The P-Site Loop of the Universally Conserved Bacterial Ribosomal Protein L5 Is Required for Maintaining Both Translation Rate and Fidelity
Abstract
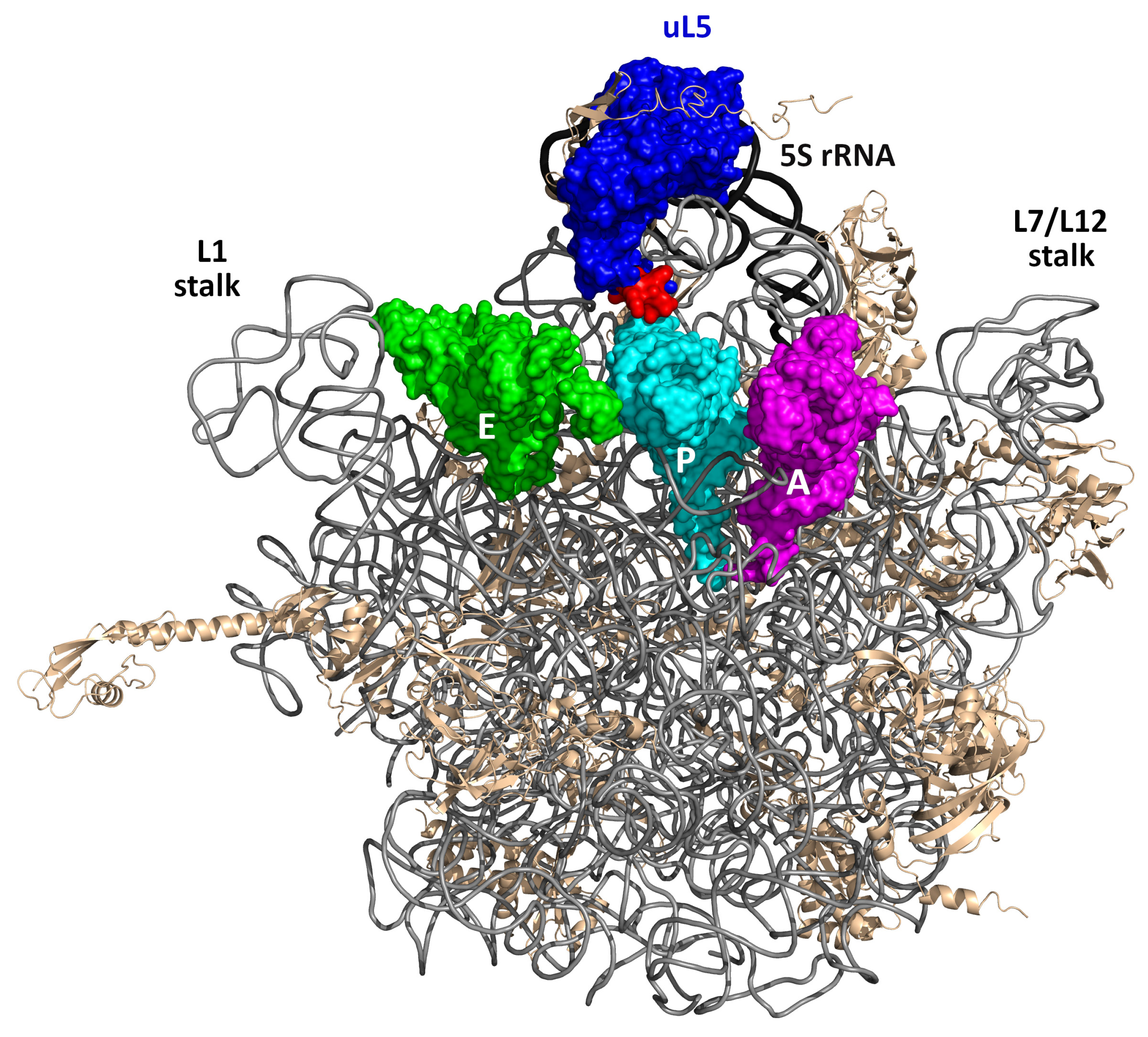
1. Introduction
2. Results
2.1. Deletion of Residues 73–80 of uL5 Reduces Growth Rate and Causes Cold Sensitivity
2.2. Ribosomal Profiles and Protein Composition of Ribosomes Are Virtually the Same in the ΔPSL Mutant and the Control Strain
2.3. The Rate of Protein Synthesis Is Reduced in the ΔPSL Mutant Cells
2.4. Translation Fidelity Is Reduced in the ΔPSL Mutant
3. Discussion
4. Materials and Methods
4.1. Introducing the Deletion in the Chromosomal rplE Gene
4.2. Constructing Reporter Chromosomal Lac Fusions for Dissecting Translation Fidelity In Vivo
4.3. Cell Growth and Experiments on Protein Synthesis and Translation Fidelity
4.4. Analysis of Ribosomal Profiles and Protein Composition of Ribosomes
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ban, N.; Beckmann, R.; Cate, J.H.D.; Dinman, J.D.; Dragon, F.; Ellis, S.R.; Lafontaine, D.L.J.; Lindahl, L.; Liljas, A.; Lipton, J.M.; et al. A new system for naming ribosomal proteins. Curr. Opin. Struct. Biol. 2014, 24, 165–169. [Google Scholar] [CrossRef] [PubMed]
- Korepanov, A.P.; Gongadze, G.M.; Garber, M.B.; Court, D.L.; Bubunenko, M.G. Importance of the 5 S rRNA-binding ribosomal proteins for cell viability and translation in Escherichia coli. J. Mol. Biol. 2007, 366, 1199–1208. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Korepanov, A.P.; Korobeinikova, A.V.; Shestakov, S.A.; Garber, M.B.; Gongadze, G.M. Protein L5 is crucial for in vivo assembly of the bacterial 50S ribosomal subunit central protuberance. Nucleic Acids Res. 2012, 40, 9153–9159. [Google Scholar] [CrossRef] [PubMed]
- Yusupov, M.M.; Yusupova, G.Z.; Baucom, A.; Lieberman, K.; Earnest, T.N.; Cate, J.H.; Noller, H.F. Crystal structure of the ribosome at 5.5 Å resolution. Science 2001, 292, 883–896. [Google Scholar] [CrossRef]
- Selmer, M.; Dunham, C.M.; Murphy, F.V., 4th; Weixlbaumer, A.; Petry, S.; Kelley, A.C.; Weir, J.R.; Ramakrishnan, V. Structure of the 70S ribosome complexed with mRNA and tRNA. Science 2006, 313, 1935–1942. [Google Scholar] [CrossRef]
- Polikanov, Y.S.; Starosta, A.L.; Juette, M.F.; Altman, R.B.; Terry, D.S.; Lu, W.; Burnett, B.J.; Dinos, G.; Reynolds, K.A.; Blanchard, S.C.; et al. Distinct tRNA Accommodation Intermediates Observed on the Ribosome with the Antibiotics Hygromycin A and A201A. Mol. Cell 2015, 58, 832–844. [Google Scholar] [CrossRef]
- O’Connor, M.; Thomas, C.L.; Zimmermann, R.A.; Dahlberg, A.E. Decoding fidelity at the ribosomal A and P sites: Influence of mutations in three different regions of the decoding domain in 16S rRNA. Nucleic Acids Res. 1997, 25, 1185–1193. [Google Scholar] [CrossRef]
- Shoji, S.; Dambacher, C.M.; Shajani, Z.; Williamson, J.R.; Schultz, P.G. Systematic Chromosomal Deletion of Bacterial Ribosomal Protein Genes. J. Mol. Biol. 2011, 413, 751–761. [Google Scholar] [CrossRef]
- Rhodin, M.H.J.; Dinman, J.D. A flexible loop in yeast ribosomal protein L11 coordinates P-site tRNA binding. Nucleic Acids Res. 2010, 38, 8377–8389. [Google Scholar] [CrossRef][Green Version]
- Bubunenko, M.; Baker, T.; Court, D.L. Essentiality of Ribosomal and Transcription Antitermination Proteins Analyzed by Systematic Gene Replacement in Escherichia coli. J. Bacteriol. 2007, 189, 2844–2853. [Google Scholar] [CrossRef]
- Cukras, A.R.; Green, R. Multiple effects of S13 in modulating the strength of intersubunit interactions in the ribosome during translation. J. Mol. Biol. 2005, 349, 47–59. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Paci, M.; Fox, G.E. Major centers of motion in the large ribosomal RNAs. Nucleic Acids Res. 2015, 43, 4640–4649. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bogdanov, A.A.; Dontsova, O.A.; Dokudovskaya, S.S.; Lavrik, I.N. Structure and function of 5S rRNA in the ribosome. Biochem. Cell Biol. Biochim. Biol. Cell 1995, 73, 869–876. [Google Scholar] [CrossRef] [PubMed]
- Harger, J.W.; Meskauskas, A.; Dinman, J.D. An “integrated model” of programmed ribosomal frameshifting. Trends Biochem. Sci. 2002, 27, 448–454. [Google Scholar] [CrossRef] [PubMed]
- Baranov, P.V.; Gesteland, R.F.; Atkins, J.F. P-site tRNA is a crucial initiator of ribosomal frameshifting. RNA 2004, 10, 221–230. [Google Scholar] [CrossRef]
- Atkins, J.F.; Björk, G.R. A gripping tale of ribosomal frameshifting: Extragenic suppressors of frameshift mutations spotlight P-site realignment. Microbiol. Mol. Biol. Rev. MMBR 2009, 73, 178–210. [Google Scholar] [CrossRef]
- Herr, A.J.; Atkins, J.F.; Gesteland, R.F. Mutations which alter the elbow region of tRNA2Gly reduce T4 gene 60 translational bypassing efficiency. EMBO J. 1999, 18, 2886. [Google Scholar] [CrossRef]
- Karimi, R.; Ehrenberg, M. Dissociation rates of peptidyl-tRNA from the P-site of E. coli ribosomes. EMBO J. 1996, 15, 1149–1154. [Google Scholar] [CrossRef]
- Karimi, R.; Ehrenberg, M. Dissociation rate of cognate peptidyl-tRNA from the A-site of hyper-accurate and error-prone ribosomes. Eur. J. Biochem. 1994, 226, 355–360. [Google Scholar] [CrossRef]
- Burakovsky, D.E.; Sergiev, P.V.; Steblyanko, M.A.; Kubarenko, A.V.; Konevega, A.L.; Bogdanov, A.A.; Rodnina, M.V.; Dontsova, O.A. Mutations at the accommodation gate of the ribosome impair RF2-dependent translation termination. RNA 2010, 16, 1848–1853. [Google Scholar] [CrossRef][Green Version]
- Arkov, A.L.; Murgola, E.J. Ribosomal RNAs in translation termination: Facts and hypotheses. Biochemistry 1999, 64, 1354–1359. [Google Scholar]
- Rodnina, M.V.; Wintermeyer, W. Protein Elongation, Co-translational Folding and Targeting. J. Mol. Biol. 2016, 428, 2165–2185. [Google Scholar] [CrossRef]
- Yu, D.; Ellis, H.M.; Lee, E.C.; Jenkins, N.A.; Copeland, N.G.; Court, D.L. An efficient recombination system for chromosome engineering in Escherichia coli. Proc. Natl. Acad. Sci. USA 2000, 97, 5978–5983. [Google Scholar] [CrossRef]
- Miller, J.H. Experiments in Molecular Genetics; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 1972; p. 466. [Google Scholar]
- Olins, P.O.; Nomura, M. Translational regulation by ribosomal protein S8 in Escherichia coli: Structural homology between rRNA binding site and feedback target on mRNA. Nucleic Acids Res. 1981, 9, 1757–1764. [Google Scholar] [CrossRef]
- Chang, A.C.; Cohen, S.N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J. Bacteriol. 1978, 134, 1141–1156. [Google Scholar] [CrossRef]
- Court, D.L.; Swaminathan, S.; Yu, D.; Wilson, H.; Baker, T.; Bubunenko, M.; Sawitzke, J.; Sharan, S.K. Mini-lambda: A tractable system for chromosome and BAC engineering. Gene 2003, 315, 63–69. [Google Scholar] [CrossRef]
- Blattner, F.R.; Plunkett, G.; Bloch, C.A.; Perna, N.T.; Burland, V.; Riley, M.; Collado-Vides, J.; Glasner, J.D.; Rode, C.K.; Mayhew, G.F.; et al. The Complete Genome Sequence of Escherichia coli K-12. Science 1997, 277, 1453–1462. [Google Scholar] [CrossRef] [PubMed]
- Gorski, K.; Roch, J.M.; Prentki, P.; Krisch, H.M. The stability of bacteriophage T4 gene 32 mRNA: A 5′ leader sequence that can stabilize mRNA transcripts. Cell 1985, 43, 461–469. [Google Scholar] [CrossRef] [PubMed]
- Duval, M.; Korepanov, A.; Fuchsbauer, O.; Fechter, P.; Haller, A.; Fabbretti, A.; Choulier, L.; Micura, R.; Klaholz, B.P.; Romby, P.; et al. Escherichia coli Ribosomal Protein S1 Unfolds Structured mRNAs Onto the Ribosome for Active Translation Initiation. PLoS Biol. 2013, 11, e1001731. [Google Scholar] [CrossRef] [PubMed]
- Andersson, D.I.; Bohman, K.; Isaksson, L.A.; Kurland, C.G. Translation rates and misreading characteristics of rpsD mutants in Escherichia coli. Mol. Gen. Genet. MGG 1982, 187, 467–472. [Google Scholar] [CrossRef] [PubMed]
- Staehelin, T.; Maglott, D.M.; Monro, R.E. On the catalytic center of peptidyl transfer: A part of the 50 S ribosome structure. Cold Spring Harb. Symp. Quant. Biol. 1969, 34, 39–48. [Google Scholar] [CrossRef] [PubMed]
- Hardy, S.J.; Kurland, C.G.; Voynow, P.; Mora, G. The ribosomal proteins of Escherichia coli. I. Purification of the 30S ribosomal proteins. Biochemistry 1969, 8, 2897–2905. [Google Scholar] [CrossRef] [PubMed]
- Madjar, J.J.; Michel, S.; Cozzone, A.J.; Reboud, J.P. A method to identify individual proteins in four different two-dimensional gel electrophoresis systems: Application to Escherichia coli ribosomal proteins. Anal. Biochem. 1979, 92, 174–182. [Google Scholar] [CrossRef] [PubMed]






Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bubunenko, M.G.; Korepanov, A.P. The P-Site Loop of the Universally Conserved Bacterial Ribosomal Protein L5 Is Required for Maintaining Both Translation Rate and Fidelity. Int. J. Mol. Sci. 2023, 24, 14285. https://doi.org/10.3390/ijms241814285
Bubunenko MG, Korepanov AP. The P-Site Loop of the Universally Conserved Bacterial Ribosomal Protein L5 Is Required for Maintaining Both Translation Rate and Fidelity. International Journal of Molecular Sciences. 2023; 24(18):14285. https://doi.org/10.3390/ijms241814285
Chicago/Turabian StyleBubunenko, Mikhail G., and Alexey P. Korepanov. 2023. "The P-Site Loop of the Universally Conserved Bacterial Ribosomal Protein L5 Is Required for Maintaining Both Translation Rate and Fidelity" International Journal of Molecular Sciences 24, no. 18: 14285. https://doi.org/10.3390/ijms241814285
APA StyleBubunenko, M. G., & Korepanov, A. P. (2023). The P-Site Loop of the Universally Conserved Bacterial Ribosomal Protein L5 Is Required for Maintaining Both Translation Rate and Fidelity. International Journal of Molecular Sciences, 24(18), 14285. https://doi.org/10.3390/ijms241814285





