Multi-Omics Analysis Reveals the Pathogenesis of Growth-Disordered Raccoon Dog
Abstract
1. Introduction
2. Results
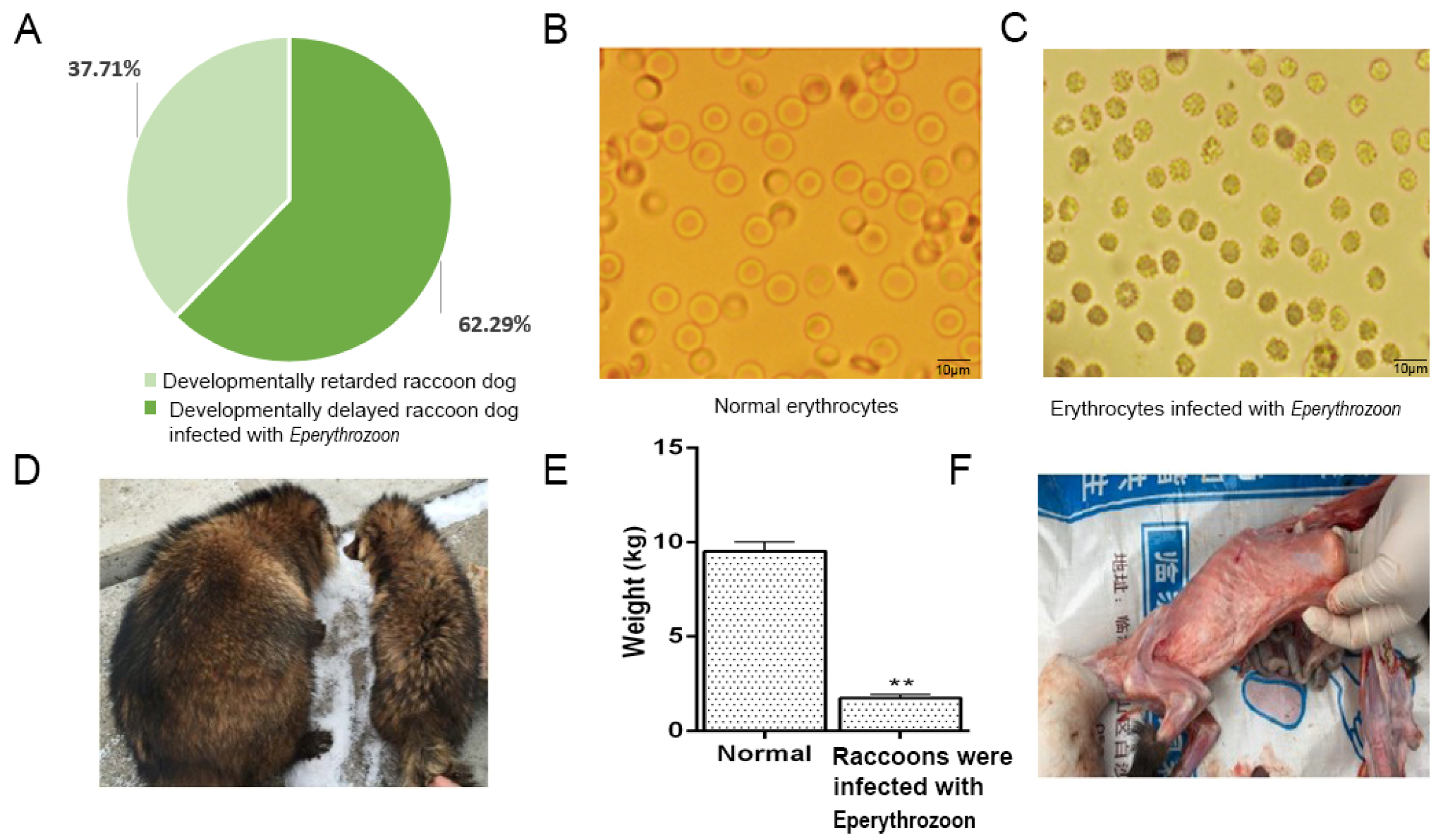
2.1. Clinical Symptoms of Raccoons with Growth Disorders
2.2. Blood Test Analysis
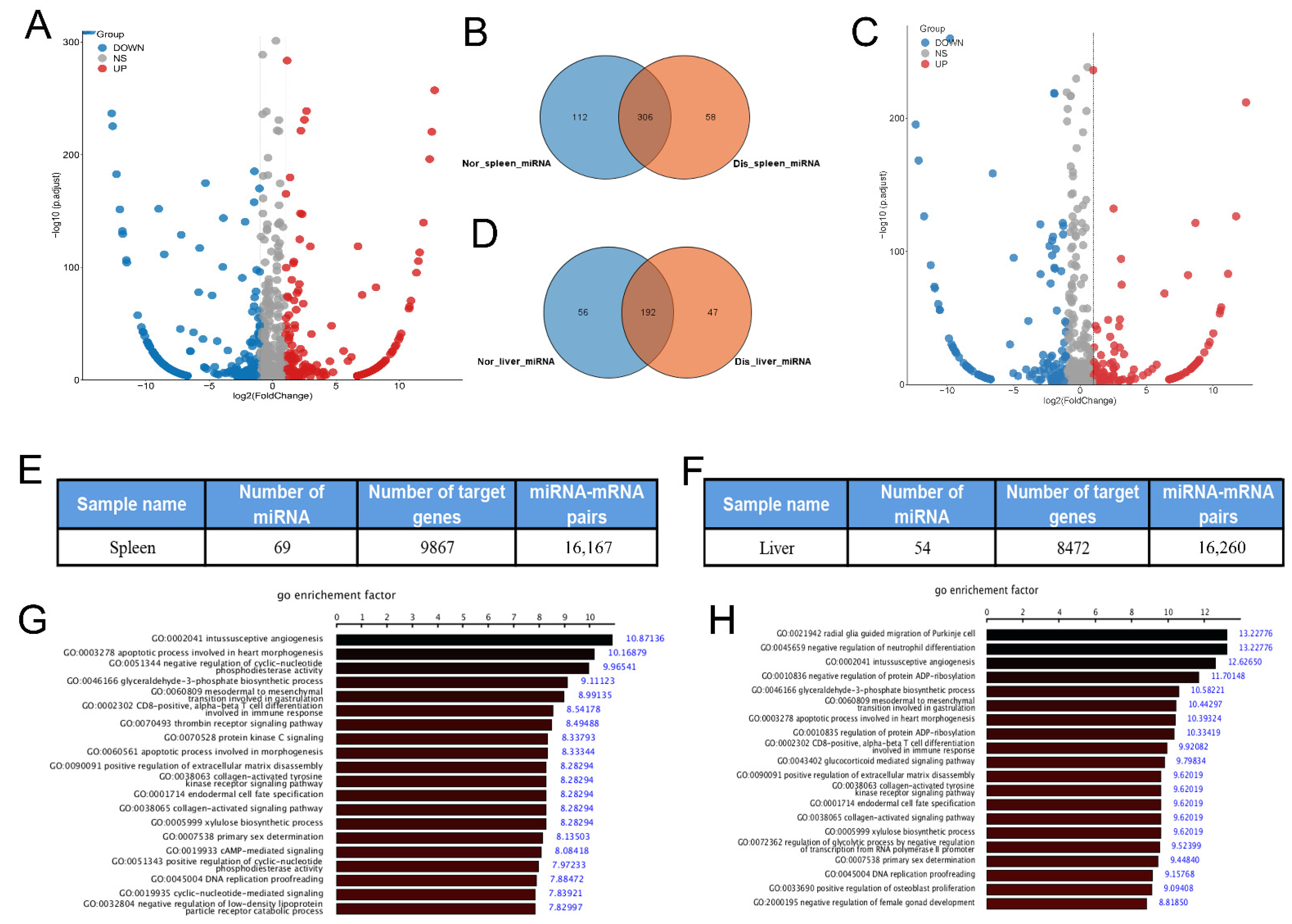
2.3. Expression Analysis and Functional Prediction of Differential mRNAs in Spleen
2.4. Expression Analysis and Functional Prediction of Differential mRNAs in the Liver
2.5. Analysis and Function Prediction of Differential miRNA
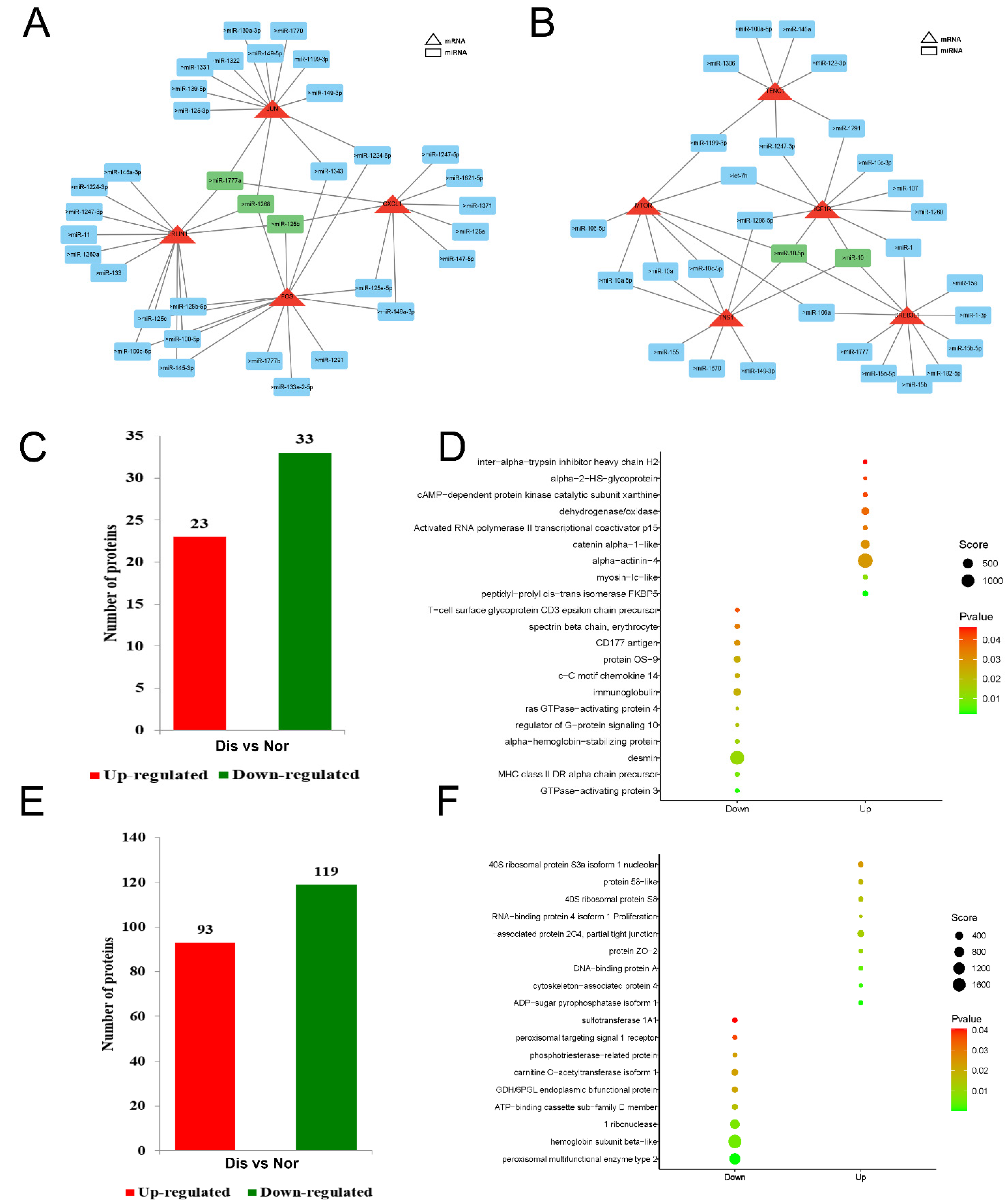
2.6. Analysis of Key mRNA-miRNA Interaction Networks
2.7. Differential Protein Analysis
2.8. GO and KEGG Enrichment Analysis of Differential Proteins
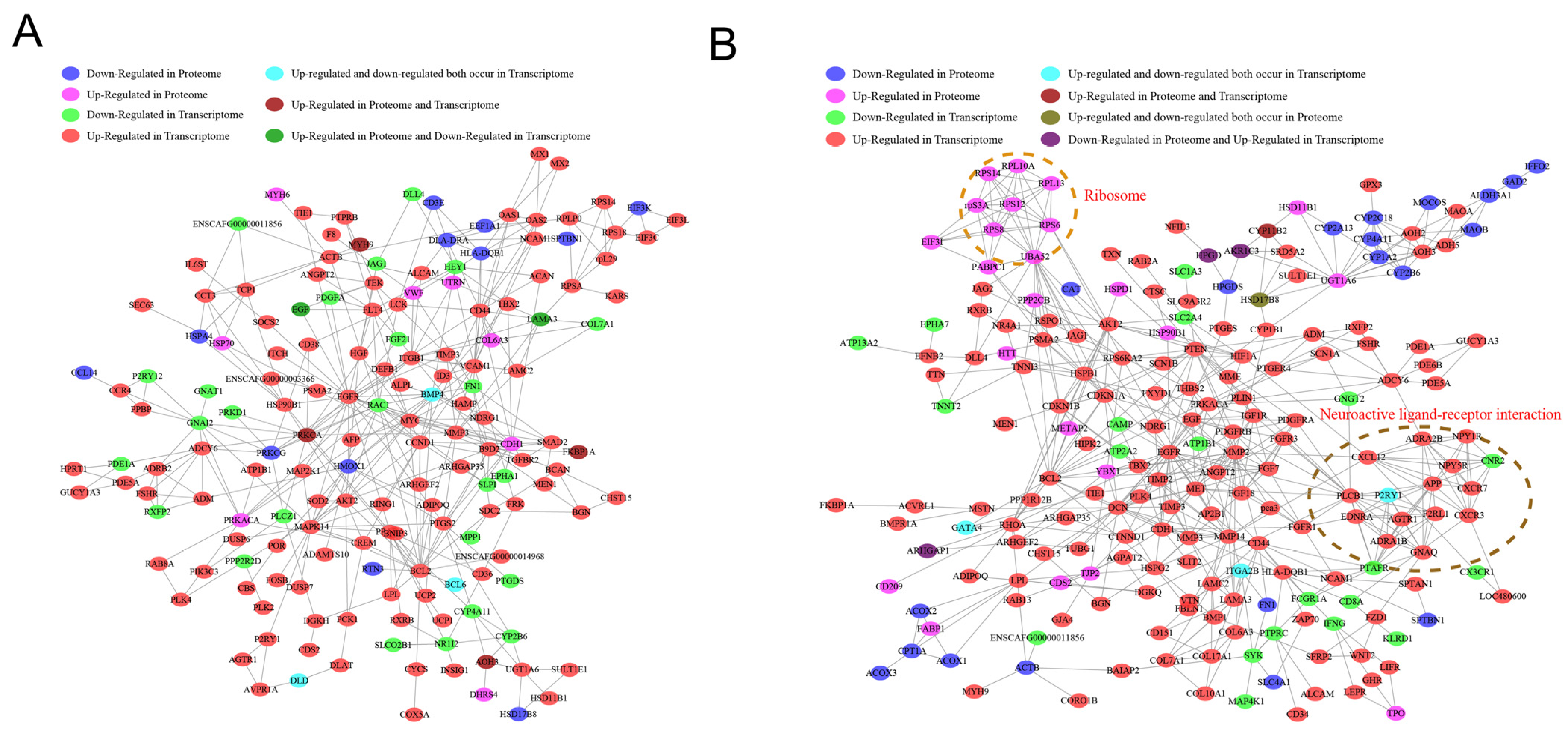
2.9. Construction of Differential Gene and Protein Regulatory Networks
3. Discussion
4. Materials and Methods
4.1. Sample Collection
4.2. Blood Test
4.3. Library Construction and RNA Sequencing
4.4. miRNA Sequencing and Analysis
4.5. Proteomic Analysis
4.6. LC-MS/MS Proteome Quantitative Analysis
4.7. GO and KEGG Analysis
4.8. Combined Transcriptome and Proteome Analysis
4.9. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Niiranen, L.; Makela, K.A.; Dona, A.; Krumsiek, J.; Karhu, T.; Makinen, M.J.; Thalmann, O.; Saarela, S.; Herzig, K.H. Seasonal Regulation of Metabolism: The Effect of Wintertime Fasting and Autumnal Fattening on Key Central Regulators of Metabolism and the Metabolic Profile of the Raccoon Dog (Nyctereutes procyonoides). Int. J. Mol. Sci. 2021, 22, 4965. [Google Scholar] [CrossRef] [PubMed]
- Mustonen, A.J.; Finnila, M.; Puukka, K.S.; Jamsa, T.J.; Saarakkala, S.; Tuukkanen, J.K.; Nieminen, T.P. Raccoon dog model shows preservation of bone during prolonged catabolism and reduced physical activity. J. Exp. Biol. 2017, 220, 2196–2202. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; He, Y.; Chen, X.; Kalim, U.; Wang, Y.; Yang, S.; Qi, H.; Cheng, H.; Lu, X.; Wang, X.; et al. Viral Metagenomics Reveals Diverse Viruses in the Feces Samples of Raccoon Dogs. Front. Vet. Sci. 2021, 8, 693564. [Google Scholar] [CrossRef] [PubMed]
- Duscher, T.; Hodzic, A.; Glawischnig, W.; Duscher, G.G. The raccoon dog (Nyctereutes procyonoides) and the raccoon (Procyon lotor)-their role and impact of maintaining and transmitting zoonotic diseases in Austria, Central Europe. Parasitol. Res. 2017, 116, 1411–1416. [Google Scholar] [CrossRef]
- Maca, O. Molecular identification of Sarcocystis lutrae (Apicomplexa: Sarcocystidae) from the raccoon dog, Nyctereutes procyonoides, and the common raccoon, Procyon lotor, in the Czech Republic. Parasites Vectors 2020, 13, 231. [Google Scholar] [CrossRef] [PubMed]
- Han, J.I.; Lee, S.J.; Jang, H.J.; Na, K.J. Asymptomatic Babesia microti-like parasite infection in wild raccoon dogs (Nyctereutes procyonoides) in South Korea. J. Wildl. Dis. 2010, 46, 632–635. [Google Scholar] [CrossRef][Green Version]
- Lan, T.; Li, H.; Yang, S.; Shi, M.; Han, L.; Sahu, S.K.; Lu, Y.; Wang, J.; Zhou, M.; Liu, H.; et al. The chromosome-scale genome of the raccoon dog: Insights into its evolutionary characteristics. iScience 2022, 25, 105117. [Google Scholar] [CrossRef]
- Mustonen, A.M.; Asikainen, J.; Kauhala, K.; Paakkonen, T.; Nieminen, P. Seasonal rhythms of body temperature in the free-ranging raccoon dog (Nyctereutes procyonoides) with special emphasis on winter sleep. Chronobiol. Int. 2007, 24, 1095–1107. [Google Scholar] [CrossRef]
- Kauhala, K.; Kowalczyk, R. Invasion of the raccoon dog Nyctereutes procyonoides in Europe: History of colonization, features behind its success, and threats to native fauna. Curr. Zool. 2011, 57, 584–598. [Google Scholar] [CrossRef]
- Liu, H.; Li, Z.; Si, H.; Zhong, W.; Fan, Z.; Li, G. Comparative analysis of the gut microbiota of the blue fox (Alopex lagopus) and raccoon dog (Nyctereutes procyonoides). Arch. Microbiol. 2020, 202, 135–142. [Google Scholar] [CrossRef]
- Mustonen, A.M.; Nieminen, P. A review of the physiology of a survival expert of big freeze, deep snow, and an empty stomach: The boreal raccoon dog (Nyctereutes procyonoides). J. Comp. Physiol. B Biochem. Syst. Environ. Physiol. 2018, 188, 15–25. [Google Scholar] [CrossRef] [PubMed]
- Wu, I.L.; Yu, J.H.; Lin, C.C.; Seak, C.J.; Olson, K.R.; Chen, H.Y. Fatal cardiac glycoside poisoning due to mistaking foxglove for comfrey. Clin. Toxicol. 2017, 55, 670–673. [Google Scholar] [CrossRef] [PubMed]
- Guimaraes, A.M.; Biondo, A.W.; Lara, A.C.; Messick, J.B. Exploratory study of Mycoplasma suis (Eperythrozoon suis) on four commercial pig farms in southern Brazil. Vet. Rec. 2007, 160, 50–53. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.S.; Guan, P.; Wu, W.; Shen, T.F.; Liu, H.L.; Cao, S.; Zhou, H. Infection rate of Eperythrozoon spp. in Chinese population: A systematic review and meta-analysis since the first Chinese case reported in 1991. BMC Infect. Dis. 2012, 12, 171. [Google Scholar] [CrossRef] [PubMed]
- Urie, N.J.; Highland, M.A.; Knowles, D.P.; Branan, M.A.; Herndon, D.R.; Marshall, K.L. Mycoplasma ovis infection in domestic sheep (Ovis aries) in the United States: Prevalence, distribution, associated risk factors, and associated outcomes. Prev. Vet. Med. 2019, 171, 104750. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.A.; Thrall, M.A.; Smith, J.L.; Salman, M.D.; Ching, S.V.; Collins, J.K. Eperythrozoon wenyonii infection in dairy cattle. J. Am. Vet. Med. Assoc. 1990, 196, 1244–1250. [Google Scholar] [PubMed]
- Yuan, C.L.; Liang, A.B.; Yao, C.B.; Yang, Z.B.; Zhu, J.G.; Cui, L. Prevalence of Mycoplasma suis (Eperythrozoon suis) infection in swine and swine-farm workers in Shanghai, China. Am. J. Vet. Res. 2009, 70, 890–894. [Google Scholar] [CrossRef]
- Zhao, Y.; Wang, Z.; Hou, Y.; Zhang, K.; Peng, X. gga-miR-99a targets SMARCA5 to regulate Mycoplasma gallisepticum (HS strain) infection by depressing cell proliferation in chicken. Gene 2017, 627, 239–247. [Google Scholar] [CrossRef]
- Yan, S.Q.; Li, Y.M.; Bai, C.Y.; Ding, X.M.; Li, W.J.; Hou, J.N.; Zhao, Z.H.; Sun, J.H. Development and characterization of polymorphic microsatellite markers for Chinese raccoon dog (Nyctereutes procyonoides procyonoides). Genet. Mol. Res. 2013, 12, 6351–6355. [Google Scholar] [CrossRef]
- Breedveld, A.; Groot, K.T.; van Egmond, M.; de Jong, E.C. Granulocytes as modulators of dendritic cell function. J. Leukoc. Biol. 2017, 102, 1003–1016. [Google Scholar] [CrossRef] [PubMed]
- Song, F.; Zhou, X.X.; Hu, Y.; Li, G.; Wang, Y. The Roles of Insulin-Like Growth Factor Binding Protein Family in Development and Diseases. Adv. Ther. 2021, 38, 885–903. [Google Scholar] [CrossRef] [PubMed]
- Franz, A.C.; Faass, O.; Kollner, B.; Shved, N.; Link, K.; Casanova, A.; Wenger, M.; D’Cotta, H.; Baroiller, J.F.; Ullrich, O.; et al. Endocrine and Local IGF-I in the Bony Fish Immune System. Biology 2016, 5, 9. [Google Scholar] [CrossRef] [PubMed]
- Jozefiak, A.; Larska, M.; Pomorska-Mol, M.; Ruszkowski, J.J. The IGF-1 Signaling Pathway in Viral Infections. Viruses 2021, 13, 1488. [Google Scholar] [CrossRef] [PubMed]
- Forbes, B.E.; Blyth, A.J.; Wit, J.M. Disorders of IGFs and IGF-1R signaling pathways. Mol. Cell. Endocrinol. 2020, 518, 111035. [Google Scholar] [CrossRef] [PubMed]
- Du, H.; Zhou, Y.; Du, X.; Zhang, P.; Cao, Z.; Sun, Y. Insulin-like growth factor binding protein 5b of Trachinotus ovatus and its heparin-binding motif play a critical role in host antibacterial immune responses via NF-κB pathway. Front. Immunol. 2023, 14, 1126843. [Google Scholar] [CrossRef] [PubMed]
- Akkiprik, M.; Hu, L.; Sahin, A.; Hao, X.; Zhang, W. The subcellular localization of IGFBP5 affects its cell growth and migration functions in breast cancer. BMC Cancer 2009, 9, 103. [Google Scholar] [CrossRef]
- Jang, J.C.; Li, J.; Gambini, L.; Batugedara, H.M.; Sati, S.; Lazar, M.A.; Fan, L.; Pellecchia, M.; Nair, M.G. Human resistin protects against endotoxic shock by blocking LPS-TLR4 interaction. Proc. Natl. Acad. Sci. USA 2017, 114, E10399–E10408. [Google Scholar] [CrossRef]
- Liu, H.; Zhao, H.; Lin, H.; Li, Z.; Xue, H.; Zhang, Y.; Lu, J. Relationship of COL9A1 and SOX9 Genes with Genetic Susceptibility of Postmenopausal Osteoporosis. Calcif. Tissue Int. 2020, 106, 248–255. [Google Scholar] [CrossRef]
- Tunca, C.; Akcimen, F.; Coskun, C.; Gundogdu-Eken, A.; Kocoglu, C.; Cevik, B.; Bekircan-Kurt, C.E.; Tan, E.; Basak, A.N. ERLIN1 mutations cause teenage-onset slowly progressive ALS in a large Turkish pedigree. Eur. J. Hum. Genet. 2018, 26, 745–748. [Google Scholar] [CrossRef]
- Huang, S.; Toufiq, M.; Saraiva, L.R.; Van Panhuys, N.; Chaussabel, D.; Garand, M. Transcriptome and Literature Mining Highlight the Differential Expression of ERLIN1 in Immune Cells during Sepsis. Biology 2021, 10, 755. [Google Scholar] [CrossRef] [PubMed]
- Riedemann, J.; Macaulay, V.M. IGF1R signalling and its inhibition. Endocr.-Relat. Cancer 2006, 13 (Suppl. S1), S33–S43. [Google Scholar] [CrossRef]
- Wang, Z.; Ye, J.; Dong, F.; Cao, L.; Wang, M.; Sun, G. TNS1: Emerging Insights into Its Domain Function, Biological Roles, and Tumors. Biology 2022, 11, 1571. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Yang, S.; Song, W. Prazosin inhibits the proliferation and survival of acute myeloid leukaemia cells through down-regulating TNS1. Biomed. Pharmacother. 2020, 124, 109731. [Google Scholar] [CrossRef] [PubMed]
- You, G.R.; Chang, J.T.; Li, Y.L.; Huang, C.W.; Tsai, Y.L.; Fan, K.H.; Kang, C.J.; Huang, S.F.; Chang, P.H.; Cheng, A.J. MYH9 Facilitates Cell Invasion and Radioresistance in Head and Neck Cancer via Modulation of Cellular ROS Levels by Activating the MAPK-Nrf2-GCLC Pathway. Cells 2022, 11, 2855. [Google Scholar] [CrossRef] [PubMed]
- Zhao, P.; Han, H.; Wu, X.; Wu, J.; Ren, Z. ARP2/3 Regulates Fatty Acid Synthesis by Modulating Lipid Droplets’ Motility. Int. J. Mol. Sci. 2022, 23, 8730. [Google Scholar] [CrossRef]
- Gao, S.; Wang, S.; Zhao, Z.; Zhang, C.; Liu, Z.; Ye, P.; Xu, Z.; Yi, B.; Jiao, K.; Naik, G.A.; et al. TUBB4A interacts with MYH9 to protect the nucleus during cell migration and promotes prostate cancer via GSK3β/β-catenin signalling. Nat. Commun. 2022, 13, 2792. [Google Scholar] [CrossRef] [PubMed]
- Pecci, A.; Ma, X.; Savoia, A.; Adelstein, R.S. MYH9: Structure, functions and role of non-muscle myosin IIA in human disease. Gene 2018, 664, 152–167. [Google Scholar] [CrossRef]
- Annett, S.; Moore, G.; Robson, T. FK506 binding proteins and inflammation related signalling pathways; basic biology, current status and future prospects for pharmacological intervention. Pharmacol. Ther. 2020, 215, 107623. [Google Scholar] [CrossRef]
- Ghartey-Kwansah, G.; Li, Z.; Feng, R.; Wang, L.; Zhou, X.; Chen, F.Z.; Xu, M.M.; Jones, O.; Mu, Y.; Chen, S.; et al. Comparative analysis of FKBP family protein: Evaluation, structure, and function in mammals and Drosophila melanogaster. BMC Dev. Biol. 2018, 18, 7. [Google Scholar] [CrossRef]
- Tong, M.; Jiang, Y. FK506-Binding Proteins and Their Diverse Functions. Curr. Molec. Pharmacol. 2015, 9, 48–65. [Google Scholar] [CrossRef]
- Cai, S.; Chen, Z.; Tang, H.; Meng, S.; Tao, L.; Wang, Q. Upregulated FKBP1A Suppresses Glioblastoma Cell Growth via Apoptosis Pathway. Int. J. Mol. Sci. 2022, 23, 4935. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Zhang, L.; Binkley, P.F.; Sadee, W.; Wang, D. Regulatory Variants Modulate Protein Kinase C α (PRKCA) Gene Expression in Human Heart. Pharm. Res. 2017, 34, 1648–1657. [Google Scholar] [CrossRef] [PubMed]
- Faingelernt, Y.; Hershkovitz, E.; Abu-Libdeh, B.; Abedrabbo, A.; Abu-Rmaileh Amro, S.; Zarivach, R.; Zangen, D.; Lavi, E.; Haim, A.; Parvari, R.; et al. Aldosterone synthase (CYP11B2) deficiency among Palestinian infants: Three novel variants and genetic heterogeneity. Am. J. Med. Genet. Part A 2021, 185, 1033–1038. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Genome Biol. 2010, 11, R106. [Google Scholar] [CrossRef] [PubMed]
- Bouyssie, D.; Gonzalez, D.P.A.; Mouton, E.; Albigot, R.; Roussel, L.; Ortega, N.; Cayrol, C.; Burlet-Schiltz, O.; Girard, J.P.; Monsarrat, B. Mascot file parsing and quantification (MFPaQ), a new software to parse, validate, and quantify proteomics data generated by ICAT and SILAC mass spectrometric analyses: Application to the proteomics study of membrane proteins from primary human endothelial cells. Mol. Cell. Proteom. 2007, 6, 1621–1637. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Mao, X.; Cai, T.; Luo, J.; Wei, L. KOBAS server: A web-based platform for automated annotation and pathway identification. Nucleic Acids Res. 2006, 34, W720–W724. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Sato, Y.; Furumichi, M.; Morishima, K.; Tanabe, M. New approach for understanding genome variations in KEGG. Nucleic Acids Res. 2019, 47, D590–D595. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y.; Kawashima, M.; Furumichi, M.; Tanabe, M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016, 44, D457–D462. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [PubMed]



| Item | Normal | Disease |
|---|---|---|
| WBC (×10−9/L) | 15.3 ± 8.91 | 18.06 ± 12.41 |
| Lymph (×10−9/L) | 0.93 ± 1.32 | 3.57 ± 4.97 |
| Gran (×10−9/L) | - | 4.567 ± 2.83 * |
| Gran (×10−9/L) | - | 17.4 ± 13.86 |
| Lymph % | 0.089 ± 0.1258 | 27.43 ± 43.36 |
| Mid % | - | 16.27 ± 4.91 ** |
| Gran % | - | 56.3 ± 38.45 * |
| RBC (×10−9/L) | 7.43 ± 0.96 | 5.41 ± 1.74 |
| HGB (g/L) | 130.6 ± 39.39 | 102 ± 34.29 |
| HCT (L/L) | 45 ± 6.09 | 31.93 ± 10.43 * |
| MCV (fL) | 60.57 ± 1.087 | 59.13 ± 3.14 |
| MCH (pg) | 19.7 ± 0.67 | 18.76 ± 1.21 |
| MCHC (g/L) | 325.34 ± 4.92 | 318.29 ± 8.04 |
| RDW-CV% | 14.88 ± 0.29 | 14.94 ± 1.35 |
| RDW-SD (fL) | 33.37 ± 1.08 | 33.06 ± 2.58 |
| PLT (×10−9/L) | 525.67 ± 136.76 | 660.71 ± 305.30 |
| MPV (fL) | 9.7 ± 0.37 | 9 ± 0.68 |
| PDW % | 15.3 ± 0.51 | 15.76 ± 0.23 |
| PCT % | 0.51 ± 0.12 | 0.61 ± 0.31 |
| Item | Normal | Disease |
|---|---|---|
| glutamic-pyruvic transaminase (IU/L) | 33.0 ± 2.6 | 570.5 ± 22.6 ** |
| glutamic oxalacetic transaminase (IU/L) | 45 ± 4.6 | 161 ± 9.8 ** |
| Total protein (g/L) | 67.8 ± 5.9 | 116.4 ± 13.8 ** |
| albumin (g/L) | 31.7 ± 6.3 | 50.6 ± 7.1 ** |
| globulin (g/L) | 36.1 ± 5.5 | 65.8 ± 6.8 ** (g/L) |
| Alkaline phosphatase (IU/L) | 59.1 ± 7.7 | 29.3 ± 6.4 * (IU/L) |
| triglyceride (mmol/L) | 1.43 ± 0.32 | 0.61 ± 0.24 * |
| Cholesterol (mmol/L) | 3.84 ± 0.55 | 5.29 ± 0.78 * |
| High density lipoprotein cholesterol (mmol/L) | 3.25 ± 0.32 | 4.63 ± 0.56 * |
| Low density lipoprotein cholesterol (mmol/L) | 0.07 ± 0.03 | 0.89 ± 0.22 ** |
| Creatine kinase (IU/L) | 293.0 ± 32.1 | 856.0 ± 44.2 ** |
| Lactate dehydrogenase (IU/L) | 153 ± 22 | 502 ± 43 ** |
| Blood sugar (mmol/L) | 4.49 ± 0.88 | 2.39 ± 0.69 ** |
| Serum potassium (mmol/L) | 5.72 ± 0.64 | 15.04 ± 1.23 ** |
| Blood sodium (mmol/L) | 143 ± 22 | 158 ± 18 |
| Blood chlorine (mmol/L) | 102 ± 9 | 117 ± 11 |
| Carbon dioxide binding power (mmol/L) | 26.3 ± 1.2 | 10.1 ± 1.4 ** |
| Blood calcium (mmol/L) | 2.84 ± 0.21 | 2.94 ± 0.32 |
| Total bilirubin (μmol/L) | - | 1.2 ± 0.1 |
| Direct bilirubin (μmol/L) | - | 1.0 ± 0.2 |
| Indirect bilirubin (μmol/L) | - | 0.2 ± 0.01 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, D.; Guo, X.; Wang, K.; Zhao, W.; Chang, Z.; Wang, Q.; Xu, C. Multi-Omics Analysis Reveals the Pathogenesis of Growth-Disordered Raccoon Dog. Int. J. Mol. Sci. 2023, 24, 14237. https://doi.org/10.3390/ijms241814237
Chen D, Guo X, Wang K, Zhao W, Chang Z, Wang Q, Xu C. Multi-Omics Analysis Reveals the Pathogenesis of Growth-Disordered Raccoon Dog. International Journal of Molecular Sciences. 2023; 24(18):14237. https://doi.org/10.3390/ijms241814237
Chicago/Turabian StyleChen, Danyang, Xiaolan Guo, Kaiying Wang, Weigang Zhao, Zhongjuan Chang, Quankai Wang, and Chao Xu. 2023. "Multi-Omics Analysis Reveals the Pathogenesis of Growth-Disordered Raccoon Dog" International Journal of Molecular Sciences 24, no. 18: 14237. https://doi.org/10.3390/ijms241814237
APA StyleChen, D., Guo, X., Wang, K., Zhao, W., Chang, Z., Wang, Q., & Xu, C. (2023). Multi-Omics Analysis Reveals the Pathogenesis of Growth-Disordered Raccoon Dog. International Journal of Molecular Sciences, 24(18), 14237. https://doi.org/10.3390/ijms241814237







