Salivary Lipids of Patients with Non-Small Cell Lung Cancer Show Perturbation with Respect to Plasma
Abstract
:1. Introduction
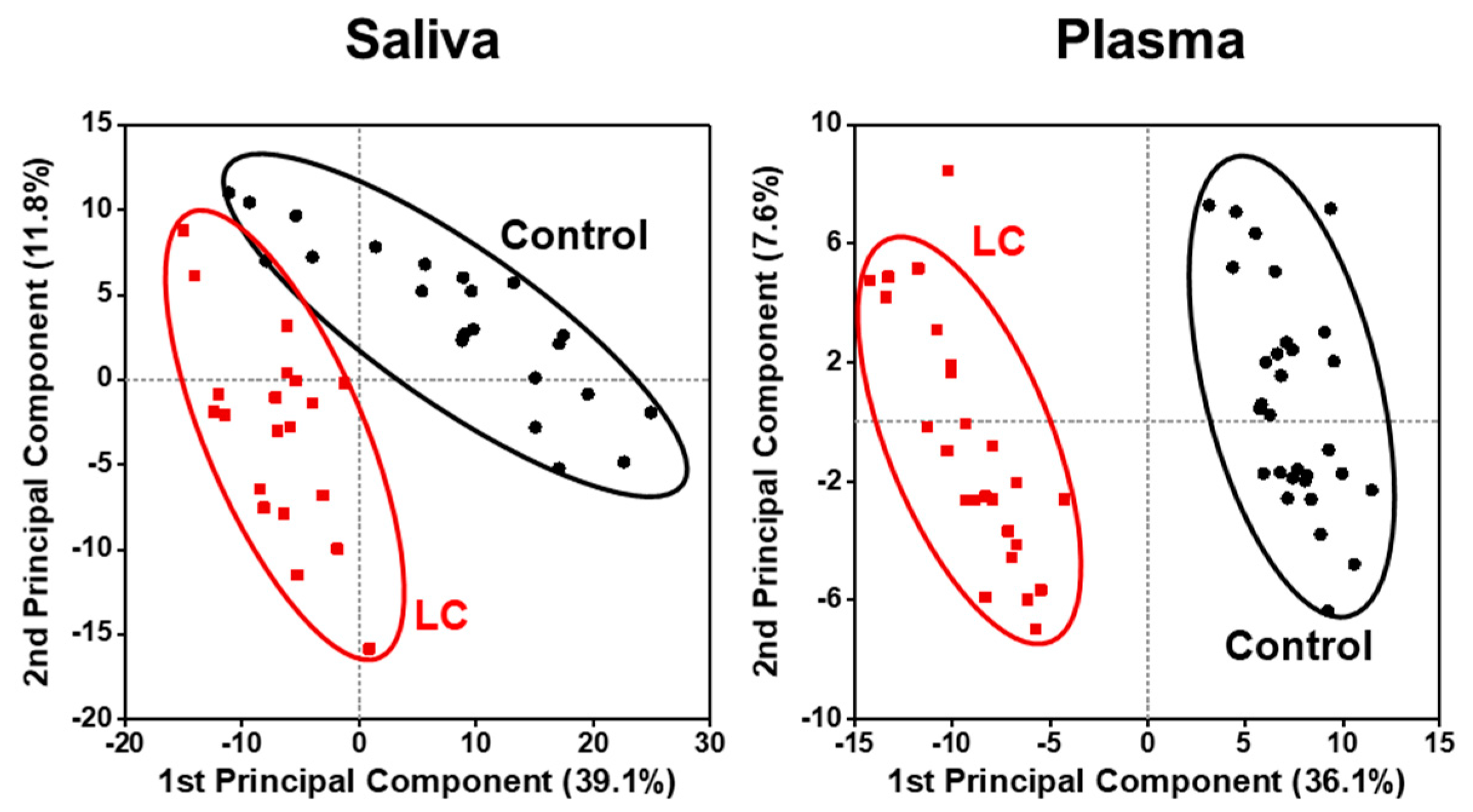
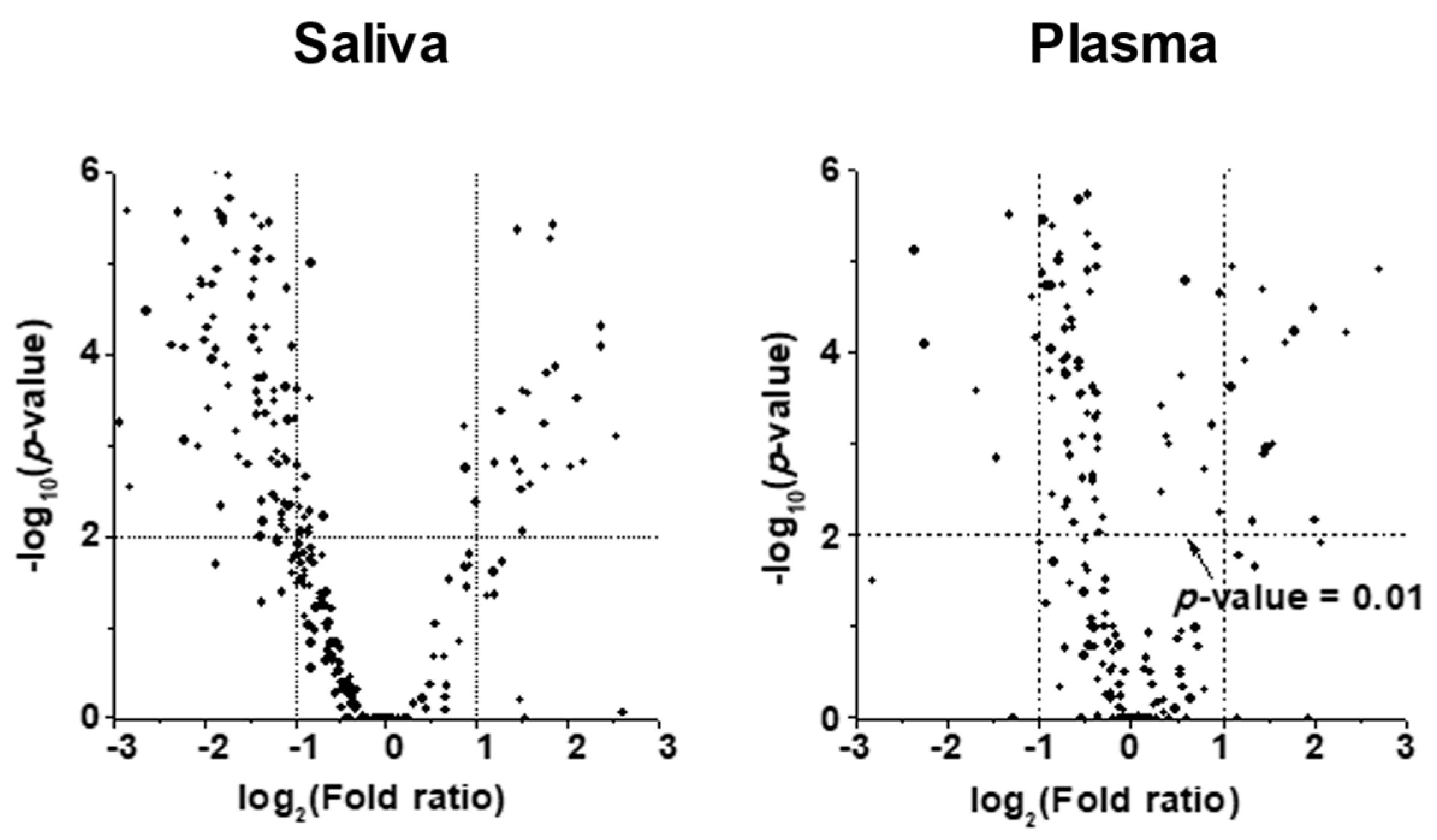
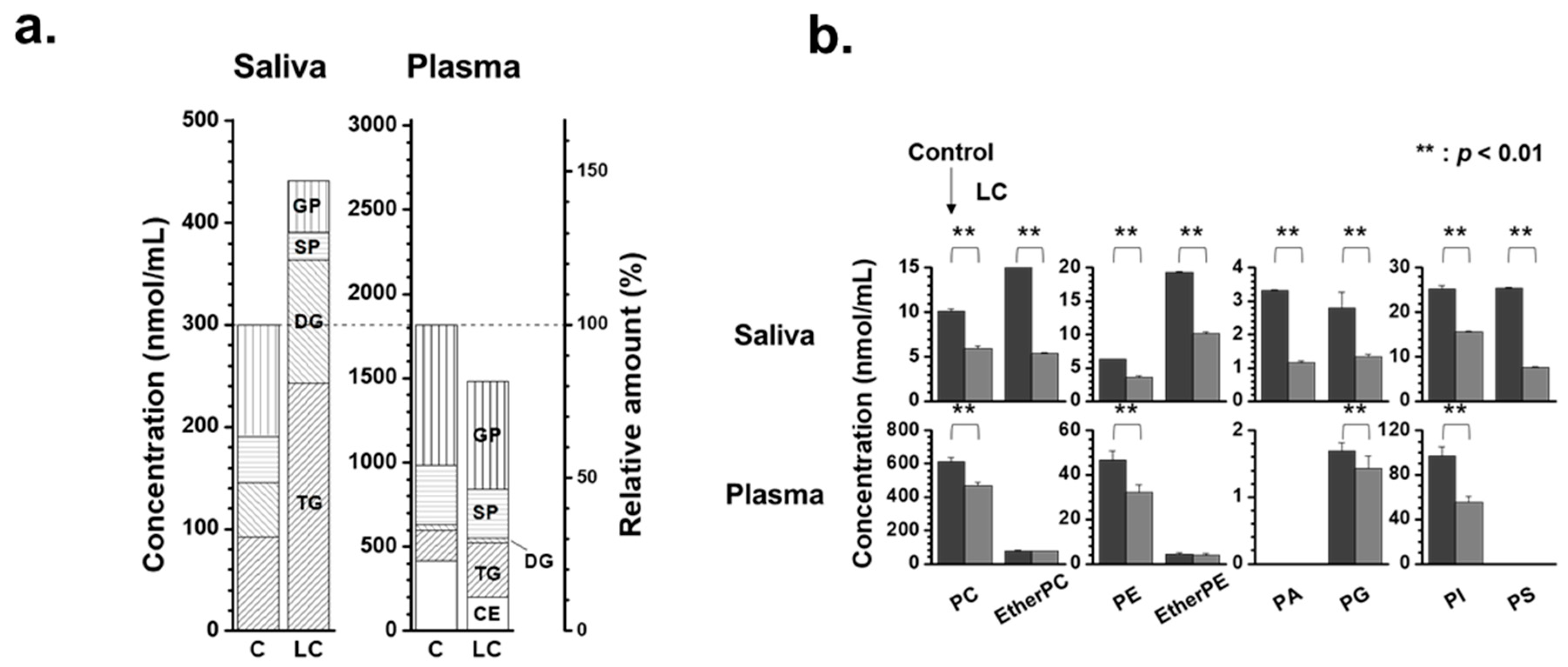
2. Results
2.1. Identification and Target-Based Lipid Quantification
2.2. Alterations in Lipid Profiles of Saliva and Plasma Samples in NSCLC Patients
3. Discussion
4. Materials and Methods
4.1. Materials and Reagents
4.2. Human Samples
4.3. Lipid Extraction
4.4. Lipid Analysis with nUHPLC-ESI-MS/MS
4.5. Method Validation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Thai, A.A.; Solomon, B.J.; Sequist, L.V.; Gainor, J.F.; Heist, R.S. Lung cancer. Lancet 2021, 398, 535–554. [Google Scholar] [CrossRef] [PubMed]
- Knight, S.B.; Crosbie, P.A.; Balata, H.; Chudziak, J.; Hussell, T.; Dive, C. Progress and prospects of early detection in lung cancer. Open Biol. 2017, 7, 170070. [Google Scholar] [CrossRef]
- Holčapek, M.; Liebisch, G.; Ekroos, K. Lipidomic Analysis. Anal. Chem. 2018, 90, 4249–4257. [Google Scholar] [CrossRef]
- Knittelfelder, O.L.; Weberhofer, B.P.; Eichmann, T.O.; Kohlwein, S.D.; Rechberger, G.N. A versatile ultra-high performance LC-MS method for lipid profiling. J. Chromatogr. B 2014, 951–952, 119–128. [Google Scholar] [CrossRef]
- García-Cañaveras, J.C.; Donato, M.T.; Castell, J.V.; Lahoz, A. A Comprehensive Untargeted Metabonomic Analysis of Human Steatotic Liver Tissue by RP and HILIC Chromatography Coupled to Mass Spectrometry Reveals Important Metabolic Alterations. J. Proteome Res. 2011, 10, 4825–4834. [Google Scholar] [CrossRef]
- Zhao, Y.Y.; Miao, H.; Cheng, X.L.; Wei, F. Lipidomics: Novel insight into the biochemical mechanism of lipid metabolism and dysregulation-associated disease. Chem. Biol. Interact. 2015, 240, 220–238. [Google Scholar] [CrossRef]
- Santos, C.R.; Schulze, A. Lipid metabolism in cancer. FEBS J. 2012, 279, 2610–2623. [Google Scholar] [CrossRef]
- Lee, G.B.; Lee, J.C.; Moon, M.H. Plasma lipid profile comparison of five different cancers by nanoflow ultrahigh performance liquid chromatography-tandem mass spectrometry. Anal. Chim. Acta 2019, 1063, 117–126. [Google Scholar] [CrossRef]
- Marien, E.; Meister, M.; Muley, T.; Fieuws, S.; Bordel, S.; Derua, R.; Spraggins, J.; Van de Plas, R.; Dehairs, J.; Wouters, J.; et al. Non-small cell lung cancer is characterized by dramatic changes in phospholipid profiles. Int. J. Cancer 2015, 137, 1539–1548. [Google Scholar] [CrossRef]
- Noreldeen, H.A.A.; Du, L.; Li, W.; Liu, X.; Wang, Y.; Xu, G. Serum lipidomic biomarkers for non-small cell lung cancer in nonsmoking female patients. J. Pharm. Biomed. Anal. 2020, 185, 113220. [Google Scholar] [CrossRef] [PubMed]
- Siqueira, W.L.; Zhang, W.; Helmerhorst, E.J.; Gygi, S.P.; Oppenheim, F.G. Identification of Protein Components in in vivo Human Acquired Enamel Pellicle Using LC−ESI−MS/MS. J. Proteome Res. 2007, 6, 2152–2160. [Google Scholar] [CrossRef] [PubMed]
- Freitas, D.; Le Feunteun, S. Oro-gastro-intestinal digestion of starch in white bread, wheat-based and gluten-free pasta: Unveiling the contribution of human salivary α-amylase. Food Chem. 2019, 274, 566–573. [Google Scholar] [CrossRef] [PubMed]
- Agatonovic-Kustrin, S.; Morton, D.W.; Smirnov, V.; Petukhov, A.; Gegechkori, V.; Kuzina, V.; Gorpinchenko, N.; Ramenskaya, G. Analytical Strategies in Lipidomics for Discovery of Functional Biomarkers from Human Saliva. Dis. Markers 2019, 2019, 6741518. [Google Scholar] [CrossRef]
- Turunen, S.; Puurunen, J.; Auriola, S.; Kullaa, A.M.; Kärkkäinen, O.; Lohi, H.; Hanhineva, K. Metabolome of canine and human saliva: A non-targeted metabolomics study. Metabolomics 2020, 16, 90. [Google Scholar] [CrossRef]
- de Oliveira, L.R.P.; Martins, C.; Fidalgo, T.K.S.; Freitas-Fernandes, L.B.; de Oliveira Torres, R.; Soares, A.L.; Almeida, F.C.L.; Valente, A.P.; de Souza, I.P.R. Salivary Metabolite Fingerprint of Type 1 Diabetes in Young Children. J. Proteome Res. 2016, 15, 2491–2499. [Google Scholar] [CrossRef]
- Sun, Y.; Liu, S.; Qiao, Z.; Shang, Z.; Xia, Z.; Niu, X.; Qian, L.; Zhang, Y.; Fan, L.; Cao, C.-X.; et al. Systematic comparison of exosomal proteomes from human saliva and serum for the detection of lung cancer. Anal. Chim. Acta 2017, 982, 84–95. [Google Scholar] [CrossRef]
- Lee, G.B.; Caner, A.; Moon, M.H. Optimisation of saliva volumes for lipidomic analysis by nanoflow ultrahigh performance liquid chromatography-tandem mass spectrometry. Anal. Chim. Acta 2022, 1193, 339318. [Google Scholar] [CrossRef]
- Cífková, E.; Brumarová, R.; Ovčačíková, M.; Dobešová, D.; Mičová, K.; Kvasnička, A.; Vaňková, Z.; Šiller, J.; Sákra, L.; Friedecký, D.; et al. Lipidomic and metabolomic analysis reveals changes in biochemical pathways for non-small cell lung cancer tissues. Biochim. Et Biophys. Acta (BBA)—Mol. Cell Biol. Lipids 2022, 1867, 159082. [Google Scholar] [CrossRef]
- Eggers, L.F.; Müller, J.; Marella, C.; Scholz, V.; Watz, H.; Kugler, C.; Rabe, K.F.; Goldmann, T.; Schwudke, D. Lipidomes of lung cancer and tumour-free lung tissues reveal distinct molecular signatures for cancer differentiation, age, inflammation, and pulmonary emphysema. Sci. Rep. 2017, 7, 11087. [Google Scholar] [CrossRef]
- Fan, Y.; Noreldeen, H.A.A.; You, L.; Liu, X.; Pan, X.; Hou, Z.; Li, Q.; Li, X.; Xu, G. Lipid alterations and subtyping maker discovery of lung cancer based on nontargeted tissue lipidomics using liquid chromatography-mass spectrometry. J. Pharm. Biomed. Anal. 2020, 190, 113520. [Google Scholar] [CrossRef] [PubMed]
- Ros-Mazurczyk, M.; Jelonek, K.; Marczyk, M.; Binczyk, F.; Pietrowska, M.; Polanska, J.; Dziadziuszko, R.; Jassem, J.; Rzyman, W.; Widlak, P. Serum lipid profile discriminates patients with early lung cancer from healthy controls. Lung Cancer 2017, 112, 69–74. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Xu, H.; Liu, R.; Gao, P.; Yang, X.; Jin, W.; Zhang, Y.; Bi, K.; Li, Q. A Novel Strategy for Targeted Lipidomics Based on LC-Tandem-MS Parameters Prediction, Quantification, and Multiple Statistical Data Mining: Evaluation of Lysophosphatidylcholines as Potential Cancer Biomarkers. Anal. Chem. 2019, 91, 3389–3396. [Google Scholar] [CrossRef]
- van der Veen, J.N.; Kennelly, J.P.; Wan, S.; Vance, J.E.; Vance, D.E.; Jacobs, R.L. The critical role of phosphatidylcholine and phosphatidylethanolamine metabolism in health and disease. Biochim. Biophys. Acta Biomembr. 2017, 1859, 1558–1572. [Google Scholar] [CrossRef] [PubMed]
- Gao, L.; Wen, Z.; Wu, C.; Wen, T.; Ong, C.N. Metabolic profiling of plasma from benign and malignant pulmonary nodules patients using mass spectrometry-based metabolomics. Metabolites 2013, 3, 539–551. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Song, Z.; Chen, Z.; Guo, Z.; Jin, H.; Ding, C.; Hong, Y.; Cai, Z. Metabolic and lipidomic characterization of malignant pleural effusion in human lung cancer. J. Pharm. Biomed. Anal. 2020, 180, 113069. [Google Scholar] [CrossRef]
- Yang, Y.; Ma, L.; Qiao, X.; Zhang, X.; Dong, S.F.; Wu, M.T.; Zhai, K.; Shi, H.Z. Salivary microRNAs show potential as biomarkers for early diagnosis of malignant pleural effusion. Transl. Lung Cancer Res. 2020, 9, 1247–1257. [Google Scholar] [CrossRef]
- Zhou, X.; Tian, C.; Cao, Y.; Zhao, M.; Wang, K. The role of serine metabolism in lung cancer: From oncogenesis to tumor treatment. Front. Genet. 2022, 13, 1084609. [Google Scholar] [CrossRef]
- Numata, M.; Voelker, D.R. Anti-inflammatory and anti-viral actions of anionic pulmonary surfactant phospholipids. Biochim. Biophys. Acta Mol. Cell. Biol. Lipids 2022, 1867, 159139. [Google Scholar] [CrossRef]
- van Dieren, J.M.; Simons-Oosterhuis, Y.; Raatgeep, H.C.; Lindenbergh-Kortleve, D.J.; Lambers, M.E.; van der Woude, C.J.; Kuipers, E.J.; Snoek, G.T.; Potman, R.; Hammad, H.; et al. Anti-inflammatory actions of phosphatidylinositol. Eur. J. Immunol. 2011, 41, 1047–1057. [Google Scholar] [CrossRef]
- Zhang, Y.; He, C.; Qiu, L.; Wang, Y.; Zhang, L.; Qin, X.; Liu, Y.; Zhang, D.; Li, Z. Serum unsaturated free Fatty acids: Potential biomarkers for early detection and disease progression monitoring of non-small cell lung cancer. J. Cancer 2014, 5, 706–714. [Google Scholar] [CrossRef] [PubMed]
- Piccinini, M.; Scandroglio, F.; Prioni, S.; Buccinna, B.; Loberto, N.; Aureli, M.; Chigorno, V.; Lupino, E.; DeMarco, G.; Lomartire, A.; et al. Deregulated sphingolipid metabolism and membrane organization in neurodegenerative disorders. Mol. Neurobiol. 2010, 41, 314–340. [Google Scholar] [CrossRef] [PubMed]
- Ogretmen, B.; Hannun, Y.A. Biologically active sphingolipids in cancer pathogenesis and treatment. Nat. Rev. Cancer 2004, 4, 604–616. [Google Scholar] [CrossRef] [PubMed]
- Hoppstadter, J.; Dembek, A.; Horing, M.; Schymik, H.S.; Dahlem, C.; Sultan, A.; Wirth, N.; Al-Fityan, S.; Diesel, B.; Gasparoni, G.; et al. Dysregulation of cholesterol homeostasis in human lung cancer tissue and tumour-associated macrophages. EBioMedicine 2021, 72, 103578. [Google Scholar] [CrossRef]
- Hall, Z.; Wilson, C.H.; Burkhart, D.L.; Ashmore, T.; Evan, G.I.; Griffin, J.L. Myc linked to dysregulation of cholesterol transport and storage in nonsmall cell lung cancer. J. Lipid Res. 2020, 61, 1390–1399. [Google Scholar] [CrossRef]
- Yu, Z.; Chen, H.; Zhu, Y.; Ai, J.; Li, Y.; Gu, W.; Borgia, J.A.; Zhang, J.; Jiang, B.; Chen, W.; et al. Global lipidomics reveals two plasma lipids as novel biomarkers for the detection of squamous cell lung cancer: A pilot study. Oncol. Lett 2018, 16, 761–768. [Google Scholar] [CrossRef]
- Eltayeb, K.; La Monica, S.; Tiseo, M.; Alfieri, R.; Fumarola, C. Reprogramming of Lipid Metabolism in Lung Cancer: An Overview with Focus on EGFR-Mutated Non-Small Cell Lung Cancer. Cells 2022, 11, 413. [Google Scholar] [CrossRef]
- Loftfield, E.; Vogtmann, E.; Sampson, J.N.; Moore, S.C.; Nelson, H.; Knight, R.; Chia, N.; Sinha, R. Comparison of Collection Methods for Fecal Samples for Discovery Metabolomics in Epidemiologic Studies. Cancer Epidemiol. Biomark. Prev. 2016, 25, 1483–1490. [Google Scholar] [CrossRef]
- Koelmel, J.P.; Kroeger, N.M.; Ulmer, C.Z.; Bowden, J.A.; Patterson, R.E.; Cochran, J.A.; Beecher, C.W.W.; Garrett, T.J.; Yost, R.A. LipidMatch: An automated workflow for rule-based lipid identification using untargeted high-resolution tandem mass spectrometry data. BMC Bioinform. 2017, 18, 331. [Google Scholar] [CrossRef]
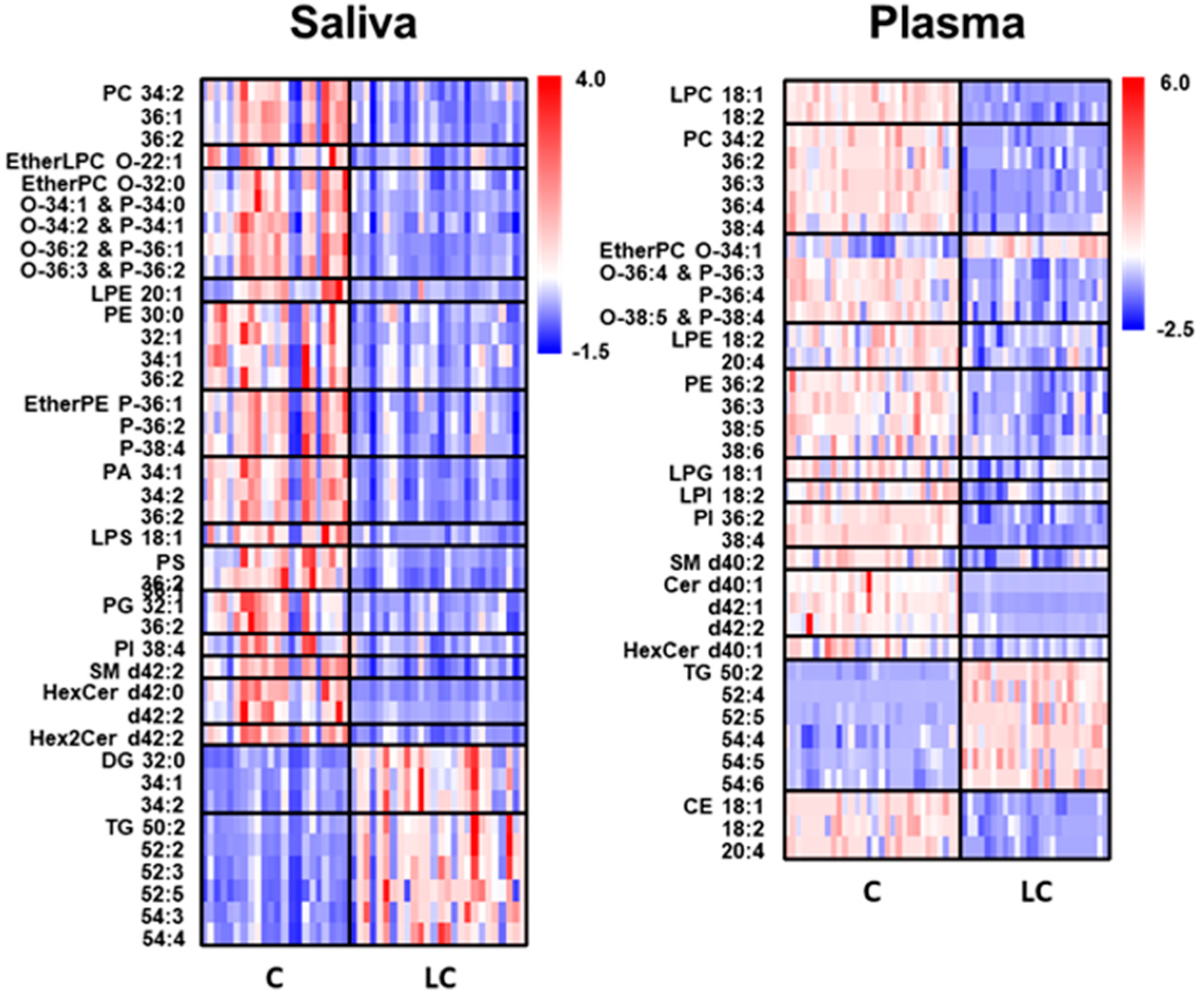
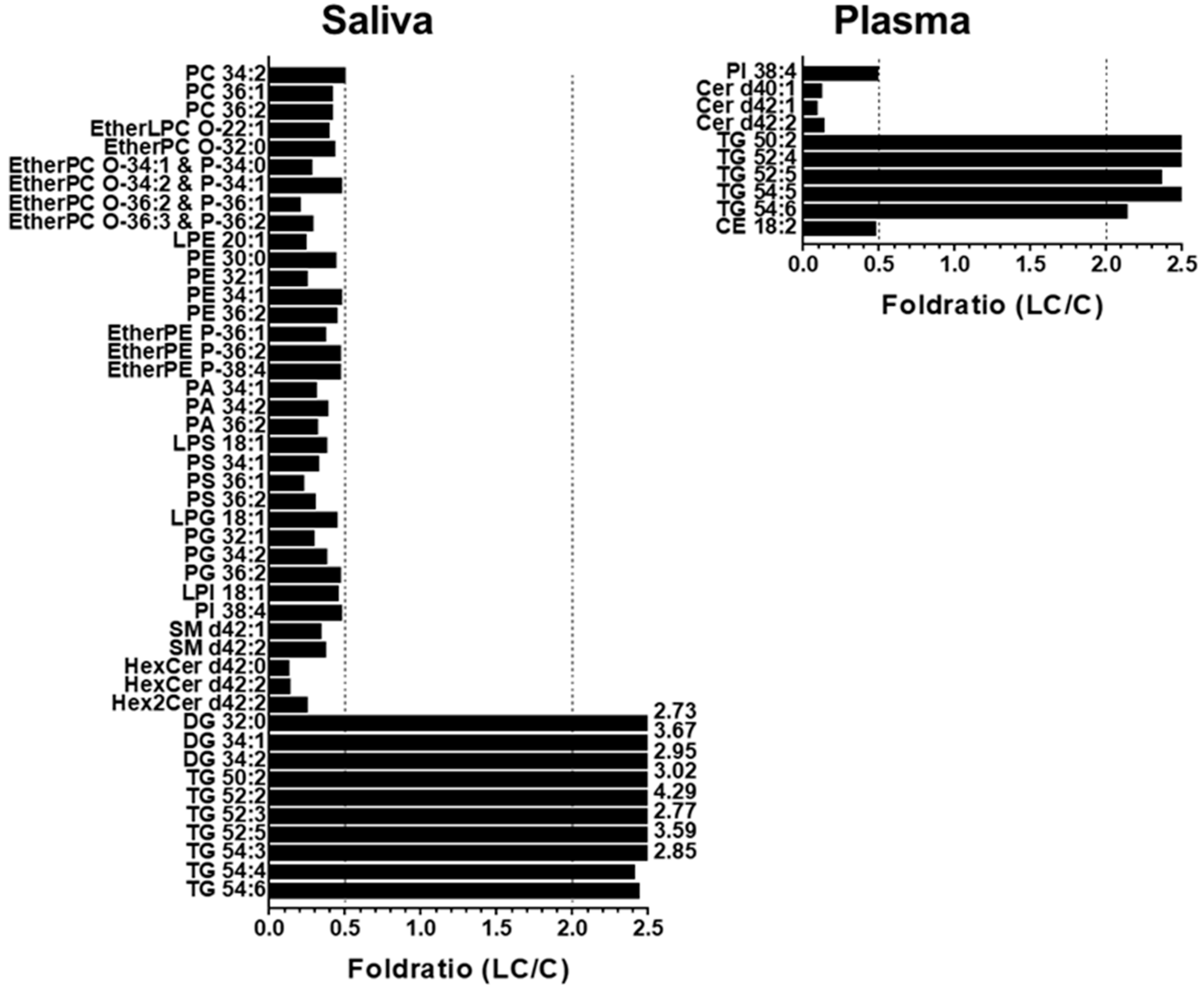
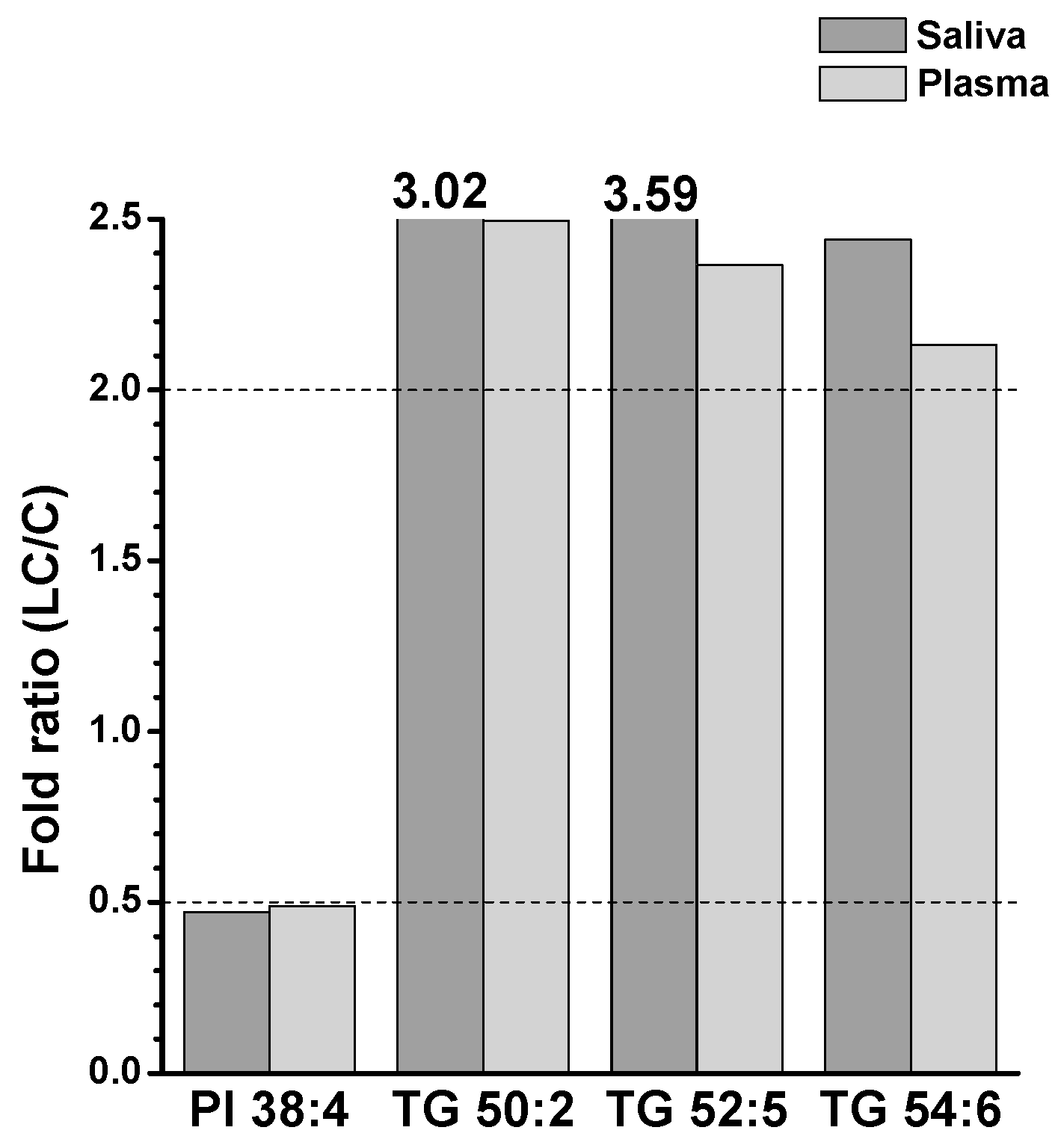
| Saliva | Plasma | ||||
|---|---|---|---|---|---|
| Molecular Species | AUC | Molecular Species | AUC | Molecular Species | AUC |
| PC 36:1 | 0.840 | PS 36:1 | 0.902 | PI 38:4 | 1.000 |
| PC 36:2 | 0.831 | PS 36:2 | 0.928 | Cer d40:1 | 1.000 |
| EtherPC O-34:1 & P-34:0 | 0.885 | PG 32:1 | 0.815 | Cer d42:1 | 1.000 |
| EtherPC O-34:2 & P-34:1 | 0.817 | SM d42:1 | 0.881 | Cer d42:2 | 0.948 |
| EtherPC O-36:2 & P-36:1 | 0.913 | SM d42:2 | 0.837 | TG 50:2 | 1.000 |
| EtherPC O-36:3 & P-36:2 | 0.841 | HexCer d42:0 | 0.940 | TG 52:4 | 0.969 |
| PE 32:1 | 0.866 | HexCer d42:2 | 0.872 | TG 52:5 | 0.927 |
| EtherPE P-36:1 | 0.835 | Hex2Cer d42:2 | 0.861 | TG 54:5 | 0.912 |
| PA 34:1 | 0.911 | DG 32:0 | 0.928 | TG 54:6 | 0.864 |
| PA 34:2 | 0.872 | DG 34:1 | 0.872 | CE 18:2 | 0.930 |
| PA 36:2 | 0.914 | DG 34:2 | 0.810 | ||
| LPS 18:1 | 0.815 | TG 52:2 | 0.847 | ||
| PS 34:1 | 0.936 | TG 52:3 | 0.806 | ||
| TG 52:5 | 0.844 | ||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hwang, B.Y.; Seo, J.W.; Muftuoglu, C.; Mert, U.; Guldaval, F.; Asadi, M.; Karakus, H.S.; Goksel, T.; Veral, A.; Caner, A.; et al. Salivary Lipids of Patients with Non-Small Cell Lung Cancer Show Perturbation with Respect to Plasma. Int. J. Mol. Sci. 2023, 24, 14264. https://doi.org/10.3390/ijms241814264
Hwang BY, Seo JW, Muftuoglu C, Mert U, Guldaval F, Asadi M, Karakus HS, Goksel T, Veral A, Caner A, et al. Salivary Lipids of Patients with Non-Small Cell Lung Cancer Show Perturbation with Respect to Plasma. International Journal of Molecular Sciences. 2023; 24(18):14264. https://doi.org/10.3390/ijms241814264
Chicago/Turabian StyleHwang, Bo Young, Jae Won Seo, Can Muftuoglu, Ufuk Mert, Filiz Guldaval, Milad Asadi, Haydar Soydaner Karakus, Tuncay Goksel, Ali Veral, Ayse Caner, and et al. 2023. "Salivary Lipids of Patients with Non-Small Cell Lung Cancer Show Perturbation with Respect to Plasma" International Journal of Molecular Sciences 24, no. 18: 14264. https://doi.org/10.3390/ijms241814264
APA StyleHwang, B. Y., Seo, J. W., Muftuoglu, C., Mert, U., Guldaval, F., Asadi, M., Karakus, H. S., Goksel, T., Veral, A., Caner, A., & Moon, M. H. (2023). Salivary Lipids of Patients with Non-Small Cell Lung Cancer Show Perturbation with Respect to Plasma. International Journal of Molecular Sciences, 24(18), 14264. https://doi.org/10.3390/ijms241814264





