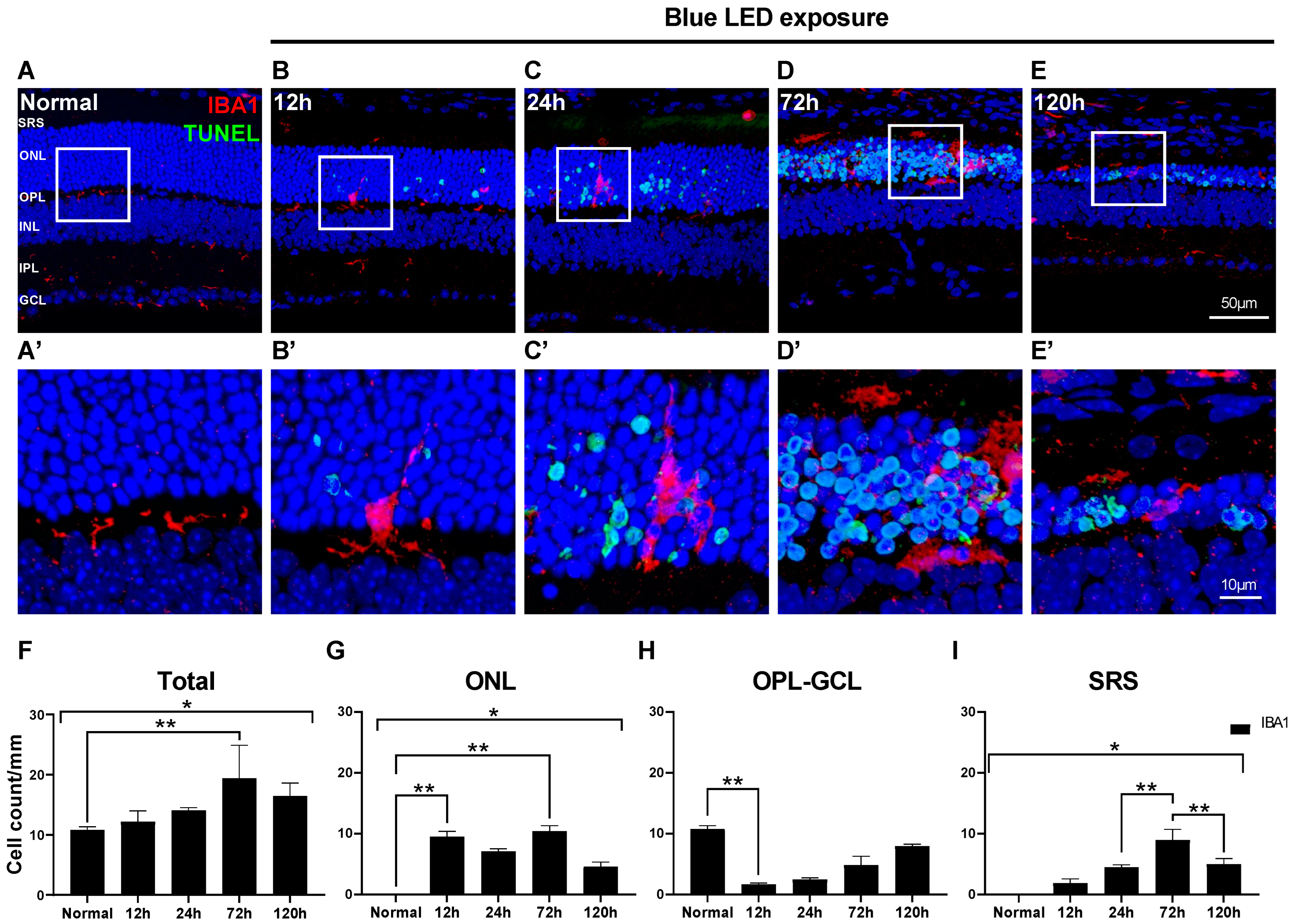
2.1. Changes in Morphology and Distribution of Microglial Cells in Blue LED-Induced RD
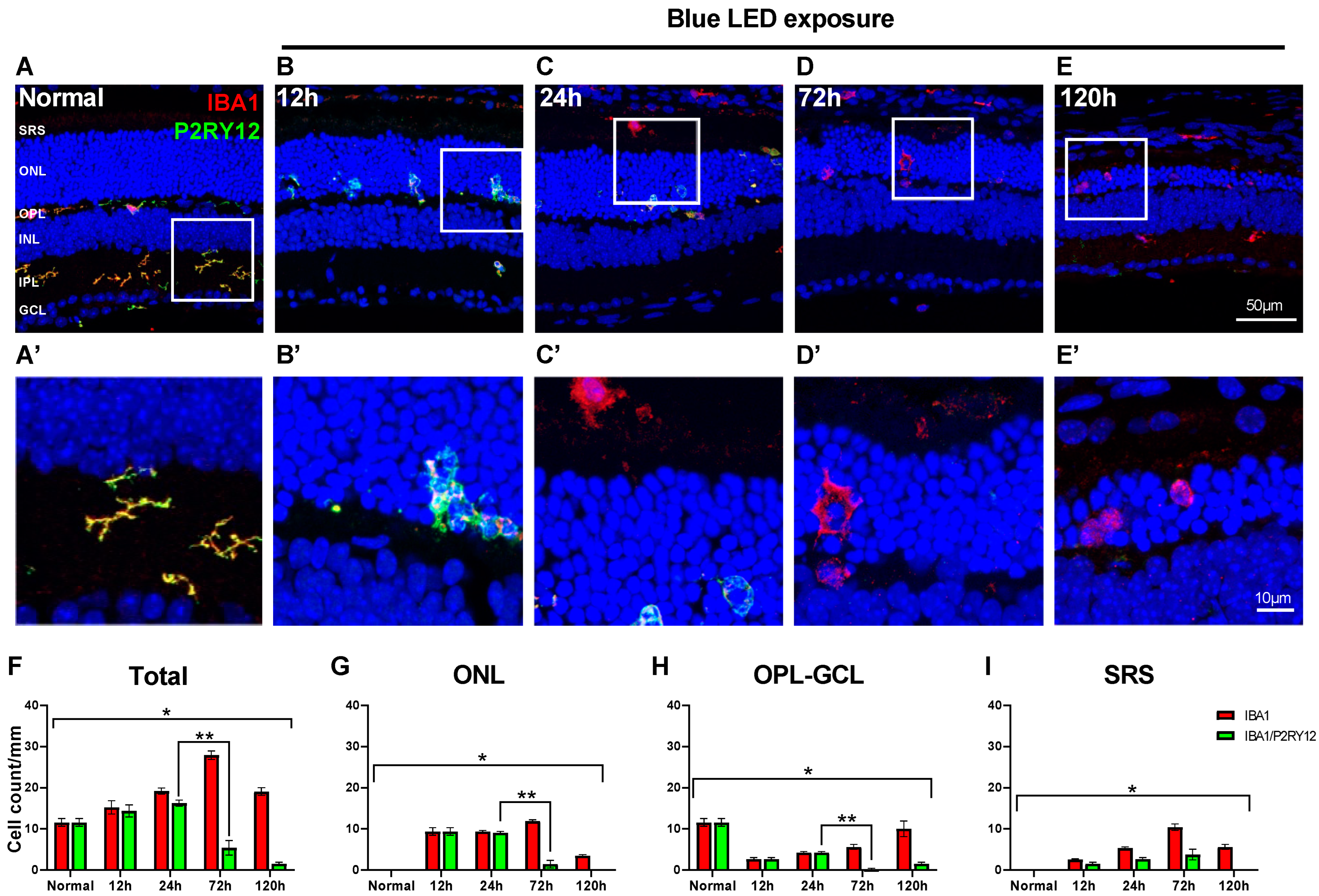
2.2. Innate Microglial Cell Response in Blue LED-Induced RD
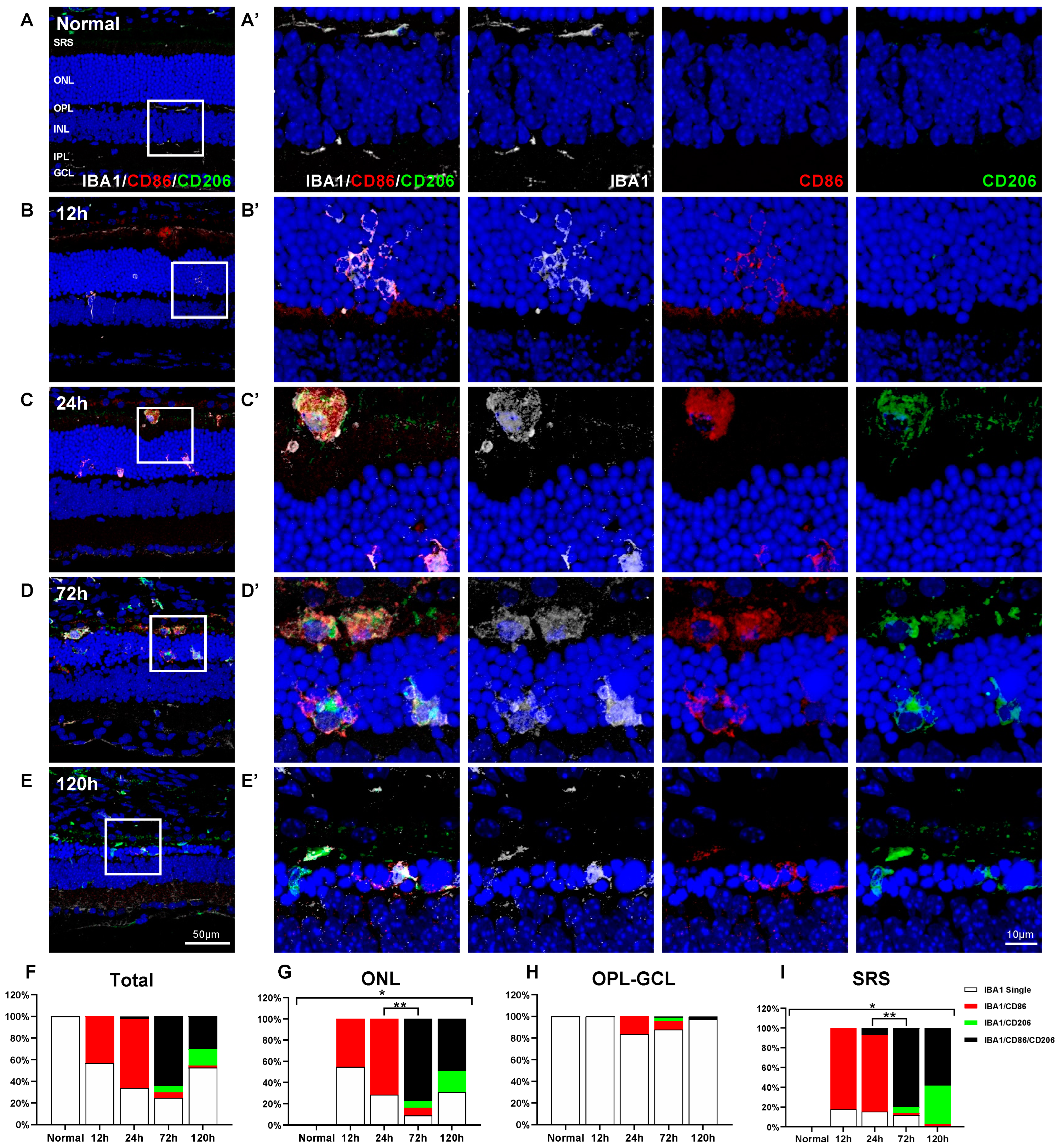
2.3. Spatiotemporal Change of CD86 and CD206 Expression in Retinal Microglial Cells in Blue LED-Induced RD
2.4. Characterization of IBA1/CD86/CD206-Triple-Labeled Microglial Population in Blue LED-Induced RD
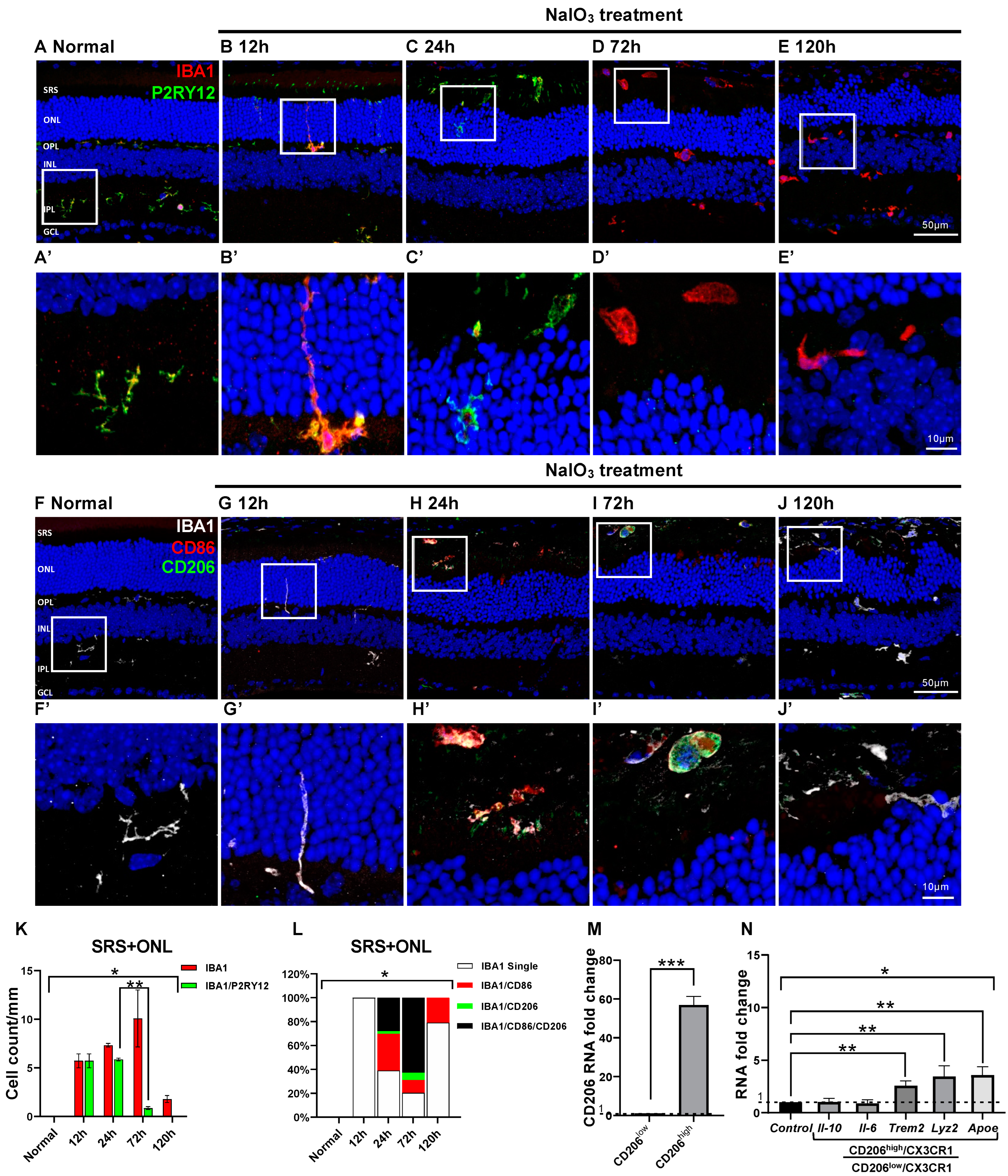
2.5. Microglial Cell Response and Character in the NaIO3-Induced RD
3. Discussion
4. Materials and Methods
4.1. Animals
4.2. Animal Models
4.3. Tissue Preparation
4.4. Terminal Deoxynucleotidyl Transferase dUTP Nick End Labeling (TUNEL) Assay
4.5. Immunohistochemistry
4.6. Fluorescence-Activated Cell Sorting (FACS)
4.7. One-Step Real-Time Polymerase Chain Reaction (qRT-PCR)
4.8. Immuno-Electron Microscopy (EM)
4.9. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lannes, N.; Eppler, E.; Etemad, S.; Yotovski, P.; Filgueira, L. Microglia at center stage: A comprehensive review about the versatile and unique residential macrophages of the central nervous system. Oncotarget 2017, 8, 114393–114413. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.; Guo, L.; Cordeiro, M.F. Retinal and Brain Microglia in Multiple Sclerosis and Neurodegeneration. Cells 2021, 10, 1507. [Google Scholar] [CrossRef]
- Ransohoff, R.M.; Brown, M.A. Innate immunity in the central nervous system. J. Clin. Investig. 2012, 122, 1164–1171. [Google Scholar] [CrossRef]
- Wake, H.; Fields, R.D. Physiological function of microglia. Neuron Glia Biol. 2011, 7, 1–3. [Google Scholar] [CrossRef][Green Version]
- Kierdorf, K.; Prinz, M. Microglia in steady state. J. Clin. Investig. 2017, 127, 3201–3209. [Google Scholar] [CrossRef]
- Kabba, J.A.; Xu, Y.; Christian, H.; Ruan, W.; Chenai, K.; Xiang, Y.; Zhang, L.; Saavedra, J.M.; Pang, T. Microglia: Housekeeper of the Central Nervous System. Cell Mol. Neurobiol. 2018, 38, 53–71. [Google Scholar] [CrossRef]
- Bachiller, S.; Jiménez-Ferrer, I.; Paulus, A.; Yang, Y.; Swanberg, M.; Deierborg, T.; Boza-Serrano, A. Microglia in Neurological Diseases: A Road Map to Brain-Disease Dependent-Inflammatory Response. Front. Cell. Neurosci. 2018, 12, 488. [Google Scholar] [CrossRef] [PubMed]
- Hansen, D.V.; Hanson, J.E.; Sheng, M. Microglia in Alzheimer’s disease. J. Cell Biol. 2018, 217, 459–472. [Google Scholar] [CrossRef]
- Leng, F.; Edison, P. Neuroinflammation and microglial activation in Alzheimer disease: Where do we go from here? Nat. Rev. Neurol. 2021, 17, 157–172. [Google Scholar] [CrossRef] [PubMed]
- Cherry, J.D.; Olschowka, J.A.; O’Banion, M.K. Neuroinflammation and M2 microglia: The good, the bad, and the inflamed. J. Neuroinflamm. 2014, 11, 98. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wang, J.; Wang, J.; Yang, B.; Weng, Q.; He, Q. Targeting Microglia and Macrophages: A Potential Treatment Strategy for Multiple Sclerosis. Front. Pharmacol. 2019, 10, 286. [Google Scholar] [CrossRef]
- Anwar, S.; Pons, V.; Rivest, S. Microglia Purinoceptor P2Y6: An Emerging Therapeutic Target in CNS Diseases. Cells 2020, 9, 1595. [Google Scholar] [CrossRef] [PubMed]
- Wong, W.L.; Su, X.; Li, X.; Cheung, C.M.; Klein, R.; Cheng, C.Y.; Wong, T.Y. Global prevalence of age-related macular degeneration and disease burden projection for 2020 and 2040: A systematic review and meta-analysis. Lancet Glob. Health 2014, 2, e106–e116. [Google Scholar] [CrossRef]
- Pennington, K.L.; DeAngelis, M.M. Epidemiology of age-related macular degeneration (AMD): Associations with cardiovascular disease phenotypes and lipid factors. Eye Vis. 2016, 3, 34. [Google Scholar] [CrossRef] [PubMed]
- Kauppinen, A.; Paterno, J.J.; Blasiak, J.; Salminen, A.; Kaarniranta, K. Inflammation and its role in age-related macular degeneration. Cell Mol. Life Sci. 2016, 73, 1765–1786. [Google Scholar] [CrossRef]
- Parmeggiani, F.; Romano, M.R.; Costagliola, C.; Semeraro, F.; Incorvaia, C.; D’Angelo, S.; Perri, P.; De Palma, P.; De Nadai, K.; Sebastiani, A. Mechanism of inflammation in age-related macular degeneration. Mediat. Inflamm. 2012, 2012, 546786. [Google Scholar] [CrossRef] [PubMed]
- Patel, M.; Chan, C.C. Immunopathological aspects of age-related macular degeneration. Semin. Immunopathol. 2008, 30, 97–110. [Google Scholar] [CrossRef]
- Levy, O.; Calippe, B.; Lavalette, S.; Hu, S.J.; Raoul, W.; Dominguez, E.; Housset, M.; Paques, M.; Sahel, J.A.; Bemelmans, A.P.; et al. Apolipoprotein E promotes subretinal mononuclear phagocyte survival and chronic inflammation in age-related macular degeneration. EMBO Mol. Med. 2015, 7, 211–226. [Google Scholar] [CrossRef]
- Gupta, N.; Brown, K.E.; Milam, A.H. Activated microglia in human retinitis pigmentosa, late-onset retinal degeneration, and age-related macular degeneration. Exp. Eye Res. 2003, 76, 463–471. [Google Scholar] [CrossRef]
- Combadiere, C.; Feumi, C.; Raoul, W.; Keller, N.; Rodero, M.; Pezard, A.; Lavalette, S.; Houssier, M.; Jonet, L.; Picard, E.; et al. CX3CR1-dependent subretinal microglia cell accumulation is associated with cardinal features of age-related macular degeneration. J. Clin. Investig. 2007, 117, 2920–2928. [Google Scholar] [CrossRef]
- Makabe, K.; Sugita, S.; Mandai, M.; Futatsugi, Y.; Takahashi, M. Microglia dynamics in retinitis pigmentosa model: Formation of fundus whitening and autofluorescence as an indicator of activity of retinal degeneration. Sci. Rep. 2020, 10, 14700. [Google Scholar] [CrossRef]
- Guo, Y.; Gao, M.; Wan, X.; Li, X.; Wang, Y.; Sun, M.; Li, T.; Jiang, M.; Luo, X.; Sun, X. An improved method for establishment of murine retinal detachment model and its 3D vascular evaluation. Exp. Eye Res. 2020, 193, 107949. [Google Scholar] [CrossRef]
- Chang, S.W.; Kim, H.I.; Kim, G.H.; Park, S.J.; Kim, I.B. Increased Expression of Osteopontin in Retinal Degeneration Induced by Blue Light-Emitting Diode Exposure in Mice. Front. Mol. Neurosci. 2016, 9, 58. [Google Scholar] [CrossRef]
- Zhao, L.; Zabel, M.K.; Wang, X.; Ma, W.; Shah, P.; Fariss, R.N.; Qian, H.; Parkhurst, C.N.; Gan, W.B.; Wong, W.T. Microglial phagocytosis of living photoreceptors contributes to inherited retinal degeneration. EMBO Mol. Med. 2015, 7, 1179–1197. [Google Scholar] [CrossRef]
- Scholz, R.; Sobotka, M.; Caramoy, A.; Stempfl, T.; Moehle, C.; Langmann, T. Minocycline counter-regulates pro-inflammatory microglia responses in the retina and protects from degeneration. J. Neuroinflamm. 2015, 12, 209. [Google Scholar] [CrossRef]
- Lu, Z.; Zhang, H.; Zhang, X.; Gao, Y.; Yin, Z.Q. Lipoxin A4 delays the progression of retinal degeneration via the inhibition of microglial overactivation. Biochem. Biophys. Res. Commun. 2019, 516, 900–906. [Google Scholar] [CrossRef]
- Hu, Z.; Zhang, Y.; Wang, J.; Mao, P.; Lv, X.; Yuan, S.; Huang, Z.; Ding, Y.; Xie, P.; Liu, Q. Knockout of Ccr2 alleviates photoreceptor cell death in rodent retina exposed to chronic blue light. Cell Death Dis. 2016, 7, e2468. [Google Scholar] [CrossRef] [PubMed]
- Okunuki, Y.; Mukai, R.; Pearsall, E.A.; Klokman, G.; Husain, D.; Park, D.H.; Korobkina, E.; Weiner, H.L.; Butovsky, O.; Ksander, B.R.; et al. Microglia inhibit photoreceptor cell death and regulate immune cell infiltration in response to retinal detachment. Proc. Natl. Acad. Sci. USA 2018, 115, E6264–E6273. [Google Scholar] [CrossRef]
- Chong, D.Y.; Boehlke, C.S.; Zheng, Q.D.; Zhang, L.; Han, Y.; Zacks, D.N. Interleukin-6 as a photoreceptor neuroprotectant in an experimental model of retinal detachment. Investig. Ophthalmol. Vis. Sci. 2008, 49, 3193–3200. [Google Scholar] [CrossRef] [PubMed]
- Besirli, C.G.; Chinskey, N.D.; Zheng, Q.D.; Zacks, D.N. Inhibition of retinal detachment-induced apoptosis in photoreceptors by a small peptide inhibitor of the fas receptor. Investig. Ophthalmol. Vis. Sci. 2010, 51, 2177–2184. [Google Scholar] [CrossRef] [PubMed]
- Apte, R.S.; Richter, J.; Herndon, J.; Ferguson, T.A. Macrophages inhibit neovascularization in a murine model of age-related macular degeneration. PLoS Med. 2006, 3, e310. [Google Scholar] [CrossRef]
- Tang, Y.; Le, W. Differential Roles of M1 and M2 Microglia in Neurodegenerative Diseases. Mol. Neurobiol. 2016, 53, 1181–1194. [Google Scholar] [CrossRef]
- Li, J.; Yu, S.; Lu, X.; Cui, K.; Tang, X.; Xu, Y.; Liang, X. The phase changes of M1/M2 phenotype of microglia/macrophage following oxygen-induced retinopathy in mice. Inflamm. Res. 2021, 70, 183–192. [Google Scholar] [CrossRef]
- Jiang, C.-T.; Wu, W.-F.; Deng, Y.-H.; Ge, J.-W. Modulators of microglia activation and polarization in ischemic stroke (Review). Mol. Med. Rep. 2020, 21, 2006–2018. [Google Scholar] [CrossRef]
- Stratoulias, V.; Venero, J.L.; Tremblay, M.-È.; Joseph, B. Microglial subtypes: Diversity within the microglial community. EMBO J. 2019, 38, e101997. [Google Scholar] [CrossRef]
- Zhou, T.; Huang, Z.; Sun, X.; Zhu, X.; Zhou, L.; Li, M.; Cheng, B.; Liu, X.; He, C. Microglia Polarization with M1/M2 Phenotype Changes in rd1 Mouse Model of Retinal Degeneration. Front. Neuroanat. 2017, 11, 77. [Google Scholar] [CrossRef] [PubMed]
- Ransohoff, R.M. A polarizing question: Do M1 and M2 microglia exist? Nat. Neurosci. 2016, 19, 987–991. [Google Scholar] [CrossRef]
- Devanney, N.A.; Stewart, A.N.; Gensel, J.C. Microglia and macrophage metabolism in CNS injury and disease: The role of immunometabolism in neurodegeneration and neurotrauma. Exp. Neurol. 2020, 329, 113310. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Luo, C.; Zhao, J.; Devarajan, G.; Xu, H. Immune regulation in the aging retina. Prog. Retin. Eye Res. 2019, 69, 159–172. [Google Scholar] [CrossRef]
- McMenamin, P.G.; Saban, D.R.; Dando, S.J. Immune cells in the retina and choroid: Two different tissue environments that require different defenses and surveillance. Prog. Retin. Eye Res. 2019, 70, 85–98. [Google Scholar] [CrossRef] [PubMed]
- Ronning, K.E.; Karlen, S.J.; Miller, E.B.; Burns, M.E. Molecular profiling of resident and infiltrating mononuclear phagocytes during rapid adult retinal degeneration using single-cell RNA sequencing. Sci. Rep. 2019, 9, 4858. [Google Scholar] [CrossRef]
- O’Koren, E.G.; Yu, C.; Klingeborn, M.; Wong, A.Y.W.; Prigge, C.L.; Mathew, R.; Kalnitsky, J.; Msallam, R.A.; Silvin, A.; Kay, J.N.; et al. Microglial Function Is Distinct in Different Anatomical Locations during Retinal Homeostasis and Degeneration. Immunity 2019, 50, 723–737. [Google Scholar] [CrossRef]
- Karlen, S.J.; Miller, E.B.; Wang, X.; Levine, E.S.; Zawadzki, R.J.; Burns, M.E. Monocyte infiltration rather than microglia proliferation dominates the early immune response to rapid photoreceptor degeneration. J. Neuroinflamm. 2018, 15, 344. [Google Scholar] [CrossRef]
- Caicedo, A.; Espinosa-Heidmann, D.G.; Pina, Y.; Hernandez, E.P.; Cousins, S.W. Blood-derived macrophages infiltrate the retina and activate Muller glial cells under experimental choroidal neovascularization. Exp. Eye Res. 2005, 81, 38–47. [Google Scholar] [CrossRef] [PubMed]
- Jurga, A.M.; Paleczna, M.; Kuter, K.Z. Overview of General and Discriminating Markers of Differential Microglia Phenotypes. Front. Cell Neurosci. 2020, 14, 198. [Google Scholar] [CrossRef] [PubMed]
- Orihuela, R.; McPherson, C.A.; Harry, G.J. Microglial M1/M2 polarization and metabolic states. Br. J. Pharmacol. 2016, 173, 649–665. [Google Scholar] [CrossRef] [PubMed]
- Wolf, Y.; Yona, S.; Kim, K.W.; Jung, S. Microglia, seen from the CX3CR1 angle. Front. Cell Neurosci. 2013, 7, 26. [Google Scholar] [CrossRef]
- Lambert, J.C.; Ibrahim-Verbaas, C.A.; Harold, D.; Naj, A.C.; Sims, R.; Bellenguez, C.; DeStafano, A.L.; Bis, J.C.; Beecham, G.W.; Grenier-Boley, B.; et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat. Genet. 2013, 45, 1452–1458. [Google Scholar] [CrossRef]
- Deczkowska, A.; Keren-Shaul, H.; Weiner, A.; Colonna, M.; Schwartz, M.; Amit, I. Disease-Associated Microglia: A Universal Immune Sensor of Neurodegeneration. Cell 2018, 173, 1073–1081. [Google Scholar] [CrossRef] [PubMed]
- Anderson, S.R.; Roberts, J.M.; Zhang, J.; Steele, M.R.; Romero, C.O.; Bosco, A.; Vetter, M.L. Developmental Apoptosis Promotes a Disease-Related Gene Signature and Independence from CSF1R Signaling in Retinal Microglia. Cell Rep. 2019, 27, 2002–2013.e5. [Google Scholar] [CrossRef]
- Painter, M.M.; Atagi, Y.; Liu, C.-C.; Rademakers, R.; Xu, H.; Fryer, J.D.; Bu, G. TREM2 in CNS homeostasis and neurodegenerative disease. Mol. Neurodegener 2015, 10, 43. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, C.L.; Koike, M.; Spusta, S.C.; Niemi, E.C.; Yenari, M.; Nakamura, M.C.; Seaman, W.E. A role for TREM2 ligands in the phagocytosis of apoptotic neuronal cells by microglia. J. Neurochem. 2009, 109, 1144–1156. [Google Scholar] [CrossRef] [PubMed]
- Chowers, G.; Cohen, M.; Marks-Ohana, D.; Stika, S.; Eijzenberg, A.; Banin, E.; Obolensky, A. Course of Sodium Iodate-Induced Retinal Degeneration in Albino and Pigmented Mice. Investig. Ophthalmol. Vis. Sci. 2017, 58, 2239–2249. [Google Scholar] [CrossRef] [PubMed]
- Moriguchi, M.; Nakamura, S.; Inoue, Y.; Nishinaka, A.; Nakamura, M.; Shimazawa, M.; Hara, H. Irreversible Photoreceptors and RPE Cells Damage by Intravenous Sodium Iodate in Mice Is Related to Macrophage Accumulation. Investig. Ophthalmol. Vis. Sci. 2018, 59, 3476–3487. [Google Scholar] [CrossRef]
- O’Koren, E.G.; Mathew, R.; Saban, D.R. Fate mapping reveals that microglia and recruited monocyte-derived macrophages are definitively distinguishable by phenotype in the retina. Sci. Rep. 2016, 6, 20636. [Google Scholar] [CrossRef]
- Ramirez, A.I.; de Hoz, R.; Fernandez-Albarral, J.A.; Salobrar-Garcia, E.; Rojas, B.; Valiente-Soriano, F.J.; Aviles-Trigueros, M.; Villegas-Perez, M.P.; Vidal-Sanz, M.; Trivino, A.; et al. Time course of bilateral microglial activation in a mouse model of laser-induced glaucoma. Sci. Rep. 2020, 10, 4890. [Google Scholar] [CrossRef] [PubMed]
- Wieghofer, P.; Hagemeyer, N.; Sankowski, R.; Schlecht, A.; Staszewski, O.; Amann, L.; Gruber, M.; Koch, J.; Hausmann, A.; Zhang, P.; et al. Mapping the origin and fate of myeloid cells in distinct compartments of the eye by single-cell profiling. EMBO J. 2021, 40, e105123. [Google Scholar] [CrossRef]
- Arroba, A.I.; Alcalde-Estevez, E.; García-Ramírez, M.; Cazzoni, D.; de la Villa, P.; Sánchez-Fernández, E.M.; Mellet, C.O.; García Fernández, J.M.; Hernández, C.; Simó, R.; et al. Modulation of microglia polarization dynamics during diabetic retinopathy in db/db mice. Biochim. Biophys. Acta 2016, 1862, 1663–1674. [Google Scholar] [CrossRef]
- Paolicelli, R.C.; Sierra, A.; Stevens, B.; Tremblay, M.E.; Aguzzi, A.; Ajami, B.; Amit, I.; Audinat, E.; Bechmann, I.; Bennett, M.; et al. Microglia states and nomenclature: A field at its crossroads. Neuron 2022, 110, 3458–3483. [Google Scholar] [CrossRef]
- Arroba, A.I.; Valverde, Á.M. Modulation of microglia in the retina: New insights into diabetic retinopathy. Acta Diabetol. 2017, 54, 527–533. [Google Scholar] [CrossRef]
- Yu, C.; Saban, D.R. Identification of a Unique Subretinal Microglia Type in Retinal Degeneration Using Single Cell RNA-Seq. Adv. Exp. Med. Biol. 2019, 1185, 181–186. [Google Scholar] [CrossRef]
- Du, X.; Byrne, E.M.; Chen, M.; Xu, H. Minocycline Inhibits Microglial Activation and Improves Visual Function in a Chronic Model of Age-Related Retinal Degeneration. Biomedicines 2022, 10, 3222. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Diaz, V.; Huang, H. The Role of Interferon Regulatory Factor 1 in Regulating Microglial Activation and Retinal Inflammation. Int. J. Mol. Sci. 2022, 23, 14664. [Google Scholar] [CrossRef]
- Gervois, P.; Lambrichts, I. The Emerging Role of Triggering Receptor Expressed on Myeloid Cells 2 as a Target for Immunomodulation in Ischemic Stroke. Front. Immunol. 2019, 10, 1668. [Google Scholar] [CrossRef] [PubMed]
- Jay, T.R.; von Saucken, V.E.; Landreth, G.E. TREM2 in Neurodegenerative Diseases. Mol. Neurodegener. 2017, 12, 56. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharjee, S.; Zhao, Y.; Dua, P.; Rogaev, E.I.; Lukiw, W.J. microRNA-34a-Mediated Down-Regulation of the Microglial-Enriched Triggering Receptor and Phagocytosis-Sensor TREM2 in Age-Related Macular Degeneration. PLoS ONE 2016, 11, e0150211. [Google Scholar] [CrossRef] [PubMed]
- Saadane, A.; Petrov, A.; Mast, N.; El-Darzi, N.; Dao, T.; Alnemri, A.; Song, Y.; Dunaief, J.L.; Pikuleva, I.A. Mechanisms that minimize retinal impact of apolipoprotein E absence. J. Lipid. Res. 2018, 59, 2368–2382. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Mahley, R.W. Apolipoprotein E: Structure and function in lipid metabolism, neurobiology, and Alzheimer’s diseases. Neurobiol. Dis. 2014, 72 Pt A, 3–12. [Google Scholar] [CrossRef]
- Krasemann, S.; Madore, C.; Cialic, R.; Baufeld, C.; Calcagno, N.; El Fatimy, R.; Beckers, L.; O’Loughlin, E.; Xu, Y.; Fanek, Z.; et al. The TREM2-APOE Pathway Drives the Transcriptional Phenotype of Dysfunctional Microglia in Neurodegenerative Diseases. Immunity 2017, 47, 566–581. [Google Scholar] [CrossRef]
- Venezie, R.D.; Toews, A.D.; Morell, P. Macrophage recruitment in different models of nerve injury: Lysozyme as a marker for active phagocytosis. J. Neurosci. Res. 1995, 40, 99–107. [Google Scholar] [CrossRef]
- Nielsen, M.C.; Hvidbjerg Gantzel, R.; Clària, J.; Trebicka, J.; Møller, H.J.; Grønbæk, H. Macrophage Activation Markers, CD163 and CD206, in Acute-on-Chronic Liver Failure. Cells 2020, 9, 1175. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, N.; Quintana, J.A.; García-Silva, S.; Mazariegos, M.; González de la Aleja, A.; Nicolás-Ávila, J.A.; Walter, W.; Adrover, J.M.; Crainiciuc, G.; Kuchroo, V.K.; et al. Phagocytosis imprints heterogeneity in tissue-resident macrophages. J. Exp. Med. 2017, 214, 1281–1296. [Google Scholar] [CrossRef] [PubMed]
- Park, H.J.; Oh, S.H.; Kim, H.N.; Jung, Y.J.; Lee, P.H. Mesenchymal stem cells enhance alpha-synuclein clearance via M2 microglia polarization in experimental and human parkinsonian disorder. Acta Neuropathol. 2016, 132, 685–701. [Google Scholar] [CrossRef] [PubMed]
- Shinoe, T.; Kuribayashi, H.; Saya, H.; Seiki, M.; Aburatani, H.; Watanabe, S. Identification of CD44 as a cell surface marker for Müller glia precursor cells. J. Neurochem. 2010, 115, 1633–1642. [Google Scholar] [CrossRef]
- Too, L.K.; Gracie, G.; Hasic, E.; Iwakura, J.H.; Cherepanoff, S. Adult human retinal Muller glia display distinct peripheral and macular expression of CD117 and CD44 stem cell-associated proteins. Acta Histochem. 2017, 119, 142–149. [Google Scholar] [CrossRef] [PubMed]
- Yanuck, S.F. Microglial Phagocytosis of Neurons: Diminishing Neuronal Loss in Traumatic, Infectious, Inflammatory, and Autoimmune CNS Disorders. Front. Psychiatry 2019, 10, 712. [Google Scholar] [CrossRef]
- Janda, E.; Boi, L.; Carta, A.R. Microglial Phagocytosis and Its Regulation: A Therapeutic Target in Parkinson’s Disease? Front. Mol. Neurosci. 2018, 11, 144. [Google Scholar] [CrossRef]
- Nan, F.; Sun, Y.; Liang, H.; Zhou, J.; Ma, X.; Zhang, D. Mannose: A Sweet Option in the Treatment of Cancer and Inflammation. Front. Pharmacol. 2022, 13, 877543. [Google Scholar] [CrossRef]
- Kim, G.H.; Paik, S.S.; Park, Y.S.; Kim, H.G.; Kim, I.B. Amelioration of Mouse Retinal Degeneration After Blue LED Exposure by Glycyrrhizic Acid-Mediated Inhibition of Inflammation. Front. Cell Neurosci. 2019, 13, 319. [Google Scholar] [CrossRef] [PubMed]
- Kim, G.H.; Kim, H.I.; Paik, S.S.; Jung, S.W.; Kang, S.; Kim, I.B. Functional and morphological evaluation of blue light-emitting diode-induced retinal degeneration in mice. Graefes Arch. Clin. Exp. Ophthalmol. 2016, 254, 705–716. [Google Scholar] [CrossRef]

| Target | Species | Catalog | Company | Concentration |
|---|---|---|---|---|
| anti-IBA1 | Rabbit | 019-19741 | Wako Pure Chemical Industries | 1:500 |
| anti-IBA1 | Goat | ab5076 | Abcam | 1:500 |
| anti-CD206 | Goat | AF2535 | R&D Systems | 1:250 |
| anti-P2RY12 | Rabbit | AS-55043A | AnaSpec | 1:500 |
| anti-CD86 | Rat | ab119857 | Abcam | 1:250 |
| anti-CD44 | Rat | 103002 | BioLegend | 1:250 |
| anti-IL-10 | Mouse | 505002 | BioLegend | 1:100 |
| anti-TREM2 | Sheep | AF1729 | R&D Systems | 1:50 |
| Gene | Species | Forward (5′→3′) | Reverse (5′→3′) |
|---|---|---|---|
| Actin | Mouse | CAT TGC TGA CAG GAT GCA GAA GG | TGC TGG AAG GTG GAC AGT GAG G |
| CD206 | Mouse | TCA TCC CTG TCT CTG TTC AGC | ATG GCA CTT AGA GCG TCC AC |
| Apoe | Mouse | GGG ACA GGG GGA GTC CTA TAA T | TTT GCC ACT CGA GCT GAT CT |
| Trem2 | Mouse | GAC CTC TCC ACC AGT TTC TCC | TAC ATG ACA CCC TCA AGG ACT G |
| Lyz2 | Mouse | TGA ACG TTG TGA GTT TGC CAG | CAG CAG AGC ACT GCA ATT GAT |
| IL-10 | Mouse | CGG GAA GAC AAT AAC TGC ACC C | CGG TTA GCA GTA TGT TGT CCA GC |
| IL-6 | Mouse | TAC CAC TTC ACA AGT CGG AGG C | CTG CAA GTG CAT CAT CGT TGT TC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.; Park, Y.S.; Kim, I.-B. A Distinct Microglial Cell Population Expressing Both CD86 and CD206 Constitutes a Dominant Type and Executes Phagocytosis in Two Mouse Models of Retinal Degeneration. Int. J. Mol. Sci. 2023, 24, 14236. https://doi.org/10.3390/ijms241814236
Zhang Y, Park YS, Kim I-B. A Distinct Microglial Cell Population Expressing Both CD86 and CD206 Constitutes a Dominant Type and Executes Phagocytosis in Two Mouse Models of Retinal Degeneration. International Journal of Molecular Sciences. 2023; 24(18):14236. https://doi.org/10.3390/ijms241814236
Chicago/Turabian StyleZhang, Yan, Yong Soo Park, and In-Beom Kim. 2023. "A Distinct Microglial Cell Population Expressing Both CD86 and CD206 Constitutes a Dominant Type and Executes Phagocytosis in Two Mouse Models of Retinal Degeneration" International Journal of Molecular Sciences 24, no. 18: 14236. https://doi.org/10.3390/ijms241814236
APA StyleZhang, Y., Park, Y. S., & Kim, I.-B. (2023). A Distinct Microglial Cell Population Expressing Both CD86 and CD206 Constitutes a Dominant Type and Executes Phagocytosis in Two Mouse Models of Retinal Degeneration. International Journal of Molecular Sciences, 24(18), 14236. https://doi.org/10.3390/ijms241814236







