Morphometric and Nanomechanical Screening of Peripheral Blood Cells with Atomic Force Microscopy for Label-Free Assessment of Alzheimer’s Disease, Parkinson’s Disease, and Amyotrophic Lateral Sclerosis
Abstract
:1. Introduction
2. Current Biomarkers for the Diagnosis of Alzheimer’s Disease, Parkinson’s Disease, and Amyotrophic Lateral Sclerosis
2.1. Fluid Biomarkers in Alzheimer’s Disease
2.2. Fluid Biomarkers in Parkinson’s Disease
2.3. Fluid Biomarkers in Amyotrophic Lateral Sclerosis
3. Platelets and Red Blood Cells in Alzheimer’s Disease, Parkinson’s Disease, and Amyotrophic Lateral Sclerosis
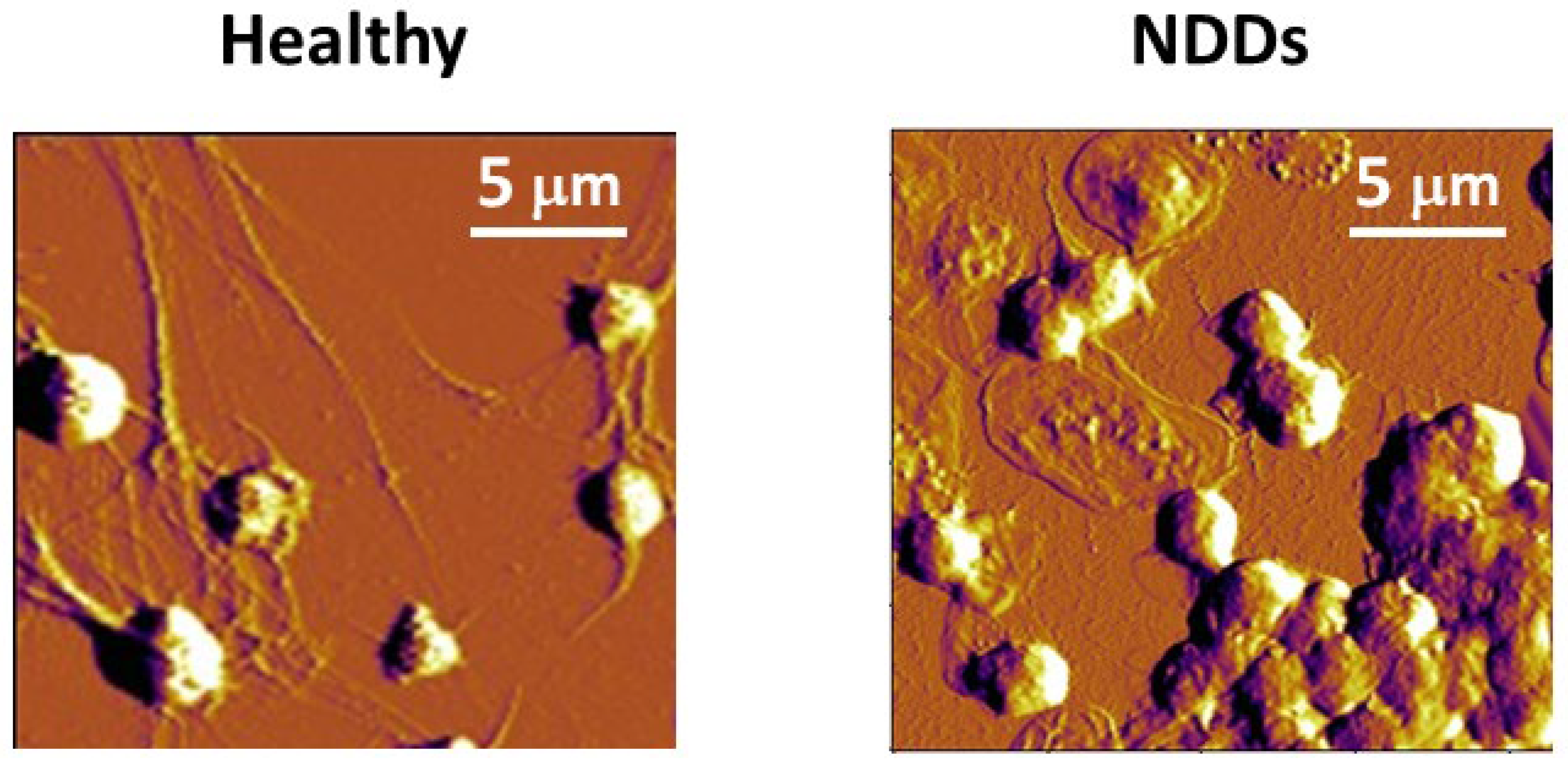
3.1. Platelets
3.2. Red Blood Cells
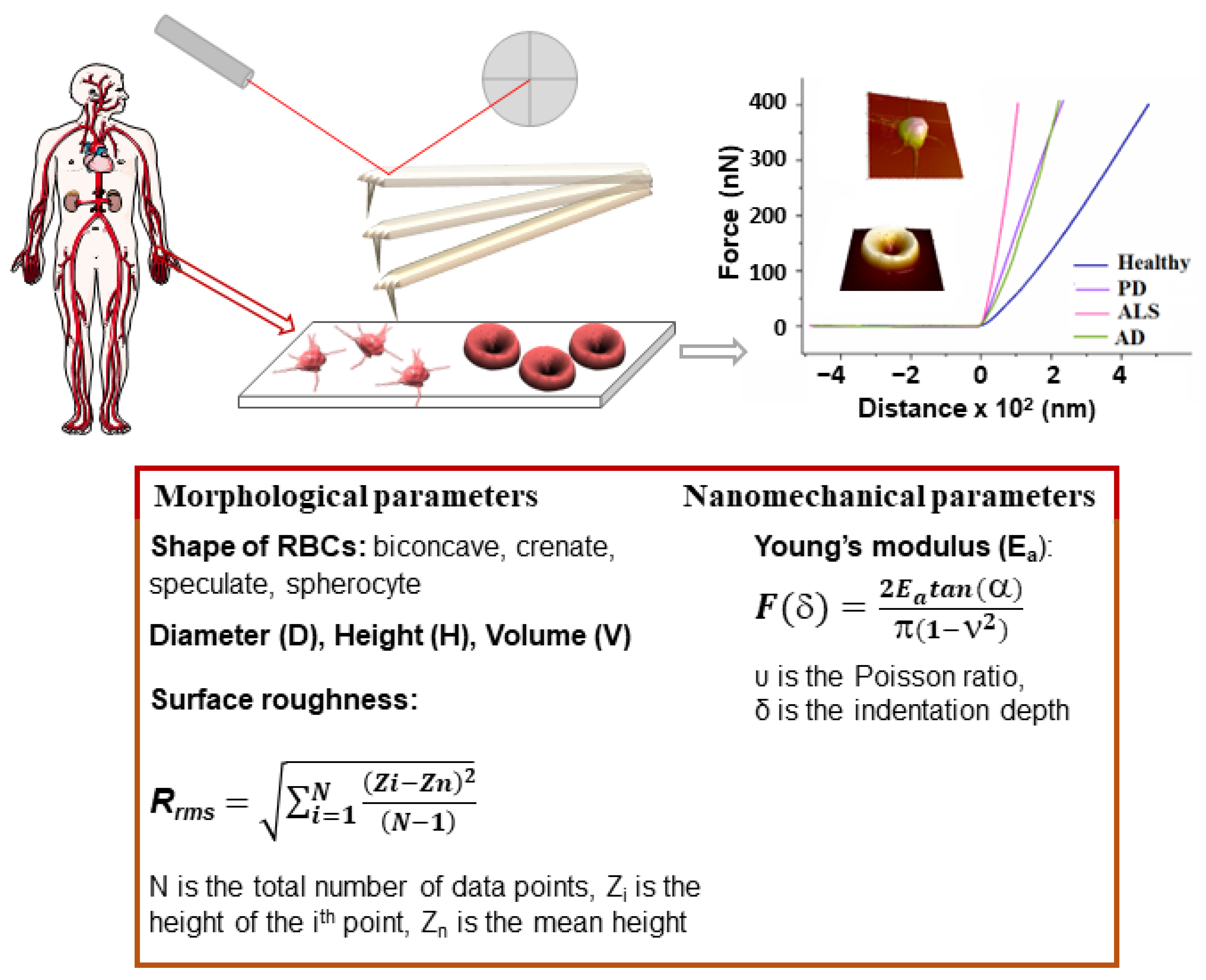
4. Atomic Force Microscopy: Morphometric and Nanomechanical Parameters in Neurodegenerative Pathologies
4.1. Atomic Force Microscopy as a Diagnostic Tool
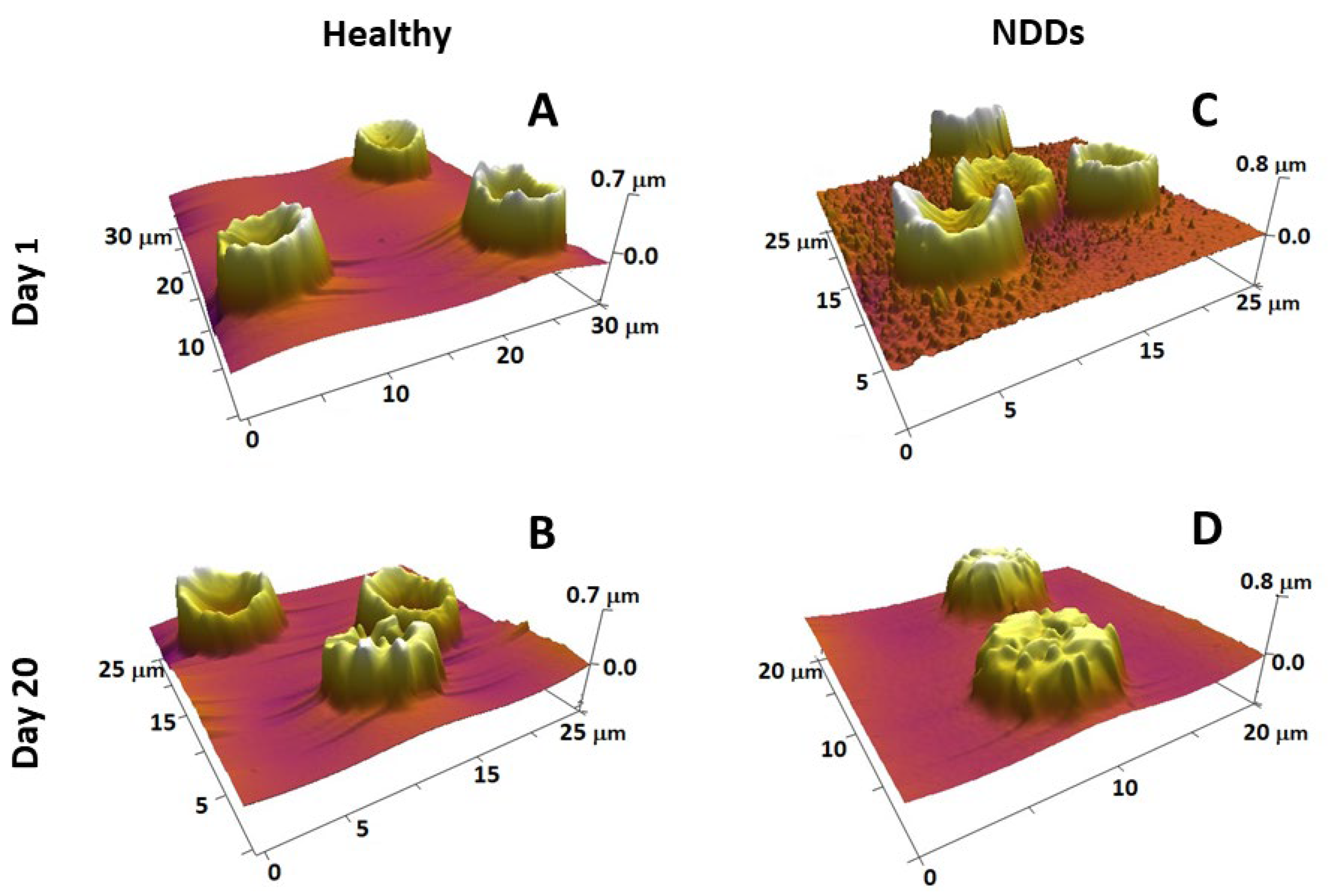
4.2. AFM Studies of Platelets and Red Blood Cells in Healthy and Pathological Conditions
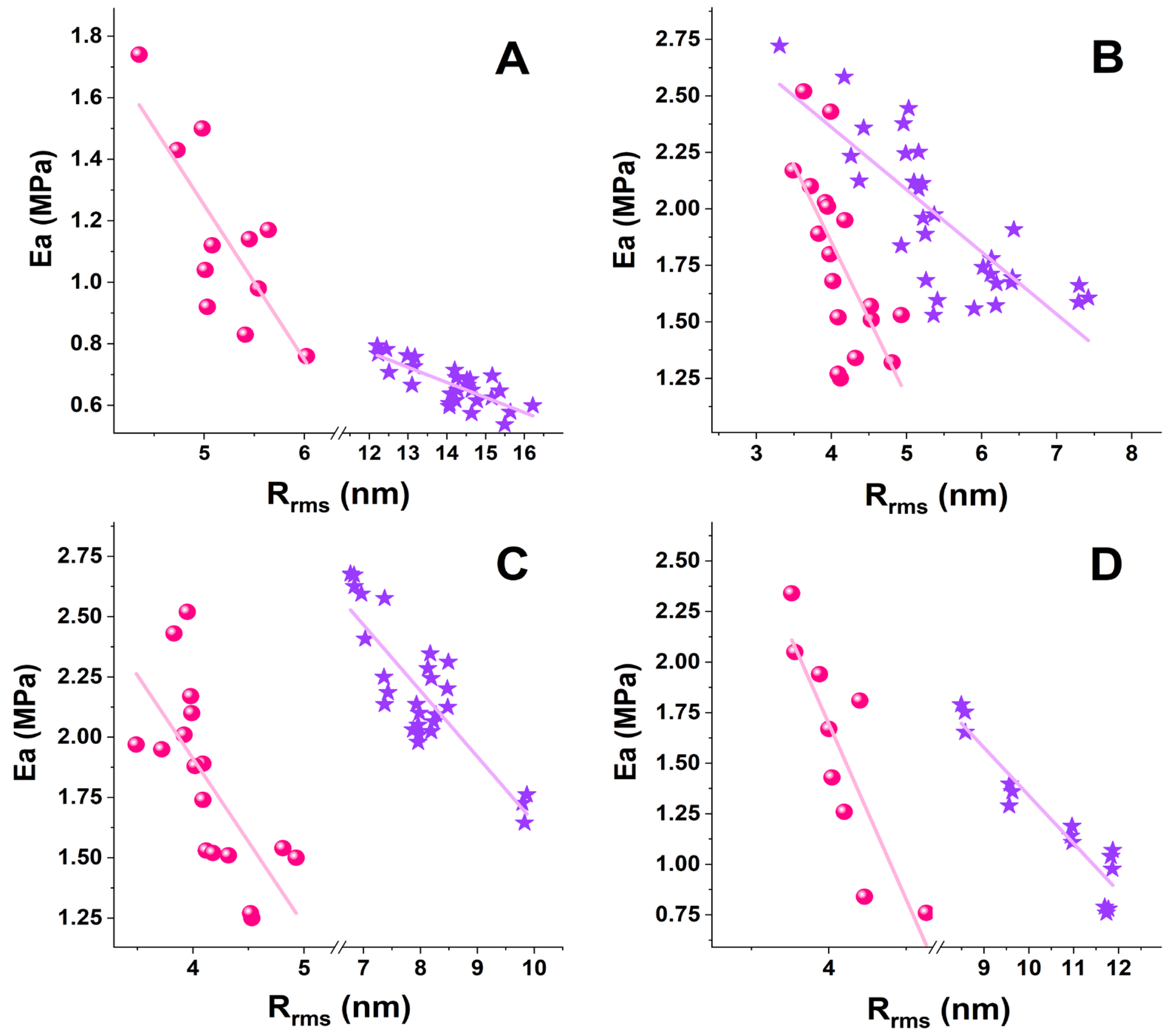
4.3. Morphometric and Nanomechanical Parameters of Red Blood Cells and Platelets in Neurodegenerative Disorders
4.4. Nanoscale Structural Features and Dynamics of Amyloidogenic Proteins
4.5. AFM versus Other Imaging Techniques
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Hou, Y.; Dan, X.; Babbar, M.; Wei, Y.; Hasselbalch, S.G.; Croteau, D.L.; Bohr, V.A. Ageing as a risk factor for neurodegenerative disease. Nat. Rev. Neurol. 2019, 15, 565–581. [Google Scholar] [CrossRef] [PubMed]
- Ciccone, S.; Maiani, E.; Bellusci, G.; Diederich, M.; Gonfloni, S. Parkinson’s Disease: A Complex Interplay of Mitochondrial DNA Alterations and Oxidative Stress. Int. J. Mol. Sci. 2013, 14, 2388–2409. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Chen, M.; Jiang, J. Mitochondrial dysfunction in neurodegenerative diseases and drug targets via apoptotic signaling. Mitochondrion 2019, 49, 35–45. [Google Scholar] [CrossRef] [PubMed]
- Profaci, C.P.; Munji, R.N.; Pulido, R.S.; Daneman, R. The blood–brain barrier in health and disease: Important unanswered questions. J. Exp. Med. 2020, 217, e20190062. [Google Scholar] [CrossRef]
- Younes-Mhenni, S.; Frih-Ayed, M.; Kerkeni, A.; Bost, M.; Chazot, G. Peripheral blood markers of oxidative stress in Parkinson’s disease. Eur. Neurol. 2007, 58, 78–83. [Google Scholar] [CrossRef]
- Singh, A.; Kukreti, R.; Saso, L.; Kukreti, S. Oxidative Stress: A Key Modulator in Neurodegenerative Diseases. Molecules 2019, 24, 1583. [Google Scholar] [CrossRef]
- Jenner, P.; Olanow, C.W. Oxidative stress and the pathogenesis of Parkinson’s disease. Neurology 1996, 47, 161–170. [Google Scholar] [CrossRef]
- Betzer, C.; Jensen, P.H. Reduced Cytosolic Calcium as an Early Decisive Cellular State in Parkinson’s Disease and Synucleinopathies. Front. Neurosci. 2018, 12, 819. [Google Scholar] [CrossRef]
- Wang, X.; Zheng, W. Ca2+ homeostasis dysregulation in Alzheimer’s disease: A focus on plasma membrane and cell organelles. FASEB J. 2019, 33, 6697–6712. [Google Scholar] [CrossRef]
- Meszlényi, V.; Patai, R.; Polgár, T.F.; Nógrádi, B.; Körmöczy, L.; Kristóf, R.; Spisák, K.; Tripolszki, K.; Széll, M.; Obál, I.; et al. Passive Transfer of Sera from ALS Patients with Identified Mutations Evokes an Increased Synaptic Vesicle Number and Elevation of Calcium Levels in Motor Axon Terminals, Similar to Sera from Sporadic Patients. Int. J. Mol. Sci. 2020, 21, 5566. [Google Scholar] [CrossRef]
- Piscopo, P.; Bellenghi, M.; Manzini, V.; Crestini, A.; Pontecorvi, G.; Corbo, M.; Ortona, E.; Carè, A.; Confaloni, A. A Sex Perspective in Neurodegenerative Diseases: MicroRNAs as Possible Peripheral Biomarkers. Int. J. Mol. Sci. 2021, 22, 4423. [Google Scholar] [CrossRef] [PubMed]
- Goedert, M. NEURODEGENERATION. Alzheimer’s and Parkinson’s diseases: The prion concept in relation to assembled Aβ, tau, and α-synuclein. Science 2015, 349, 1255555. [Google Scholar] [CrossRef] [PubMed]
- Jellinker, K.A. Basic mechanisms of neurodegeneration: Acritical update. J. Cell Mol. Med. 2010, 14, 457–487. [Google Scholar] [CrossRef] [PubMed]
- Amor, S.; Peferoen, L.A.N.; Vogel, D.Y.S.; Breur, M.; van der Valk, P.; Baker, D.; van Noort, J.M. Inflammation in neurodegenerative diseases—An update. Immunology 2014, 142, 151–166. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, S. Oxidative Stress, Inflammation, and Disease. In Oxidative stress and Biomaterials; Dziubla, T., Butterfield, D.A., Eds.; Elsevier Inc./Academic Press: Cambridge, MA, USA, 2016; pp. 35–58. [Google Scholar]
- Migliore, L.; Coppedè, F. Environmental-induced oxidative stress in neurodegenerative disorders and aging. Mutat. Genet. Toxicol. Environ. Mutagen. 2009, 674, 73–84. [Google Scholar] [CrossRef] [PubMed]
- Thompson, T.B.; Chaggar, P.; Kuhl, E.; Goriely, A. Protein-protein interactions in neurodegenerative diseases: A conspiracy theory. PLoS Comput. Biol. 2020, 16, e1008267. [Google Scholar] [CrossRef]
- Giacomelli, C.; Daniele, S.; Martini, C. Potential biomarkers and novel pharmacological targets in protein aggregation-related neurodegenerative diseases. Biochem. Pharmacol. 2017, 131, 1–15. [Google Scholar] [CrossRef]
- Meldolesi, J. News about the Role of Fluid and Imaging Biomarkers in Neurodegenerative Diseases. Biomedicines 2021, 9, 252. [Google Scholar] [CrossRef]
- Wojsiat, J.; Prandelli, C.; Laskowska-Kaszub, K.; Martín-Requero, A.; Wojda, U. Oxidative Stress and Aberrant Cell Cycle in Alzheimer’s Disease Lymphocytes: Diagnostic Prospects. J. Alzheimer’s Dis. 2015, 46, 329–350. [Google Scholar] [CrossRef]
- Bertolotti, A. Importance of the subcellular location of protein deposits in neurodegenerative diseases. Curr. Opin. Neurobiol. 2018, 51, 127–133. [Google Scholar] [CrossRef]
- Cuanalo-Contreras, K.; Mukherjee, A.; Soto, C. Role of protein misfolding and proteostasis deficiency in protein misfolding diseases and aging. Int. J. Cell Biol. 2013, 2013, 638083. [Google Scholar] [CrossRef] [PubMed]
- Kikis, E.A.; Gidalevitz, T.; Morimoto, R.I. Protein homeostasis in models of aging and age-related conformational disease. Adv. Exp. Med. Biol. 2010, 694, 138–159. [Google Scholar] [PubMed]
- Soto, C.; Pritzkow, S. Unfolding the role of protein misfolding in neurodegenerative diseases. Nat. Rev. Neurosci. 2018, 4, 49–60. [Google Scholar] [CrossRef] [PubMed]
- Baldacci, F.; Daniele, S.; Piccarducci, R.; Giampietri, L.; Pietrobono, D.; Giorgi, F.S.; Nicoletti, V.; Frosini, D.; Libertini, P.; Lo Gerfo, A.; et al. Potential Diagnostic Value of Red Blood Cells α-Synuclein Heteroaggregates in Alzheimer’s Disease. Mol. Neurobiol. 2019, 56, 6451–6459. [Google Scholar] [CrossRef] [PubMed]
- Kiko, T.; Nakagawa, K.; Satoh, A.; Tsuduki, T.; Furukawa, K.; Arai, H.; Miyazawa, T. Amyloid β levels in human red blood cells. PLoS ONE 2012, 7, e49620. [Google Scholar] [CrossRef]
- Andersen, A.D.; Binzer, M.; Stenager, E.; Gramsbergen, J.B. Cerebrospinal fluid biomarkers for Parkinson’s disease: A systematic review. Acta Neurol. Scand. 2017, 135, 34–56. [Google Scholar] [CrossRef]
- Beach, T.G. A Review of Biomarkers for Neurodegenerative Disease: Will They Swing Us Across the Valley? Neurol. Ther. 2017, 6, 5–13. [Google Scholar] [CrossRef]
- Bridel, C.; van Wieringen, W.N.; Zetterberg, H.; Tijms, B.M.; Teunissen, C.E.; The NFL Group. Diagnostic value of cerebrospinalfluid neurofilament light protein in neurology: A systematic review and meta-analysis. JAMA Neurol. 2019, 76, 1035–1048. [Google Scholar] [CrossRef]
- Olsson, B.; Lautner, R.; Andreasson, U.; Öhrfelt, A.; Portelius, E.; Bjerke, M.; Hölttä, M.; Rosén, C. CSF and blood biomarkers for the diagnosis of Alzheimer’s disease: A systematic review and meta-analysis. Lancet Neurol. 2016, 15, 673–684. [Google Scholar] [CrossRef]
- Nakamura, A.; Kaneko, N.; Villemagne, V.L.; Kato, T.; Doecke, J.; Doré, V.; Fowler, C.; Li, Q.-X.; Martins, R.; Rowe, C.; et al. High performance plasma amyloid-β biomarkers for Alzheimer’s disease. Nature 2018, 554, 249–254. [Google Scholar] [CrossRef]
- Seelaar, H.; Van Swieten, J.C. In vivo PET imaging of neuroinflammation in familial frontotemporal dementia. J. Neurol. Neurosurg. Psychiatry 2021, 92, 319–322. [Google Scholar] [CrossRef] [PubMed]
- Hansson, O. Biomarkers for neurodegenerative diseases. Nat. Med. 2021, 27, 954–963. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Río, M.; Caballero, M.M.; Górriz Sáez, J.M.; Mínguez-Castellanos, A. Diagnosis of Neurodegenerative Diseases: The Clinical Approach. Curr. Alzheimer Res. 2016, 13, 469–474. [Google Scholar] [CrossRef] [PubMed]
- Shusharina, N.; Yukhnenko, D.; Botman, S.; Sapunov, V.; Savinov, V.; Kamyshov, G.; Sayapin, D.; Voznyuk, I. Modern Methods of Diagnostics and Treatment of Neurodegenerative Diseases and Depression. Diagnostics 2023, 13, 573. [Google Scholar] [CrossRef]
- Jack, C.R., Jr.; Petersen, R.C.; Xu, Y.C.; O’Brien, P.C.; Smith, G.E.; Ivnik, R.J.; Boeve, B.F.; Waring, S.C.; Tangalos, E.G.; Kokmen, E. Prediction of AD with MRI-based hippocampal volume in mild cognitive impairment. Neurology 1999, 52, 1397–1403. [Google Scholar] [CrossRef]
- Kamagata, K.; Andica, C.; Kato, A.; Saito, Y.; Uchida, W.; Hatano, T.; Lukies, M.; Ogawa, T.; Takeshige-Amano, H.; Akashi, T.; et al. Diffusion Magnetic Resonance Imaging-Based Biomarkers for Neurodegenerative Diseases. Int. J. Mol. Sci. 2021, 22, 5216. [Google Scholar] [CrossRef] [PubMed]
- Assaf, Y.; Pasternak, O. Diffusion Tensor Imaging (DTI)-based White Matter Mapping in Brain Research: A Review. J. Mol. Neurosci. 2008, 34, 51–61. [Google Scholar] [CrossRef] [PubMed]
- Choo, X.Y.; Alukaidey, L.; White, A.R.; Grubman, A. Neuroinflammation and Copper in Alzheimer’s Disease. Int. J. Alzheimer’s Dis. 2013, 2013, 145345. [Google Scholar] [CrossRef]
- Grimmer, T.; Wutz, C.; Alexopoulos, P.; Drzezga, A.; Förster, S.; Förstl, H.; Goldhardt, O.; Ortner, M.; Sorg, C.; Kurz, A. Visual Versus Fully Automated Analyses of β-FDG and Amyloid PET for Prediction of Dementia Due to Alzheimer Disease in Mild Cognitive Impairment. J. Nucl. Med. 2016, 57, 204–207. [Google Scholar] [CrossRef]
- Rowley, P.A.; Samsonov, A.A.; Betthauser, T.J.; Pirasteh, A.; Johnson, S.C.; Eisenmenger, L.B. Amyloid and Tau PET Imaging of Alzheimer Disease and Other Neurodegenerative Conditions. Semin. Ultrasound CT MRI 2020, 41, 572–583. [Google Scholar] [CrossRef]
- Knopman, D.S.; Jagust, J.W. Alzheimer Disease Spectrum: Syndrome and Etiology from Clinical and PET Imaging Perspectives. Neurology 2021, 96, 7. [Google Scholar] [CrossRef]
- Saeed, U.; Compagnone, J.; Aviv, R.I.; Strafella, A.P.; Black, S.E.; Lang, A.E.; Masellis, M. Imaging biomarkers in Parkinson’s disease and Parkinsonian syndromes: Current and emerging concepts. Transl. Neurodegener. 2017, 6, 8. [Google Scholar] [CrossRef] [PubMed]
- Jack, C.R., Jr.; Wiste, H.J.; Algeciras-Schimnich, A.; Figdore, D.J.; Schwarz, C.G.; Lowe, V.J.; Ramanan, V.K.; Vemuri, P.; Mielke, M.M.; Knopman, D.S.; et al. Predicting amyloid PET and tau PET stages with plasma biomarkers. Brain 2023, 146, 2029–2044. [Google Scholar] [CrossRef] [PubMed]
- Cova, I.; Priori, A. Diagnostic biomarkers for Parkinson’s disease at a glance: Where are we? J. Neural. Transm. 2018, 125, 1417–1432. [Google Scholar] [CrossRef] [PubMed]
- He, R.; Yan, X.; Guo, J.; Xu, Q.; Tang, B.; Sun, Q. Recent advances in biomarkers for Parkinson’s disease. Front. Aging Neurosci. 2018, 10, 305. [Google Scholar] [CrossRef]
- Laterza, O.F.; Hendrickson, R.C.; Wagner, J.A. Molecular Biomarkers. Ther. Innov. Regul. Sci. 2007, 41, 573–585. [Google Scholar] [CrossRef]
- Broza, Y.Y.; Zhou, X.; Yuan, M.; Qu, D.; Zheng, Y.; Vishinkin, R.; Khatib, M.; Wu, W.; Haick, H. Disease Detection with Molecular Biomarkers: From Chemistry of Body Fluids to Nature-Inspired Chemical Sensors. Chem. Rev. 2019, 119, 11761–11817. [Google Scholar] [CrossRef]
- Blennow, K.; Hampel, H.; Weiner, M.; Zetterberg, H. Cerebrospinal fluid and plasma biomarkers in Alzheimer disease. Nat. Rev. Neurol. 2010, 6, 131–144. [Google Scholar] [CrossRef]
- Lewczuk, P.; Riederer, P.; O’Bryant, S.E.; Verbeek, M.M.; Dubois, B.; Visser, P.J.; Jellinger, K.A.; Engelborghs, S.; Ramirez, A.; Parnetti, L.; et al. Cerebrospinal fluid and blood biomarkers for neurodegenerative dementias: An update of the Consensus of the Task Force on Biological Markers in Psychiatry of the World Federation of Societies of Biological Psychiatry. World J. Biol. Psychiatry 2018, 19, 244–328. [Google Scholar] [CrossRef]
- Blennow, K.; Zetterberg, H. The past and future of Alzheimer’s disease fluid biomarkers. J. Alzhimer’s Dis. 2018, 62, 1125–1140. [Google Scholar] [CrossRef]
- Wojsiat, J.; Laskowska-Kaszub, K.; Mietelska-Porowska, A.; Wojda, U. Search for Alzheimer’s disease biomarkers in blood cells: Hypotheses-driven approach. Biomark. Med. 2017, 11, 917–931. [Google Scholar] [CrossRef]
- Posavi, M.; Diaz-Ortiz, M.; Liu, B.; Swanson, C.R.; Skrinak, R.T.; Hernandez-Con, P.; Amado, D.A.; Fullard, M.; Rick, J.; Siderowf, A.; et al. Characterization of Parkinson’s disease using blood-based biomarkers: A multicohort proteomic analysis. PLoS Med. 2019, 16, e1002931. [Google Scholar] [CrossRef] [PubMed]
- Ashton, N.J.; Hye, A.; Rajkumar, A.P.; Leuzy, A.; Snowden, S.; Suárez-Calvet, M.; Karikari, T.K.; Schöll, M.; La Joie, R.; Rabinovici, G.D.; et al. An update on blood-based biomarkers for non-Alzheimer neurodegenerative disorders. Nat. Rev. Neurol. 2020, 16, 265–284. [Google Scholar] [CrossRef] [PubMed]
- Langbaum, J.B.; Fleisher, A.S.; Chen, K.; Ayutyanont, N.; Lopera, F.; Quiroz, Y.T.; Caselli, R.J.; Tariot, P.N.; Reiman, E.M. Ushering in the study and treatment of preclinical Alzheimer disease. Nat. Rev. Neurol. 2013, 9, 371–381. [Google Scholar] [CrossRef]
- Schapira, A.H. Recent developments in biomarkers in Parkinson disease. Curr. Opin. Neurol. 2013, 26, 395–400. [Google Scholar] [CrossRef] [PubMed]
- Lee, P.H.; Lee, G.; Park, H.J.; Bang, O.Y.; Joo, I.S.; Huh, K. The plasma α-synuclein levels in patients with Parkinson’s disease and multiple system atrophy. J. Neural Transm. 2006, 113, 1435–1439. [Google Scholar] [CrossRef]
- Li, Q.X.; Mok, S.S.; Laughton, K.M.; McLean, C.A.; Cappai, R.; Masters, C.L.; Culvenor, J.G.; Horne, M.K. Plasma α-synuclein is decreased in subjects with Parkinson’s disease. Exp. Neurol. 2007, 204, 583–588. [Google Scholar] [CrossRef]
- Hong, Z.; Shi, M.; Chung, K.A.; Quinn, J.F.; Peskind, E.R.; Galasko, D.; Jankovic, J.; Zabetian, C.P.; Leverenz, J.B.; Baird, G.; et al. DJ-1 and α-synuclein in human cerebrospinal fluid as biomarkers of Parkinson’s disease. Brain 2010, 133, 713–726. [Google Scholar] [CrossRef]
- Mollenhauer, B.; Locascio, J.L.; Schulz-Schaeffer, W.; Sixel-Döring, F.; Trenkwalder, C.; Schlossmacher, M.G. α-synuclein and tau concentrations in cerebrospinal fluid of patients presenting with parkinsonism: A cohort study. Lancet Neurol. 2011, 10, 230–240. [Google Scholar] [CrossRef]
- Lin, C.-H.; Yang, S.-Y.; Horng, H.-E.; Yang, C.-C.; Chieh, J.-J.; Chen, H.-H.; Liu, B.-H.; Chiu, M.-J. Plasma α-synuclein predicts cognitive decline in Parkinson’s disease. J. Neurol. Neurosurg. Psychiatry 2017, 88, 818–824. [Google Scholar] [CrossRef]
- Polimeno, L.; Asabella, A.N.; Mazzocca, A.; De Fazio, G.; Polimeno, R.; Buquicchio, R.; Lavelli, V.; Rubini, G. Plasma levels of clusterin are representative of the early phase of the neurodegenerative process in Parkinson’s disease. J. Clin. Mol. Med. 2018, 1, 1–5. [Google Scholar] [CrossRef]
- Chio, A.; Calvo, A.; Bovio, G.; Canosa, A.; Bertuzzo, D.; Galmozzi, F.; Cugnasco, P.; Clerico, M.; De Mercanti, S.; Bersano, E.; et al. Amyotrophic lateral sclerosis outcome measures and the role of albumin and creatinine. JAMA Neurol. 2014, 71, 1134–1142. [Google Scholar] [CrossRef]
- Ryberg, H.; Bowser, R. Protein biomarkers for amyotrophic lateral sclerosis. Expert Rev. Proteom. 2008, 5, 249–262. [Google Scholar] [CrossRef] [PubMed]
- Turner, M.R.; Kiernan, M.C.; Leigh, P.N.; Talbot, K. Biomarkers in amyotrophic lateral sclerosis. Lancet Neurol. 2009, 8, 94–109. [Google Scholar] [CrossRef]
- Quinn, J.F.; Patel, T.; Wong, D.; Das, S.; Freedman, J.E.; Laurent, L.C.; Carter, B.S.; Hochberg, F.; Van Keuren-Jensen, K.; Huentelman, M.; et al. Extracellular RNAs: Development as biomarkers of human disease. J. Extracell. Vesicles 2015, 4, 27495. [Google Scholar] [CrossRef] [PubMed]
- Blennow, K.; Zetterberg, H. Biomarkers for Alzheimer’s Disease: Current Status and Prospects for the Future. J. Intern. Med. 2018, 284, 643–663. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, P.; Elmi, M.; Goozee, K.; Shah, T.; Sohrabi, H.R.; Dias, C.B.; Pedrini, S.; Shen, K.; Asih, P.R.; Dave, P.; et al. Ultrasensitive Detection of Plasma Amyloid-β as a Biomarker for Cognitively Normal Elderly Individuals at Risk of Alzheimer’s Disease. J. Alzheimer’s Dis. 2019, 71, 775–783. [Google Scholar] [CrossRef]
- Álvarez-Sánchez, L.; Peña-Bautista, C.; Ferré-González, L.; Balaguer, A.; Baquero, M.; Casanova-Estruch, B.; Cháfer-Pericás, C. Assessment of Plasma and Cerebrospinal Fluid Biomarkers in Different Stages of Alzheimer’s Disease and Frontotemporal Dementia. Int. J. Mol. Sci. 2023, 24, 1226. [Google Scholar] [CrossRef]
- Cho, H.J.; Schulz, P.; Venkataraman, L.; Caselli, R.J.; Sierks, M.R. Sex-Specific Multiparameter Blood Test for the Early Diagnosis of Alzheimer’s Disease. Int. J. Mol. Sci. 2022, 23, 15670. [Google Scholar] [CrossRef]
- Cullen, N.C.; Leuzy, A.; Janelidze, S.; Palmqvist, S.; Svenningsson, A.L.; Stomrud, E.; Dage, J.L.; Mattsson-Carlgren, N.; Hansson, O. Plasma Biomarkers of Alzheimer’s Disease Improve Prediction of Cognitive Decline in Cognitively Unimpaired Elderly Populations. Nat. Commun. 2021, 12, 3555. [Google Scholar] [CrossRef]
- Leuzy, A.; Mattsson-Carlgren, N.; Palmqvist, S.; Janelidze, S.; Dage, J.L.; Hansson, O. Blood-based Biomarkers for Alzheimer’s Disease. EMBO Mol. Med. 2022, 14, e14408. [Google Scholar] [CrossRef] [PubMed]
- Janelidze, S.; Palmqvist, S.; Leuzy, A.; Stomrud, E.; Verberk, I.M.W.; Zetterberg, H.; Ashton, N.J.; Pesini, P.; Sarasa, L.; Allué, J.A.; et al. Detecting Amyloid Positivity in Early Alzheimer’s Disease Using Combinations of Plasma Aβ42/Aβ40 and P-tau. Alzheimer’s Dement. 2022, 18, 283–293. [Google Scholar] [CrossRef]
- Dubois, B.; Feldman, H.H.; Jacova, C.; Hampel, H.; Molinuevo, J.L.; Blennow, K.; DeKosky, S.T.; Gauthier, S.; Selkoe, D.; Bateman, R.; et al. Advancing research diagnostic criteria for Alzheimer’s disease: The IWG-2 criteria. Lancet Neurol. 2014, 13, 614–629. [Google Scholar] [CrossRef] [PubMed]
- Simonsen, A.H.; Herukka, S.K.; Andreasen, N.; Baldeiras, I.; Bjerke, M.; Blennow, K.; Engelborghs, S.; Frisoni, G.B.; Gabryelewicz, T.; Galluzzi, S.; et al. Recommendations for CSF AD biomarkers in the diagnostic evaluation of dementia. Alzheimer’s Dement. 2017, 13, 274–284. [Google Scholar] [CrossRef]
- McKhann, G.M.; Knopman, D.S.; Chertkow, H.; Hyman, B.T.; Jack, C.R., Jr.; Kawas, C.H.; Klunk, W.E.; Koroshetz, W.J.; Manly, J.J.; Mayeux, R.; et al. The diagnosis of dementia due to Alzheimer’s disease: Recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimer’s Dement. 2011, 7, 263–269. [Google Scholar] [CrossRef]
- Yamashita, K.; Miura, M.; Watanabe, S.; Ishiki, K.; Arimatsu, Y.; Kawahira, J.; Kubo, T.; Sasaki, K.; Arai, T.; Hagino, K.; et al. Fully Automated and Highly Specific Plasma β-Amyloid Immunoassays Predict β-Amyloid Status Defined by Amyloid Positron Emission Tomography with High Accuracy. Alzheimer’s Res. Ther. 2022, 14, 86. [Google Scholar] [CrossRef]
- Mielke, M.M.; Aakre, J.A.; Algeciras-Schimnich, A.; Proctor, N.K.; Machulda, M.M.; Eichenlaub, U.; Knopman, D.S.; Vemuri, P.; Graff-Radford, J.; Jack, C.R., Jr.; et al. Comparison of CSF Phosphorylated Tau 181 and 217 for Cognitive Decline. Alzheimer’s Dement. 2022, 18, 602–611. [Google Scholar] [CrossRef]
- Mielke, M.M.; Hagen, C.E.; Xu, J.; Chai, X.; Vemuri, P.; Lowe, V.J.; Airey, D.C.; Knopman, D.S.; Roberts, R.O.; Machulda, M.M.; et al. Plasma Phospho-Tau181 Increases with Alzheimer’s Disease Clinical Severity and Is Associated with Tau- and Amyloid-Positron Emission Tomography. Alzheimer’s Dement. 2018, 14, 989–997. [Google Scholar] [CrossRef]
- Tatebe, H.; Kasai, T.; Ohmichi, T.; Kishi, Y.; Kakeya, T.; Waragai, M.; Kondo, M.; Allsop, D.; Tokuda, T. Quantification of Plasma Phosphorylated Tau to Use as a Biomarker for Brain Alzheimer Pathology: Pilot Case-Control Studies Including Patients with Alzheimer’s Disease and down Syndrome. Mol. Neurodegener. 2017, 12, 63–74. [Google Scholar] [CrossRef]
- West, T.; Kirmess, K.M.; Meyer, M.R.; Holubasch, M.S.; Knapik, S.S.; Hu, Y.; Contois, J.H.; Jackson, E.N.; Harpstrite, S.E.; Bateman, R.J.; et al. A blood-based diagnostic test incorporating plasma Aβ42/40 ratio, ApoE proteotype, and age accurately identifies brain amyloid status: Findings from a multi cohort validity analysis. Mol. Neurodegener. 2021, 16, 30. [Google Scholar] [CrossRef]
- Tissot, C.; Therriault, J.; Kunach, P. Comparing tau status determined via plasma pTau181, pTau231 and [18F] MK6240 tau-PET. EBioMedicine 2022, 76, 103837. [Google Scholar] [CrossRef] [PubMed]
- O’Connor, A.; Karikari, T.K.; Poole, T.; Ashton, N.J.; Lantero Rodriguez, J.; Khatun, A.; Swift, I.; Heslegrave, A.J.; Abel, E.; Chung, E.; et al. Plasma Phospho-Tau181 in Presymptomatic and Symptomatic Familial Alzheimer’s Disease: A Longitudinal Cohort Study. Mol. Psychiatry 2021, 26, 5967–5976. [Google Scholar] [CrossRef] [PubMed]
- Suárez-Calvet, M.; Karikari, T.K.; Ashton, N.J.; Lantero Rodríguez, J.; Milà-Alomà, M.; Gispert, J.D.; Salvadó, G.; Minguillon, C.; Fauria, K.; Shekari, M.; et al. Novel tau biomarkers phosphorylated at T181, T217 or T231 rise in the initial stages of the preclinical Alzheimer’s continuum when only subtle changes in Aβ pathology are detected. EMBO Mol. Med. 2020, 12, e12921. [Google Scholar] [CrossRef] [PubMed]
- Barthélemy, N.R.; Li, Y.; Joseph-Mathurin, N.; Gordon, B.A.; Hassenstab, J.; Benzinger, T.L.S.; Buckles, V.; Fagan, A.M.; Perrin, R.J.; Goate, A.M.; et al. Dominantly Inherited Alzheimer Network. A soluble phosphorylated tau signature links tau, amyloid and the evolution of stages of dominantly inherited Alzheimer’s disease. Nat. Med. 2020, 26, 398–407. [Google Scholar] [CrossRef] [PubMed]
- Ashton, N.J.; Pascoal, T.A.; Karikari, T.K.; Benedet, A.L.; Lantero-Rodriguez, J.; Brinkmalm, G.; Snellman, A.; Schöll, M.; Troakes, C.; Hye, A.; et al. Plasma p-tau231: A new biomarker for incipient Alzheimer’s disease pathology. Acta Neuropathol. 2021, 141, 709–724. [Google Scholar] [CrossRef]
- Ashton, N.J.; Janelidze, S.; Mattsson-Carlgren, N.; Binette, A.P.; Strandberg, O.; Brum, W.S.; Karikari, T.K.; González-Ortiz, F.; Di Molfetta, G.; Meda, F.J.; et al. Differential roles of Aβ42/40, p-tau231 and p-tau217 for Alzheimer’s trial selection and disease monitoring. Nat. Med. 2022, 28, 2555–2562. [Google Scholar] [CrossRef]
- Milà-Alomà, M.; Ashton, N.J.; Shekari, M.; Salvadó, G.; Ortiz-Romero, P.; Montoliu-Gaya, L.; Benedet, A.L.; Karikari, T.K.; Lantero-Rodriguez, J.; Vanmechelen, E.; et al. Plasma p-tau231 and p-tau217 as state markers of amyloid-β pathology in preclinical Alzheimer’s disease. Nat. Med. 2022, 28, 1797–1801. [Google Scholar] [CrossRef]
- Atik, A.; Stewart, T.; Zhang, J. Alpha-Synuclein as a Biomarker for Parkinson’s Disease. Brain Pathol. 2016, 26, 410–418. [Google Scholar] [CrossRef]
- Shi, M.; Bradner, J.; Hancock, A.M.; Chung, K.A.; Quinn, J.F.; Peskind, E.R.; Galasko, D.; Jankovic, J.; Zabetian, C.P.; Kim, H.M.; et al. Cerebrospinal fluid biomarkers for Parkinson disease diagnosis and progression. Ann. Neurol. 2011, 69, 570–580. [Google Scholar] [CrossRef]
- Bougea, A.; Stefanis, L.; Paraskevas, G.P.; Emmanouilidou, E.; Vekrelis, K.; Kapaki, E. Plasma α-synuclein levels in patients with Parkinson’s disease: A systematic review and meta-analysis. Neurol. Sci. 2019, 40, 929–938. [Google Scholar] [CrossRef]
- El-Agnaf, O.; Salem, S.A.; Paleologou, K.E.; Curran, M.D.; Gibson, M.J.; Court, J.A.; Schlossmacher, M.G.; Allsop, D. Detection of oligomeric forms of α-synuclein protein in human plasma as a potential biomarker for Parkinson’s disease. FASEB J. 2006, 20, 419–425. [Google Scholar] [CrossRef] [PubMed]
- Kang, K.H.; Liou, H.H.; Hour, M.J.; Liou, H.C.; Fu, W.M. Protection of dopaminergic neurons by 5-lipoxygenase inhibitor. Neuropharmacology 2013, 73, 380–387. [Google Scholar] [CrossRef] [PubMed]
- Iljina, M.; Dear, A.J.; Garcia, G.A.; De, S.; Tosatto, L.; Flagmeier, P.; Whiten, D.R.; Michaels, T.C.T.; Dobson, D.F.C.M.; Knowles, T.P.J.; et al. Quantifying co-oligomer formation by α-synuclein. ACS Nano 2018, 12, 10855–10866. [Google Scholar] [CrossRef]
- Spires-Jones, T.L.; Attems, J.; Thal, D.R. Interactions of pathological proteins in neurodegenerative diseases. Acta Neuropathol. 2017, 134, 187–205. [Google Scholar] [CrossRef]
- Galpern, W.R.; Lang, A.E. Interface between tauopathies and synucleinopathies: A tale of two proteins. Ann. Neurol. 2006, 59, 449–458. [Google Scholar] [CrossRef]
- Emin, D.; Zhang, Y.P.; Lobanova, E.; Miller, A.; Li, X.; Xia, Z.; Dakin, H.; Sideris, D.I.; Lam, J.Y.L.; Ranasinghe, R.T.; et al. Small soluble α-synuclein aggregates are the toxic species in Parkinson’s disease. Nat. Commun. 2022, 13, 5512. [Google Scholar] [CrossRef] [PubMed]
- Parnetti, L.; Chiasserini, D.; Persichetti, E.; Eusebi, P.; Varghese, S.; Qureshi, M.M.; Dardis, A.; Deganuto, M.; De Carlo, C.; Castrioto, A.; et al. Cerebrospinal fluid lysosomal enzymes and α-synuclein in Parkinson’s disease. Mov. Disord. 2014, 29, 1019–1027. [Google Scholar] [CrossRef] [PubMed]
- Parnetti, L.; Farotti, L.; Eusebi, P.; Chiasserini, D.; De Carlo, C.; Giannandrea, D.; Salvadori, N.; Lisetti, V.; Tambasco, N.; Rossi, A.; et al. Differential role of CSF α-synuclein species, tau, and Aβ42 in Parkinson’s Disease. Front. Aging Neurosci. 2014, 6, 53. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Liu, X.; Zhong, Y. Interleukin-17A: The Key Cytokine in Neurodegenerative Diseases. Front. Aging Neurosci. 2020, 12, 566922. [Google Scholar] [CrossRef]
- Ng, A.S.L.; Tan, Y.J.; Yong, A.C.W.; Saffari, S.E.; Lu, Z.; Ng, E.Y.; Ng, S.Y.E.; Chia, N.S.Y.; Choi, X.; Heng, D.; et al. Utility of plasma Neurofilament light as a diagnostic and prognostic biomarker of the postural instability gait disorder motor subtype in early Parkinson’s disease. Mol. Neurodegener. 2020, 15, 33. [Google Scholar] [CrossRef]
- Hazy, E.; Bokor, M.; Kalmar, L.; Gelencser, A.; Kamasa, P.; Han, K.H.; Tompa, K.; Tompa, P. Distinct hydration properties of wild-type and familial point mutant A53T of a-synuclein associated with Parkinson’s disease. Biophys. J. 2011, 101, 2260–2266. [Google Scholar] [CrossRef] [PubMed]
- Krisko, A.; Radman, M. Protein damage, ageing and age-related diseases. Open Biol. 2019, 9, 180249. [Google Scholar] [CrossRef] [PubMed]
- Wilkins, H.M.; Dimachkie, M.M.; Agbas, A. Blood-based Biomarkers for Amyotrophic Lateral Sclerosis. In Amyotrophic Lateral Sclerosis; Araki, T., Ed.; Exon Publications: Brisbane, Australia, 2021; pp. 105–120. [Google Scholar]
- Savage, N. Calculating disease. Nature 2017, 550, S115–S117. [Google Scholar] [CrossRef] [PubMed]
- Kasai, T.; Tokuda, T.; Ishigami, N.; Sasayama, H.; Foulds, P.; Mitchell, D.J.; Mann, D.M.A. Increased TDP-43 protein in cerebrospinal fluid of patients with amyotrophic lateral sclerosis. Acta Neuropathol. 2009, 117, 55–62. [Google Scholar] [CrossRef]
- Gambino, C.M.; Ciaccio, A.M.; Lo Sasso, B.; Giglio, R.V.; Vidali, M.; Agnello, L.; Ciaccio, M. The Role of TAR DNA Binding Protein 43 (TDP-43) as a CandiDate Biomarker of Amyotrophic Lateral Sclerosis: A Systematic Review and Meta-Analysis. Diagnostics 2023, 13, 416. [Google Scholar] [CrossRef]
- Lind, A.-L.; Wu, D.; Freyhult, E.; Bodolea, C.; Ekegren, T.; Larssson, A.; Gustafsson, M.G.; Katila, L.; Bergquist, J.; Gordh, T.; et al. A multiplex protein panel applied to cerebrospinal fluid reveals three new biomarker candidates in ALS but none in neuropathic pain patients. PLoS ONE 2016, 11, e0149821. [Google Scholar] [CrossRef]
- Zetterberg, H.; Jacobsson, J.; Rosengren, L.; Blennow, K.; Andersen, P.M. Cerebrospinal fluid neurofilament light levels in amyotrophic lateral sclerosis: Impact of SOD1 genotype. Eur. J. Neurol. 2007, 14, 1329–1333. [Google Scholar] [CrossRef]
- Lu, C.H.; Macdonald-Wallis, C.; Gray, E.; Pearce, N.; Petzold, A.; Norgren, N.; Giovannoni, G.; Fratta, P.; Sidle, K.; Fish, M.; et al. Neurofilament light chain: A prognostic biomarker in amyotrophic lateral sclerosis. Neurology 2015, 84, 2247–2257. [Google Scholar] [CrossRef]
- Lanznaster, D.; Hergesheimer, R.C.; Bakkouche, S.E.; Beltran, S.; Vourch, P.; Andres, C.R.; Dufour-Rainfray, D.; Corcia, P.; Blasco, H. Aβ1-42 and Tau as Potential Biomarkers for Diagnosis and Prognosis of Amyotrophic Lateral Sclerosis. Int. J. Mol. Sci. 2020, 21, 2911. [Google Scholar] [CrossRef]
- Manzo, E.; Lorenzini, I.; Barrameda, D.; O’Conner, A.G.; Barrows, J.M.; Starr, A.; Kovalik, T.; Rabichow, B.E.; Lehmkuhl, E.M.; Shreiner, D.D.; et al. Glycolysis upregulation is neuroprotective as a compensatory mechanism in ALS. eLife 2019, 8, e45114. [Google Scholar] [CrossRef]
- Wuolikainen, A.; Jonsson, P.; Ahnlund, M.; Antti, H.; Marklund, S.L.; Moritz, T.; Forsgren, L.; Andersen, P.M.; Trupp, M. Multi-platform mass spectrometry analysis of the CSF and plasma metabolomes of rigorously matched amyotrophic lateral sclerosis, Parkinson’s disease and control subjects. Mol. Biosyst. 2016, 12, 1287–1298. [Google Scholar] [CrossRef] [PubMed]
- Neumann, M.; Sampathu, D.M.; Kwong, L.K.; Truax, A.C.; Micsenyi, M.C.; Chou, T.T.; Bruce, J.; Schuck, T.; Grossman, M.; Clark, C.M.; et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science 2006, 314, 130–133. [Google Scholar] [CrossRef] [PubMed]
- Rosen, D.R.; Siddique, T.; Patterson, D.; Figlewicz, D.A.; Sapp, P.; Hentati, A.; Donaldson, D.; Goto, J.; O’Regan, J.P.; Deng, H.X.; et al. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature 1993, 362, 59–62. [Google Scholar] [CrossRef] [PubMed]
- Verber, N.; Shaw, P.J. Biomarkers in amyotrophic lateral sclerosis: A review of new developments. Curr. Opin. Neurol. 2020, 33, 662–668. [Google Scholar] [CrossRef]
- Verber, N.S.; Shepheard, S.R.; Sassani, M.; McDonough, H.E.; Moore, S.A.; Alix, J.J.P.; Wilkinson, I.D.; Jenkins, T.M.; Shaw, P.J. Biomarkers in Motor Neuron Disease: A State of the Art Review. Front. Neurol. 2019, 10, 291. [Google Scholar] [CrossRef]
- Lunetta, C.; Moglia, C.; Lizio, A.; Caponnetto, C.; Dubbioso, R.; Giannini, F.; Matà, S.; Mazzini, L.; Sabatelli, M.; Siciliano, G.; et al. The Italian multicenter experience with edaravone in amyotrophic lateral sclerosis. J. Neurol. 2020, 267, 3258–3267. [Google Scholar] [CrossRef]
- Poesen, K.; Van Damme, P. Diagnostic and prognostic performance of neurofilaments in ALS. Front. Neurol. 2018, 9, 1167. [Google Scholar] [CrossRef]
- Barbo, M.; Ravnik-Glavač, M. Extracellular Vesicles as Potential Biomarkers in Amyotrophic Lateral Sclerosis. Genes 2023, 14, 325. [Google Scholar] [CrossRef]
- Gaiani, A.; Martinelli, I.; Bello, L.; Querin, G.; Puthenparampil, M.; Ruggero, S.; Toffanin, E.; Cagnin, A.; Briani, C.; Pegoraro, E.; et al. Diagnostic and prognostic biomarkers in amyotrophic lateral sclerosis: Neurofilament light chain levels in definite subtypes of disease. JAMA Neurol. 2017, 74, 525–532. [Google Scholar] [CrossRef]
- Verde, F.; Steinacker, P.; Weishaupt, J.H.; Kassubek, J.; Oeckl, P.; Halbgebauer, S.; Tumani, H.; von Arnim, C.A.F.; Dorst, J.; Feneberg, E.; et al. Neurofilament light chain in serum for the diagnosis of amyotrophic lateral sclerosis. J. Neurol. Neurosurg. Psychiatry 2019, 90, 157–164. [Google Scholar] [CrossRef]
- Hirano, A.; Donnenfeld, H.; Sasaki, S.; Nakano, I. Fine Structural Observations of Neurofilamentous Changes in Amyotrophic Lateral Sclerosis. J. Neuropathol. Exp. Neurol. 1984, 43, 461–470. [Google Scholar] [CrossRef]
- McCombe, P.A.; Pfluger, C.; Singh, P.; Lim, C.Y.; Airey, C.; Henderson, R.D. Serial measurements of phosphorylated neurofilament-heavy in the serum of subjects with amyotrophic lateral sclerosis. J. Neurol. Sci. 2015, 353, 122–129. [Google Scholar] [CrossRef]
- Kasai, T.; Kojima, Y.; Ohmichi, T.; Tatebe, H.; Tsuji, Y.; Noto, Y.I.; Kitani-Morii, F.; Shinomoto, M.; Allsop, D.; Mizuno, T.; et al. Combined use of CSF NfL and CSF TDP-43 improves diagnostic performance in ALS. Ann. Clin. Transl. Neurol. 2019, 6, 2489–2502. [Google Scholar] [CrossRef]
- Oeckl, P.; Jardel, C.; Salachas, F.; Lamari, F.; Andersen, P.M.; Bowser, R.; de Carvalho, M.; Costa, J.; van Damme, P.; Gray, E.; et al. Multicenter validation of CSF neurofilaments as diagnostic biomarkers for ALS. Amyotroph. Lateral Scler. Front. Degener. 2016, 17, 404–413. [Google Scholar] [CrossRef]
- Zetterberg, H.; Skillbäck, T.; Mattsson, N.; Trojanowski, J.Q.; Portelius, E.; Shaw, L.M.; Weiner, M.W.; Blennow, K. Association of Cerebrospinal Fluid Neurofilament Light Concentration with Alzheimer Disease Progression. JAMA Neurol. 2016, 73, 60–67. [Google Scholar] [CrossRef]
- Bäckström, D.C.; Domellöf, M.E.; Linder, J.; Olsson, B.; Öhrfelt, A.; Trupp, M.; Zetterberg, H.; Blennow, K.; Forsgren, L. Cerebrospinal fluid patterns and the risk of future dementia in early, incident Parkinson disease. JAMA Neurol. 2015, 72, 1175–1182. [Google Scholar] [CrossRef]
- Kojima, Y.; Kasai, T.; Noto, Y.I.; Ohmichi, T.; Tatebe, H.; Kitaoji, T.; Tsuji, Y.; Kitani-Morii, F.; Shinomoto, M.; Allsop, D.; et al. Amyotrophic lateral sclerosis: Correlations between fluid biomarkers of NfL, TDP-43, and tau, and clinical characteristics. PLoS ONE 2021, 16, e0260323. [Google Scholar] [CrossRef]
- Sun, J.; Carrero, J.J.; Zagai, U.; Evans, M.; Ingre, C.; Pawitan, Y.; Fang, F. Blood biomarkers and prognosis of amyotrophic lateral sclerosis. Eur. J. Neurol. 2020, 27, 2125–2133. [Google Scholar] [CrossRef]
- Colletti, T.; Agnello, L.; Spataro, R.; Guccione, L.; Notaro, A.; Lo Sasso, B.; Blandino, V.; Graziano, F.; Gambino, C.M.; Giglio, R.V.; et al. Prognostic Role of CSF β-amyloid 1–42/1–40 Ratio in Patients Affected by Amyotrophic Lateral Sclerosis. Brain Sci. 2021, 11, 302. [Google Scholar] [CrossRef]
- Sol, J.; Jové, M.; Povedano, M.; Sproviero, W.; Domínguez, R.; Piñol-Ripoll, G.; Romero-Guevara, R.; Hye, A.; Al-Chalabi, A.; Torres, P.; et al. Lipidomic traits of plasma and cerebrospinal fluid in amyotrophic lateral sclerosis correlate with disease progression. Brain Commun. 2021, 3, fcab143. [Google Scholar] [CrossRef]
- Area-Gomez, E.; Larrea, D.; Yun, T.; Xu, Y.; Hupf, J.; Zandkarimi, F.; Chan, R.B.; Mitsumoto, H. Lipidomics study of plasma from patients suggest that ALS and PLS are part of a continuum of motor neuron disorders. Sci. Rep. 2021, 11, 13562. [Google Scholar] [CrossRef] [PubMed]
- Mariosa, D.; Hammar, N.; Malmström, H.; Ingre, C.; Jungner, I.; Ye, W.; Fang, F.; Walldius, G. Blood biomarkers of carbohydrate, lipid, and apolipoprotein metabolisms and risk of amyotrophic lateral sclerosis: A more than 20-year followup of the Swedish AMORIS cohort. Ann. Neurol. 2017, 81, 718–728. [Google Scholar] [CrossRef] [PubMed]
- Bandres-Ciga, S.; Noyce, A.J.; Hemani, G.; Nicolas, A.; Calvo, A.; Mora, G.; ITALSGEN Consortium; International ALS Genomics Consortium; Tienari, P.J.; Stone, D.J.; et al. Shared polygenic risk and causal inferences in amyotrophic lateral sclerosis. Ann. Neurol. 2019, 85, 470–481. [Google Scholar] [CrossRef]
- Chakraborty, A.; Diwan, A. Biomarkers and molecular mechanisms of Amyotrophic Lateral Sclerosis. AIMS Neurosci. 2022, 9, 423–443. [Google Scholar] [CrossRef]
- Periayah, M.H.; Halim, A.S.; Mat Saad, A.Z. Mechanism Action of Platelets and Crucial Blood Coagulation Pathways in Hemostasis. Int. J. Hematol. Oncol. Stem Cell Res. 2017, 11, 319–327. [Google Scholar] [PubMed]
- Koçer, A.; Yaman, A.; Niftaliyev, E.; Dürüyen, H.; Eryılmaz, M.; Koçer, E. Assessment of platelet indices in patients with neurodegenerative diseases: Mean platelet volume was increased in patients with Parkinson’s disease. Curr. Gerontol. Geriatr. Res. 2013, 2013, 986254. [Google Scholar] [CrossRef]
- Behari, M.; Shrivastava, M. Role of platelets in neurodegenerative diseases: A universal pathophysiology. Int. J. Neurosci. 2013, 123, 287–299. [Google Scholar] [CrossRef]
- Holinstat, M. Normal platelet function. Cancer Metastasis Rev. 2017, 36, 195–198. [Google Scholar] [CrossRef]
- Burnouf, T.; Walker, T.L. The multifaceted role of platelets in mediating brain function. Blood 2022, 140, 815–827. [Google Scholar] [CrossRef]
- Leiter, O.; Walker, T.L. Platelets in Neurodegenerative Conditions—Friend or Foe? Front. Immun. 2020, 11, 747. [Google Scholar] [CrossRef]
- Kiktenko, A.I.; Zlobina, G.P.; Brusov, O.S.; Zakharova, M.N. Structure of peripheral blood platelets surface in patients with amyotrophic lateral sclerosis and multiple sclerosis. Zhurnal Nevrol. i Psikhiatrii Im. SS Korsakova 2005, 105, 40–42. [Google Scholar]
- Lim, K.M.; Kim, H.H.; Bae, O.N.; Noh, J.Y.; Kim, K.Y.; Kim, S.H.; Chung, S.M.; Shin, S.; Kim, H.Y.; Chung, J.H. Inhibition of platelet aggregation by 1-methyl-4-phenyl pyridinium ion (MPP+) through ATP depletion: Evidence for the reduced platelet activities in Parkinson’s disease. Platelets 2009, 20, 163–170. [Google Scholar] [CrossRef] [PubMed]
- Stellos, K.; Panagiota, V.; Kögel, A.; Leyhe, T.; Gawaz, M.; Laske, C. Predictive value of platelet activation for the rate of cognitive decline in Alzheimer’s disease patients. J. Cereb. Blood Flow Metab. 2010, 30, 1817–1820. [Google Scholar] [CrossRef] [PubMed]
- Espinosa-Parrilla, Y.; Gonzalez-Billault, C.; Fuentes, E.; Palomo, I.; Alarcón, M. Decoding the Role of Platelets and Related MicroRNAs in Aging and Neurodegenerative Disorders. Front. Aging Neurosci. 2019, 11, 151. [Google Scholar] [CrossRef] [PubMed]
- Zaninetti, C.; Sachs, L.; Palankar, R. Role of Platelet Cytoskeleton in Platelet Biomechanics: Current and Emerging Methodologies and Their Potential Relevance for the Investigation of Inherited Platelet Disorders. Hamostaseologie 2020, 40, 337–347. [Google Scholar] [CrossRef]
- Winokur, R.; Hartwig, J.H. Mechanism of shape change in chilled human platelets. Blood 1995, 85, 1796–1804. [Google Scholar] [CrossRef]
- Hartwig, J.H. Mechanisms of actin rearrangements mediating platelet activation. J. Cell Biol. 1992, 118, 1421–1442. [Google Scholar] [CrossRef]
- Hartwig, J.H. The platelet: Form and function. Semin. Hematol. 2006, 43, S94–S100. [Google Scholar] [CrossRef]
- Loftus, J.C.; Choate, J.; Albrecht, R.M. Platelet activation and cytoskeletal reorganization: High voltage electron microscopic examination of intact and Triton-extracted whole mounts. J. Cell Biol. 1984, 98, 2019–2025. [Google Scholar] [CrossRef]
- Jones, C.I. Platelet function and ageing. Mammal. Genome 2016, 27, 358–366. [Google Scholar] [CrossRef]
- Montenont, E.; Rondina, M.T.; Campbell, R.A. Altered functions of platelets during aging HHS Public Access. Curr. Opin. Hematol. 2019, 26, 336–342. [Google Scholar] [CrossRef] [PubMed]
- Segal, J.B.; Moliterno, A.R. Platelet counts differ by sex, ethnicity, and age in the United States. Ann. Epidemiol. 2006, 16, 123–130. [Google Scholar] [CrossRef] [PubMed]
- Bush, A.I.; Martins, R.N.; Rumble, B.; Moir, R.; Fuller, S.; Milward, E.; Currie, J.; Ames, D.; Weidemann, A.; Fischer, P.; et al. The amyloid precursor protein of Alzheimer’s disease is released by human platelets. J. Biol. Chem. 1990, 265, 15977–15983. [Google Scholar] [CrossRef] [PubMed]
- Catricala, S.; Torti, M.; Ricevuti, G. Alzheimer disease and platelets: How’s that relevant. Immun. Ageing 2012, 9, 20. [Google Scholar] [CrossRef]
- Mukaetova-Ladinska, E.; Abdell-All, Z.; Andrade, J.; Da Silva, J.; Boksha, I.; Burbaeva, G.; Kalaria, R.N.; Brien, J.T.O. Platelet tau protein as a potential peripheral biomarker in Alzheimer’s disease: An explorative study. Curr. Alzheimer Res. 2013, 15, 800–808. [Google Scholar] [CrossRef]
- Reinhard, C.; Hébert, S.S.; De Strooper, B. The amyloid-β precursor protein: Integrating structure with biological function. EMBO J. 2005, 24, 3996–4006. [Google Scholar] [CrossRef]
- Ferrer-Raventos, P.; Beyer, K. Alternative platelet activation pathways and their role in neurodegenerative diseases. Neurobiol. Dis. 2021, 159, 105512. [Google Scholar] [CrossRef]
- Canobbio, I.; Catricalà, S.; Di Pasqua, L.G.; Guidetti, G.; Consonni, A.; Manganaro, D.; Torti, M. Immobilized amyloid Aβ peptides support platelet adhesion and activation. FEBS Lett. 2013, 587, 2606–2611. [Google Scholar] [CrossRef]
- Donner, L.; Elvers, M. Platelets and neurodegenerative diseases. In Thrombotic and Non-Thrombotic Disorders: Pathophysiology, Pharmacology and Therapeutics: An Update; Gresele, P., Kleiman, N.S., Lopez, J.A., Page, C.P., Eds.; Springer International Publishing: Cham, Switzerland, 2017; pp. 1209–1224. [Google Scholar]
- Blass, J.P.; Gibson, G.E. Nonneural Markers in Alzheimer Disease. Alzheimer Dis. Assoc. Disord. 1992, 6, 205–224. [Google Scholar] [CrossRef]
- Briones, M.R.S.; Snyder, A.M.; Ferreira, R.C.; Neely, E.B.; Connor, J.R.; Broach, J.R. A possible role for platelet-activating factor receptor in amyotrophic lateral sclerosis treatment. Front. Neurol. 2018, 9, 39. [Google Scholar] [CrossRef]
- Dantzer, R. Neuroimmune interactions: From the brain to the immune system and vice versa. Physiol. Rev. 2018, 98, 477–504. [Google Scholar] [CrossRef] [PubMed]
- Barbour, R.; Kling, K.; Anderson, J.P.; Banducci, K.; Cole, T.; Diep, L.; Fox, M.; Goldstein, J.M.; Soriano, F.; Seubert, P.; et al. Red blood cells are the major source of α-synuclein in blood. Neurodegener. Dis. 2008, 5, 55–59. [Google Scholar] [CrossRef] [PubMed]
- Hsu, L.J.; Sagara, Y.; Arroyo, A.; Rockenstein, E.; Sisk, A.; Mallory, M.; Wong, J.; Takenouchi, T.; Hashimoto, M.; Masliah, E. α-synuclein promotes mitochondrial deficit and oxidative stress. Am. J. Pathol. 2000, 157, 401–410. [Google Scholar] [CrossRef] [PubMed]
- Pei, Y.; Maitta, R.W. Alpha synuclein in hematopoiesis and immunity. Heliyon 2019, 5, e02590. [Google Scholar] [CrossRef]
- Dupuis, L.; Spreux-Varoquaux, O.; Bensimon, G.; Jullien, P.; Lacomblez, L.; Salachas, F.; Bruneteau, G.; Pradat, P.F.; Loeffler, J.P.; Meininger, V. Platelet serotonin level predicts survival in amyotrophic lateral sclerosis. PLoS ONE 2010, 5, e13346. [Google Scholar] [CrossRef]
- Sevush, S.; Jy, W.; Horstman, L.L.; Mao, W.W.; Kolodny, L.; Ahn, Y.S. Platelet activation in Alzheimer disease. Arch. Neurol. 1998, 55, 530–536. [Google Scholar] [CrossRef] [PubMed]
- Sotnikov, I.; Veremeyko, T.; Starossom, S.C.; Barteneva, N.; Weiner, H.L.; Ponomarev, E.D. Platelets recognize brain-specific glycolipid structures, respond to neurovascular damage and promote neuroinflammation. PLoS ONE 2013, 8, e58979. [Google Scholar] [CrossRef] [PubMed]
- Leiter, O.; Walker, T.L. Platelets: The missing link between the blood and brain? Prog. Neurobiol. 2019, 183, 101695. [Google Scholar] [CrossRef] [PubMed]
- Perello, M.; Stuart, R.; Nillni, E.A. Prothyrotropin-releasing hormone targets its processing products to different vesicles of the secretory pathway. J. Biol. Chem. 2008, 283, 19936–19947. [Google Scholar] [CrossRef]
- Italiano, J.E.; Richardson, J.L.; Patel-Hett, S.; Battinelli, E.; Zaslavsky, A.; Short, S.; Ryeom, S.; Folkman, J.; Klement, G.L. Angiogenesis is regulated by a novel mechanism: Pro- and antiangiogenic proteins are organized into separate platelet α-granules and differentially released. Blood 2008, 111, 1227–1233. [Google Scholar] [CrossRef]
- Chen, Y.; Yuan, Y.; Li, W. Sorting machineries: How platelet-dense granules differ from α-granules. Biosci. Rep. 2018, 38, BSR20180458. [Google Scholar] [CrossRef] [PubMed]
- Ambrosio, A.L.; Di Pietro, S.M. Storage pool diseases illuminate platelet dense granule biogenesis. Platelets 2017, 8, 138–146. [Google Scholar] [CrossRef] [PubMed]
- Heijnen, H.F.; Schiel, A.E.; Fijnheer, R.; Geuze, H.J.; Sixma, J.J. Activated platelets release two types of membrane vesicles: Microvesicles by surface shedding and exosomes derived from exocytosis of multivesicular bodies and α-granules. Blood 1999, 94, 3791–3799. [Google Scholar] [CrossRef]
- Conway, M.E. Alzheimer’s disease: Targeting the glutamatergic system. Biogerontology 2020, 21, 257–274. [Google Scholar] [CrossRef] [PubMed]
- Lovino, L.; Tremblay, M.E.; Civiero, L. Glutamate-induced excitotoxicity in Parkinson’s disease: The role of glial cells. J. Pharm. Sci. 2020, 144, 151–164. [Google Scholar]
- Rao, J.S.; Hantaï, D.; Festoff, B.W. Thrombospondin, a platelet α-granule and matrix glycoprotein, is increased in muscle basement membrane of patients with amyotrophic lateral sclerosis. J. Neurol. Sci. 1992, 113, 99–107. [Google Scholar] [CrossRef]
- Smirnova, I.V.; Festoff, B.W. Alterations in serum thrombospondin in patients with amyotrophic lateral sclerosis. J. Neurol. Sci. 1994, 127, 207–213. [Google Scholar] [CrossRef]
- Talib, L.L.; Joaquim, H.P.; Forlenza, O.V. Platelet biomarkers in Alzheimer’s disease. World J. Psychiatry 2012, 2, 95–101. [Google Scholar] [CrossRef]
- Ponomarev, E.D. Fresh evidence for platelets as neuronal and innate immune cells: Their role in the activation, differentiation, and deactivation of Th1, Th17, and Tregs during tissue inflammation. Front. Immunol. 2018, 9, 406. [Google Scholar] [CrossRef]
- Pretini, V.; Koenen, M.H.; Kaestner, L.; Fens, M.H.A.M.; Schiffelers, R.M.; Bartels, M.; Van Wijk, R. Red Blood Cells: Chasing Interactions. Front. Physiol. 2019, 10, 945. [Google Scholar] [CrossRef]
- Goodman, S.R.; Shiffer, K. The spectrin membrane skeleton of normal and abnormal human erythrocytes: A review. Am. J. Physiol. 1983, 244, 121–141. [Google Scholar] [CrossRef]
- Mankelow, T.J.; Satchwell, T.J.; Burton, N.M. Refined views of multiprotein complexes in the erythrocyte membrane. Blood Cells Mol. Dis. 2012, 49, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Lux, S.D. Anatomy of the red cell membrane skeleton: Unanswered questions. Blood 2016, 127, 187–199. [Google Scholar] [CrossRef] [PubMed]
- Pan, L.; Yan, R.; Li, W.; Xu, K. Super-Resolution Microscopy Reveals the Native Ultrastructure of the Erythrocyte Cytoskeleton. Cell Rep. 2018, 22, 1151–1158. [Google Scholar] [CrossRef]
- Thangaraju, K.; Neerukonda, S.N.; Katneni, U.; Buehler, P.W. Extracellular Vesicles from Red Blood Cells and Their Evolving Roles in Health, Coagulopathy and Therapy. Int. J. Mol. Sci. 2021, 22, 153. [Google Scholar] [CrossRef] [PubMed]
- Aoki, T. A Comprehensive Review of Our Current Understanding of Red Blood Cell (RBC) Glycoproteins. Membranes 2017, 7, 56. [Google Scholar] [CrossRef]
- Staricoff, M.A.; Tanner, M.J.A. Role of band 3 and glycophorin C in the maintenance of the shape and mechanical properties of the human red blood cell. Cell. Mol. Biol. Lett. 1996, 2, 151–161. [Google Scholar]
- Appell, K.C.; Low, P.S. Evaluation of structural interdependence of membrane-spanning and cytoplasmic domains of band 3. Biochemistry 1982, 21, 2151–2157. [Google Scholar] [CrossRef]
- Mohandas, N.; Evans, E. Mechanical properties of the red cell membrane in relation to molecular structure and genetic defects. Annu. Rev. Biophys. Biomol. Struct. 1994, 23, 787–818. [Google Scholar] [CrossRef]
- Willekens, F.L.; Bosch, F.H.; Roerdinkholder-Stoelwinder, B.; Groenen-Dopp, Y.A.M.; Werre, J.M. Quantification of loss of haemoglobin components from the circulating red blood cell in vivo. Eur. J. Haematol. 1997, 58, 246–250. [Google Scholar] [CrossRef]
- Franco, R.S.; Puchulu-Campanella, M.E.; Barber, L.A.; Palascak, M.B.; Joiner, C.H.; Low, P.S.; Cohen, R.M. Changes in the properties of normal human red blood cells during in vivo aging. Am. J. Hematol. 2012, 88, 44–51. [Google Scholar] [CrossRef]
- Ford, J. Red blood cell morphology. Int. Soc. Lab. Hematol. 2013, 35, 351–357. [Google Scholar] [CrossRef]
- Girasole, M.; Dinarelli, S.; Boumis, G. Structure and function in native and pathological erythrocytes: A quantitative view from the nanoscale. Micron 2012, 43, 1273–1286. [Google Scholar] [CrossRef]
- Dinarelli, S.; Krumova, S.; Todinova, S.; Taneva, S.G.; Lenzi, E.; Mussi, V.; Longo, G.; Girasole, M. Insight into the Morphological Pattern Observed Along the Erythrocytes’ Aging: Coupling Quantitative AFM Data to microcalorimetry and Raman spectroscopy. J. Mol. Recognit. 2018, 31, e2732. [Google Scholar] [CrossRef]
- Geekiyanage, N.M.; Sauret, E.; Saha, S.C.; Flower, R.L.; Gu, Y.T. Deformation behaviour of stomatocyte, discocyte and echinocyte red blood cell morphologies during optical tweezers stretching. Biomech. Model. Mechanobiol. 2020, 19, 1827–1843. [Google Scholar] [CrossRef] [PubMed]
- Xing, X.; Jin, H.; Lu, Y.; Wang, Q.; Pan, Y.; Cai, J.; Wang, H. Detection of erythrocytes in patient with elliptocytosis complicating ITP using atomic force microscopy. Micron 2011, 42, 42–46. [Google Scholar] [CrossRef] [PubMed]
- Danek, A.; Walker, R.H. Neuroacanthocytosis. Curr. Opin. Neurol. 2005, 18, 386–392. [Google Scholar] [CrossRef]
- Nguyen, D.B.; Ly, T.B.T.; Bernhardt, I. Microvesicles Released from Human Red Blood Cells: Properties and Potential Application. In Novel Implications of Exosomes in Diagnosis and Treatment of Cancer and Infectious Diseases; Wang, J., Ed.; InTechOpen: London, UK, 2017; pp. 137–161. [Google Scholar]
- Malka, R.; Delgado, F.F.; Manalis, S.R.; Higgins, J.M. In vivo volume and hemoglobin dynamics of human red blood cells. PLoS Comput. Biol. 2014, 10, e1003839. [Google Scholar] [CrossRef]
- Chen, S.H.; Bu, X.L.; Jin, W.S.; Shen, L.L.; Wang, J.; Zhuang, Z.Q.; Zhang, T.; Zeng, F.; Yao, X.Q.; Zhou, H.D.; et al. Altered peripheral profile of blood cells in Alzheimer disease: A hospital-based case-control study. Medicine 2017, 96, e6843. [Google Scholar] [CrossRef]
- Deng, Q.; Zhou, X.; Chen, J.; Pan, M.; Gao, H.; Zhou, J.; Wang, D.; Chen, Q.; Zhang, X.; Wang, Q.; et al. Lower hemoglobin levels in patients with parkinson’s disease are associated with disease severity and iron metabolism. Brain Res. 2017, 1655, 145–151. [Google Scholar] [CrossRef]
- Mattson, M.P.; Begley, J.G.; Mark, R.J.; Furukawa, K. Aβ25–35 induces rapid lysis of red blood cells: Contrast with Aβ1–42 and examination of underlying mechanisms. Brain Res. 1997, 771, 147–153. [Google Scholar] [CrossRef] [PubMed]
- Mohanty, J.G.; Eckley, D.M.; Williamson, J.D.; Launer, L.J.; Rifkind, J.M. Do red blood cell-β-amyloid interactions alter oxygen delivery in Alzheimer’s disease? Adv. Exp. Med. Biol. 2008, 614, 29–35. [Google Scholar] [PubMed]
- Stevenson, A.; Lopez, D.; Khoo, P.; Kalaria, R.N.; Mukaetova-Ladinska, E.B. Exploring Erythrocytes as Blood Biomarkers for Alzheimer’s Disease. J. Alzheimer’s Dis. 2017, 60, 845–857. [Google Scholar] [CrossRef] [PubMed]
- Lan, J.; Liu, J.; Zhao, Z.; Xue, R.; Zhang, N.; Zhang, P.; Zhao, P.; Zheng, F.; Sun, X. The peripheral blood of Aβ binding RBC as a biomarker for diagnosis of Alzheimer’s disease. Age Ageing 2015, 44, 458–464. [Google Scholar] [CrossRef] [PubMed]
- Daniele, S.; Frosini, D.; Pietrobono, D.; Petrozzi, L.; Lo Gerfo, A.; Baldacci, F.; Fusi, J.; Giacomelli, C.; Siciliano, G.; Trincavelli, M.L.; et al. α-Synuclein Heterocomplexes with β-Amyloid Are Increased in Red Blood Cells of Parkinson’s Disease Patients and Correlate with Disease Severity. Front. Mol. Neurosci. 2018, 11, 53. [Google Scholar] [CrossRef]
- Wang, X.; Yu, S.; Li, F.; Feng, T. Detection of α-synuclein oligomers in red blood cells as a potential biomarker of Parkinson’s disease. Neurosci. Lett. 2015, 599, 115–119. [Google Scholar] [CrossRef]
- Tian, C.; Liu, G.; Gao, L.; Soltys, D.; Pan, C.; Stewart, T.; Shi, M.; Xie, Z.; Liu, N.; Feng, T.; et al. Erythrocytic α-Synuclein as a potential biomarker for Parkinson’s disease. Transl. Neurodegener. 2019, 8, 15. [Google Scholar] [CrossRef]
- Matsumoto, J.; Stewart, T.; Sheng, L.; Li, N.; Bullock, K.; Song, N.; Shi, M.; Banks, W.A.; Zhang, J. Transmission of α-synuclein-containing erythrocyte-derived extracellular vesicles across the blood-brain barrier via adsorptive mediated transcytosis: Another mechanism for initiation and progression of Parkinson’s disease? Acta Neuropathol. Commun. 2017, 5, 71. [Google Scholar] [CrossRef]
- Jellinger, K.A. The enigma of mixed dementia. Alzheimer’s Dement. 2007, 3, 40–53. [Google Scholar] [CrossRef]
- Lima, C.; Pinto, S.; Napoleao, P.; Pronto-Laborinho, A.C.; Barrob, M.A.; Freitas, T.; de Carvalhob, M.; Saldanha, C. Identification of erythrocyte biomarkers in amyotrophic lateral sclerosis. Clin. Hemorheol. Microcirc. 2016, 63, 423–437. [Google Scholar] [CrossRef]
- Alsteens, D.; Gaub, H.; Newton, R.; Pfreundschuh, M.G.; Christoph, M.; Daniel, J. Atomic force microscopy-based characterization and design of biointerfaces. Nat. Rev. Mater. 2017, 2, 17008. [Google Scholar] [CrossRef]
- Guan, G.; He, Y.; Mei, L. Atomic force microscopy: A nanobiotechnology for cellular research. Nano Trans. Med. 2022, 1, e9130004. [Google Scholar] [CrossRef]
- Yeow, N.; Tabor, R.F.; Garnier, G. Atomic force microscopy: From red blood cells to immunohaematology. Adv. Colloid Interface Sci. 2017, 249, 149–162. [Google Scholar] [CrossRef] [PubMed]
- Dufrêne, Y.; Ando, T.; Garcia, R.; Alsteens, D.; Martinez-Martin, D.; Engel, A.; Gerber, C.; Müller, D.J. Imaging modes of atomic force microscopy for application in molecular and cell biology. Nat. Nanotechnol. 2017, 12, 295–307. [Google Scholar] [CrossRef] [PubMed]
- García-Arribas, A.B.; Goñi, F.M.; Alonso, A. Lipid Self-Assemblies under the Atomic Force Microscope. Int. J. Mol. Sci. 2021, 22, 10085. [Google Scholar] [CrossRef]
- Trindade, G.S.; Vilela, J.M.; Ferreira, J.M.; Aguiar, P.H.; Leite, J.A.; Guedes, M.I.; Lobato, Z.I.; Madureira, M.C.; da Silva, M.I.; da Fonseca, F.G.; et al. Use of atomic force microscopy as a diagnostic tool to identify orthopoxvirus. J. Virol. Methods 2007, 141, 198–204. [Google Scholar] [CrossRef]
- Viljoen, A.; Mathelié-Guinlet, M.; Ray, A.; Strohmeyer, N.; Oh, Y.L.; Hinterdorfer, P.; Müller, D.J.; Alsteens, D.; Dufrêne, Y.F. Force spectroscopy of single cells using atomic force microscopy. Nat. Rev. Methods Primers 2021, 1, 63. [Google Scholar] [CrossRef]
- Krieg, M.; Fläschner, G.; Alsteens, D.; Gaub, B.M.; Roos, W.H.; Wuite, G.J.L.; Gaub, H.E.; Gerber, C.; Dufrêne, Y.F.; Müller, D.J. Atomic force microscopy-based mechanobiology. Nat. Rev. Phys. 2019, 1, 41–57. [Google Scholar] [CrossRef]
- Iyer, S.; Gaikwad, R.M.; Subba-Rao, V.; Woodworth, C.D.; Sokolov, I. Atomic force microscopy detects differences in the surface brush of normal and cancerous cells. Nat. Nanotechnol. 2009, 4, 389–393. [Google Scholar] [CrossRef]
- Cross, S.E.; Jin, Y.S.; Rao, J.; Gimzewski, J.K. Nanomechanical analysis of cells from cancer patients. Nat. Nanotechnol. 2007, 2, 780–783. [Google Scholar] [CrossRef]
- Plodinec, M.; Loparic, M.; Monnier, C.A.; Obermann, E.C.; Zanetti-Dallenbach, R.; Oertle, P.; Hyotyla, J.T.; Aebi, U.; Bentires-Alj, M.; Lim, R.Y.; et al. The nanomechanical signature of breast cancer. Nat. Nanotechnol. 2012, 7, 757–765. [Google Scholar] [CrossRef] [PubMed]
- Sokolov, I.; Dokukin, M.E.; Kalaparthi, V.; Miljkovic, M.; Wang, A.; Seigne, J.D.; Grivas, P.; Demidenko, E. Noninvasive diagnostic imaging using machine-learning analysis of nanoresolution images of cell surfaces: Detection of bladder cancer. Proc. Natl. Acad. Sci. USA 2018, 115, 12920–12925. [Google Scholar] [CrossRef]
- Butt, H.J.; Wolff, E.K.; Gould, S.A.; Dixon Northern, B.; Peterson, C.M.; Hansma, P.K. Imaging cells with the atomic force microscope. J. Struct. Biol. 1990, 105, 54–61. [Google Scholar] [CrossRef] [PubMed]
- Gould, S.A.C.; Drake, B.; Prater, C.B.; Weisenhorn, A.L.; Manne, S.; Hansma, H.G.; Hansma, P.K.; Masse, J.; Longmire, M.; Elings, V.; et al. From atoms to integrated-circuit chips, blood cells and bacteria with the atomic force microscope. J. Vac. Sci. Technol. 1990, 8, 369–375. [Google Scholar] [CrossRef]
- Zachée, P.; Snauwaert, J.; Vandenberghe, P.; Hellemans, L.; Boogaerts, M. Imaging red blood cells with the atomic force microscope. Br. J. Haematol. 1996, 95, 472–481. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, W.; Wang, S.; Wang, C.; Xie, J.; Chen, X.; Xu, Y.; Mao, P. Detection of human erythrocytes influenced by iron deficiency anemia and thalassemia using atomic force microscopy. Micron 2012, 43, 1287–1292. [Google Scholar] [CrossRef]
- Strijkova-Kenderova, V.; Todinova, S.; Andreeva, T.; Bogdanova, D.; Langari, A.; Danailova, A.; Krumova, S.; Zlatareva, E.; Kalaydzhiev, N.; Milanov, I.; et al. Morphometry and Stiffness of Red Blood Cells—Signatures of Neurodegenerative Diseases and Aging. Int. J. Mol. Sci. 2022, 23, 227. [Google Scholar] [CrossRef]
- Girasole, M.; Pompeo, G.; Cricenti, A.; Congiu-Castellano, A.; Andreola, F.; Serafino, A.; Frazer, B.H.; Boumis, G.; Amiconi, G. Roughness of the plasma membrane as an independent morphological parameter to study RBCs: A quantitative atomic force microscopy investigation. Biochim. Biophys. Acta 2007, 1768, 1268–1276. [Google Scholar] [CrossRef]
- Girasole, M.; Dinarelli, S.; Boumis, G. Structural, morphological and nanomechanical characterisation of intermediate states in the ageing of erythrocytes. J. Mol. Recognit. 2012, 25, 285–291. [Google Scholar] [CrossRef]
- Kamruzzahan, A.S.M.; Kienberger, F.; Stroh, C.M.; Berg, J.; Huss, R.; Ebner, A.; Zhu, R.; Rank, C.; Gruber, H.J.; Hinterdorfer, P. Imaging morphological details and pathological differences of red blood cells using tapping-mode AFM. Biol. Chem. 2004, 385, 955–960. [Google Scholar] [CrossRef]
- Dulińska, I.; Targosz, M.; Strojny, W.; Lekka, M.; Czuba, P.; Balwierz, W.; Szymoński, M. Stiffness of normal and pathological erythrocytes studied by means of atomic force microscopy. J. Biochem. Biophys. Methods 2006, 66, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Ciasca, G.; Papi, M.; Di Claudio, S.; Chiarpotto, M.; Palmieri, V.; Maulucci, G.; Nocca, G.; Rossi, C.; De Spirito, M. Mapping viscoelastic properties of healthy and pathological red blood cells at the nanoscale level. Nanoscale 2015, 7, 17030–17037. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, W.; Wang, S.; Wang, C.; Xie, J.; Chen, X.; Xu, Y.; Mao, P. Detection of erythrocytes in patients with multiple myeloma using atomic force microscopy. Scanning 2012, 34, 295–301. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Li, J. Detection of erythrocytes in patients with Waldenström macroglobulinemia using atomic force microscopy. Acta Biochim. Biophys. Sin. 2014, 46, 420–425. [Google Scholar] [CrossRef]
- Cluitmans, J.C.A.; Tomelleri, C.; Yapici, Z.; Dinkla, S.; Bovee-Geurts, P.; Chokkalingam, V.; De Franceschi, L.; Brock, R.; Bosman, G.J. Abnormal Red Cell Structure and Function in Neuroacanthocytosis. PLoS ONE 2015, 10, e0125580. [Google Scholar] [CrossRef]
- Sot, J.; García-Arribas, A.B.; Abad, B.; Arranz, S.; Portune, K.; Andrade, F.; Martín-Nieto, A.; Velasco, O.; Arana, E.; Tueros, I.; et al. Erythrocyte Membrane Nanomechanical Rigidity Is Decreased in Obese Patients. Int. J. Mol. Sci. 2022, 23, 1920. [Google Scholar] [CrossRef]
- Kozlova, E.; Sergunova, V.; Sherstyukova, E.; Gudkova, O.; Kozlov, A.; Inozemtsev, V.; Lyapunova, S.; Chernysh, A. Topological Relationships Cytoskeleton-Membrane Nanosurface-Morphology as a Basic Mechanism of Total Disorders of RBC Structures. Int. J. Mol. Sci. 2022, 23, 2045. [Google Scholar] [CrossRef]
- Korniluk, A.; Koper-Lenkiewicz, O.M.; Kamińska, J.; Kemona, H.; Dymicka-Piekarska, V. Mean Platelet Volume (MPV): New Perspectives for an Old Marker in the Course and Prognosis of Inflammatory Conditions. Mediat. Inflamm. 2019, 2019, 9213074. [Google Scholar] [CrossRef]
- Branehög, I.; Kutti, J.; Ridell, B.; Swolin, B.; Weinfeld, A. The relation of thrombokinetics to bone marrow megakaryocytes in idiopathic thrombocytopenic purpura (ITP). Blood 1975, 45, 551–562. [Google Scholar] [CrossRef]
- Chu, S.G.; Becker, R.C.; Berger, P.B.; Bhatt, D.L.; Eikelboom, J.W.; Konkle, B.; Mohler, E.R.; Reilly, M.P.; Berger, J.S. Mean platelet volume as a predictor of cardiovascular risk: A systematic review and meta-analysis. J. Thromb. Haemost. 2010, 8, 148–156. [Google Scholar] [CrossRef]
- Shrivastava, M.; Das, T.K.; Behari, M.; Pati, U.; Vivekanandhan, S. Ultrastructural Variations in Platelets and Platelet Mitochondria: A Novel Feature in Amyotrophic Lateral Sclerosis. Ultrastruct. Pathol. 2011, 35, 52–59. [Google Scholar] [CrossRef]
- Cole, M.G.; Prchal, J.F. Low serum vitamin B12 in Alzheimer-type dementia. Age Ageing 1984, 13, 101–105. [Google Scholar] [CrossRef] [PubMed]
- McCaddon, A. Vitamin B12 in neurology and ageing; Clinical and genetic aspects. Biochimie 2013, 95, 1066–1076. [Google Scholar] [CrossRef]
- Najim al-Din, A.S.; Khojali, M.; Habbosh, H.; Farah, S.; Idris, A.R.; al-Muhtasib, F. Macrocytosis in multiple sclerosis. A study in 82 de novo Arab patients. J. Neurol. Neurosurg. Psychiatry 1991, 54, 415–416. [Google Scholar] [CrossRef] [PubMed]
- Bandaru, S.S.; Killeen, R.B.; Gupta, V. Poikilocytosis. In StatPearls [Internet]; StatPearls Publishing: Treasure Island, FL, USA, 2023. Available online: https://www.ncbi.nlm.nih.gov/books/NBK562141/ (accessed on 10 August 2023).
- Kozlova, E.; Sherstyukova, E.; Sergunova, V.; Grechko, A.; Kuzovlev, A.; Lyapunova, S.; Inozemtsev, V.; Kozlov, A.; Chernysh, A. Atomic Force Microscopy and High-Resolution Spectrophotometry for Study of Anoxemia and Normoxemia in Model Experiment In Vitro. Int. J. Mol. Sci. 2023, 24, 11043. [Google Scholar] [CrossRef] [PubMed]
- Bester, J.; Buys, A.V.; Lipinski, B.; Kell, D.B.; Pretorius, E. High ferritin levels have major effects on the morphology of erythrocytes in Alzheimer’s disease. Front. Aging Neurosci. 2013, 5, 88. [Google Scholar] [CrossRef] [PubMed]
- Pretorius, E.; Swanepoel, A.C.; Buys, A.V.; Vermeulen, N.; Duim, W.; Kell, D.B. Eryptosis as a marker of Parkinson’s disease. Aging 2014, 6, 788–819. [Google Scholar] [CrossRef] [PubMed]
- Todinova, S.; Krumova, S.; Bogdanova, D.; Danailova, A.; Zlatareva, E.; Kalaydzhiev, N.; Langari, A.; Milanov, I.; Taneva, S.G. Red Blood Cells’ Thermodynamic Behavior in Neurodegenerative Pathologies and Aging. Biomolecules 2021, 11, 1500. [Google Scholar] [CrossRef]
- Strijkova, V.; Todinova, S.; Andreeva, T.; Langari, A.; Bogdanova, D.; Zlatareva, E.; Kalaydzhiev, N.; Milanov, I.; Taneva, S.G. Platelets’ Nanomechanics and Morphology in Neurodegenerative Pathologies. Biomedicines 2022, 10, 2239. [Google Scholar] [CrossRef] [PubMed]
- Li, A.; Chen, J.; Liang, Z.H.; Cai, J.; Cai, H.H.; Chen, M. Comparison of ultrastructural and nanomechanical signature of platelets from acute myocardial infarction and platelet activation. Biochem. Biophys. Res. Commun. 2017, 486, 245–251. [Google Scholar] [CrossRef]
- García-Rubio, D.; Rodríguez-Varela, M.; Martínez-Vieyra, I.; de la Mora, M.B.; Méndez-Méndez, J.V.; Durán-Álvarez, J.C.; Cerecedo, D. Alterations to the contents of plasma membrane structural lipids are associated with structural changes and compartmentalization in platelets in hypertension. Exp. Cell Res. 2019, 385, 111692. [Google Scholar] [CrossRef] [PubMed]
- Van Rooy, M.J.; Duim, W.; Ehlers, R.; Buys, A.V.; Pretorius, E. Platelet hyperactivity and fibrin clot structure in transient ischemic attack individuals in the presence of metabolic syndrome: A microscopy and thromboelastography study. Cardiovasc. Diabetol. 2015, 14, 86. [Google Scholar] [CrossRef] [PubMed]
- Andreeva, T.; Komsa-Penkova, R.; Langari, A.; Krumova, S.; Golemanov, G.; Georgieva, G.B.; Taneva, S.G.; Giosheva, I.; Mihaylova, N.; Tchorbanov, A.; et al. Morphometric and Nanomechanical Features of Platelets from Women with Early Pregnancy Loss Provide New Evidence of the Impact of Inherited Thrombophilia. Int. J. Mol. Sci. 2021, 22, 7778. [Google Scholar] [CrossRef]
- Dinarelli, S.; Girasole, M.; Misiti, F. Amyloid β peptide affects erythrocyte morphology: Role of intracellular signaling pathways. Clin. Hemorheol. Microcirc. 2019, 71, 437–449. [Google Scholar] [CrossRef] [PubMed]
- Carelli-Alinovi, C.; Dinarelli, S.; Sampaolese, B.; Misiti, F.; Girasole, M. Morphological changes induced in erythrocyte by amyloid beta peptide and glucose depletion: A combined atomic force microscopy and biochemical study. Biochim. Biophys. Acta (BBA) Biomembr. 2019, 1861, 236–244. [Google Scholar] [CrossRef] [PubMed]
- Kosenko, E.A.; Tikhonova, L.A.; Montoliu, C.; Barreto, G.E.; Aliev, G.; Kaminsky, Y.G. Metabolic Abnormalities of Erythrocytes as a Risk Factor for Alzheimer’s Disease. Front. Neurosci. 2018, 11, 728. [Google Scholar] [CrossRef] [PubMed]
- Kowalewski, T.; Holtzman, D.M. In situ atomic force microscopy study of Alzheimer’s b-amyloid peptide on different substrates: New insights into mechanism of b-sheet formation. Proc. Natl. Acad. Sci. USA 1999, 96, 3688–3693. [Google Scholar] [CrossRef]
- Banerjee, S.; Hashemi, M.; Zagorski, K.; Lyubchenko, Y.L. Interaction of Aβ42 with Membranes Triggers the Self-Assembly into Oligomers. Int. J. Mol. Sci. 2020, 21, 1129. [Google Scholar] [CrossRef]
- Feuillie, C.; Lambert, E.; Ewald, M.; Azouz, M.; Henry, S.; Marsaudon, S.; Cullin, C.; Lecomte, S.; Molinari, M. High Speed AFM and NanoInfrared Spectroscopy Investigation of Aβ1-42 Peptide Variants and Their Interaction With POPC/SM/Chol/GM1 Model Membranes. Front. Mol. Biosci. 2020, 7, 571696. [Google Scholar] [CrossRef]
- Watanabe-Nakayama, T.; Ono, K.; Itami, M.; Takahashi, R.; Teplow, D.B.; Yamada, M. High-speed atomic force microscopy reveals structural dynamics of amyloid β1-42 aggregates. Proc. Natl. Acad. Sci. USA 2016, 113, 5835–5840. [Google Scholar] [CrossRef]
- Banerjee, S.; Sun, Z.; Hayden, E.Y.; Teplow, D.B.; Lyubchenko, Y.L. Nanoscale Dynamics of Amyloid β-42 Oligomers As Revealed by High-Speed Atomic Force Microscopy. ACS Nano 2017, 11, 12202–12209. [Google Scholar] [CrossRef] [PubMed]
- Tinker-Mill, C.; Mayes, J.; Allsop, D.; Kolosov, O.V. Ultrasonic force microscopy for nanomechanical characterization of early and late-stage amyloid-β peptide aggregation. Sci. Rep. 2014, 4, 4004. [Google Scholar] [CrossRef] [PubMed]
- Ruggeri, F.S.; Benedetti, F.; Knowles, T.P.J.; Lashuel, H.A.; Sekatskii, S.; Dietler, G. Identification and nanomechanical characterization of the fundamental single-strand protofilaments of amyloid α-synuclein fibrils. Proc. Natl. Acad. Sci. USA 2018, 115, 7230–7235. [Google Scholar] [CrossRef]
- Knowles, T.P.J.; Vendruscolo, M.; Dobson, C.M. The amyloid state and its association with protein misfolding diseases. Nat. Rev. Mol. Cell Biol. 2014, 15, 384–396. [Google Scholar] [CrossRef]
- Ruggeri, F.S.; Flagmeier, P.; Kumita, J.R.; Meisl, G.; Chirgadze, D.Y.; Bongiovanni, M.N.; Knowles, T.P.J.; Dobson, C.M. The Influence of Pathogenic Mutations in α-Synuclein on Biophysical and Structural Characteristics of Amyloid Fibrils. ACS Nano 2020, 14, 5213–5222. [Google Scholar] [CrossRef]
- Camino, J.D.; Gracia, P.; Chen, S.W.; Sot, J.; de la Arada, I.; Sebastián, V.; Arrondo, J.L.R.; Goñi, F.M.; Dobson, C.M.; Cremades, N. The extent of protein hydration dictates the preference for heterogeneous or homogeneous nucleation generating either parallel or antiparallel β-sheet α-synuclein aggregates. Chem. Sci. 2020, 11, 11902–11914. [Google Scholar] [CrossRef]
- Ruggeri, F.S.; Adamcik, J.; Jeong, J.S.; Lashuel, H.A.; Mezzenga, R.; Dietler, G. Influence of the β-sheet content on the mechanical properties of aggregates during amyloid fibrillization. Angew. Chem.-Int. Ed. 2015, 54, 2462–2466. [Google Scholar] [CrossRef] [PubMed]
- Goedert, M. Alpha-synuclein and neurodegenerative diseases. Nat. Rev. Neurosci. 2001, 2, 492–501. [Google Scholar] [CrossRef]
- Watanabe, H.; Uchihashi, T.; Kobashi, T.; Shibata, M.; Nishiyama, J.; Yasuda, R.; Ando, T. Wide-area scanner for high-speed atomic force microscopy. Rev. Sci. Instrum. 2013, 84, 053702. [Google Scholar] [CrossRef]
- Suzuki, Y.; Sakai, N.; Yoshida, A.; Yoshitsugu, U.; Akira, Y.; Yuka, I.; Shuichi, I.; Koichi, K.; Kunio, T. High-speed atomic force microscopy combined with inverted optical microscopy for studying cellular events. Sci. Rep. 2013, 3, 2131. [Google Scholar] [CrossRef]
- Marchesi, A.; Umeda, K.; Komekawa, T.; Matsubara, T.; Holger Flechsig, H.; Ando, T.; Watanabe, S.; Kodera, N.; Franz, C.M. An ultra-wide scanner for large-area high-speed atomic force microscopy with megapixel resolution. Sci. Rep. 2021, 11, 13003. [Google Scholar] [CrossRef] [PubMed]
- Dazzi, A.; Prater, C.B.; Hu, Q.; Chase, D.B.; Rabolt, J.F.; Marcott, C. AFM-IR: Combining atomic force microscopy and infrared spectroscopy for nanoscale chemical characterization. Appl. Spectrosc. 2012, 66, 1365–1384. [Google Scholar] [CrossRef] [PubMed]
- Dazzi, A.; Prater, C.B. AFM-IR: Technology and applications in nanoscale infrared spectroscopy and chemical imaging. Chem. Rev. 2017, 117, 5146–5173. [Google Scholar] [CrossRef] [PubMed]
- Muller, E.A.; Pollard, B.; Raschke, M.B. Infrared chemical nano-imaging: Accessing structure, coupling, and dynamics on molecular length scales. J. Phys. Chem. Lett. 2015, 6, 1275–1284. [Google Scholar] [CrossRef] [PubMed]
- Goldsbury, C.; Green, J. Time-lapse atomic force microscopy in the characterization of amyloid-like fibril assembly and oligomeric intermediates. Methods Mol. Biol. 2005, 299, 103–128. [Google Scholar]
- Bonhommeau, S.; Lecomte, S. Tip-Enhanced Raman Spectroscopy: A Tool for Nanoscale Chemical and Structural Characterization of Biomolecules. ChemPhysChem 2018, 9, 8–18. [Google Scholar] [CrossRef]
- Shao, F.; Zenobi, R. Tip-enhanced Raman spectroscopy: Principles, practice, and applications to nanospectroscopic imaging of 2D materials. Anal. Bioanal. Chem. 2019, 411, 37–61. [Google Scholar] [CrossRef]
- Buys, A.V.; Van Rooy, M.-J.; Soma, P.; Van Papendorp, D.; Lipinski, B.; Pretorius, E. Changes in red blood cell membrane structure in type 2 diabetes: A scanning electron and atomic force microscopy study. Cardiovasc. Diabetol. 2013, 12, 25. [Google Scholar] [CrossRef]
- Alummoottil, S.; van Rooy, M.J.; Bester, J.; Grobbelaar, C.; Phulukdaree, A. Scanning Electron and Atomic Force Microscopic Analysis of Erythrocytes in a Cohort of Atopic Asthma Patients—A Pilot Study. Hemato 2023, 4, 90–99. [Google Scholar] [CrossRef]
- Uchihashi, T.; Scheuring, S. Applications of high-speed atomic force microscopy to real-time visualization of dynamic biomolecular processes, Biochim. Biophys. Acta-Gen. Subj. 2018, 1862, 229–240. [Google Scholar] [CrossRef]
- Ando, T.; Kodera, N.; Takai, E.; Maruyama, D.; Saito, K.; Toda, A. A high-speed atomic force microscope for studying biological macromolecules. Proc. Natl. Acad. Sci. USA 2001, 98, 12468–12472. [Google Scholar] [CrossRef] [PubMed]
- Milhiet, P.E.; Yamamoto, D.; Berthoumieu, O.; Dosset, P.; le Grimellec, C.; Verdier, J.M.; Marchal, S.; Ando, T. Deciphering the structure, growth and assembly of amyloid-like fibrils using high-speed atomic force microscopy. PLoS ONE 2010, 5, e13240. [Google Scholar] [CrossRef] [PubMed]
- Sadeghian, H.; Herfst, R.; Dekker, B.; Winters, J.; Bijnagte, T.; Rijnbeek, R. High-throughput atomic force microscopes operating in parallel. Rev. Sci. Instrum. 2017, 88, 033703. [Google Scholar] [CrossRef] [PubMed]
- Xia, F.; Youcef-Toumi, K.; Sattel, T.; Manske, E.; Rangelow, I.W. Active Probe Atomic Force Microscopy with Quattro-Parallel Cantilever Arrays for High-Throughput Large-Scale Sample Inspection. J. Vis. Exp. 2023, 196, e65210. [Google Scholar] [CrossRef] [PubMed]
- Tsukada, K.; Sekizuka, E.; Oshio, C.; Minamitani, H. Direct measurement of erythrocyte deformability in diabetes mellitus with a transparent microchannel capillary model and high-speed video camera system. Microvasc. Res. 2001, 61, 231–239. [Google Scholar] [CrossRef] [PubMed]
- Tomaiuolo, G. Biomechanical properties of red blood cells in health and disease towards microfluidics. Biomicrofluidics 2014, 8, 051501. [Google Scholar] [CrossRef]
- Faivre, M.; Renoux, C.; Bessaa, A.; Da Costa, L.; Joly, J.; Gauthier, A.; Connes, P. Mechanical Signature of Red Blood Cells Flowing Out of a Microfluidic Constriction Is Impacted by Membrane Elasticity, Cell Surface-to-Volume Ratio and Diseases. Front. Physiol. 2020, 11, 576. [Google Scholar] [CrossRef]
- Zheng, Y.; Nguyen, J.; Wai, Y.; Sun, Y. Recent advances in microfluidic techniques for single-cell biophysical characterization. Lab Chip 2013, 13, 2464–2483. [Google Scholar] [CrossRef]
- Faustino, V.; Pinho, D.; Yaginuma, T.; Calhelha, R.C.; Ferreira, I.C.F.R.; Lima, R. Extensional flow-based microfluidic device: Deformability assessment of red blood cells in contact with tumor cells. Biochip J. 2014, 8, 42–47. [Google Scholar] [CrossRef]
- Faustino, V.; Rodrigues, R.O.; Pinho, D.; Costa, E.; Santos-Silva, A.; Miranda, V.; Amaral, J.S.; Lima, R. A microfluidic deformability assessment of pathological red blood cells flowing in a hyperbolic converging microchannel. Micromachines 2019, 10, 645. [Google Scholar] [CrossRef]
- Tomaiuolo, G.; Guido, S. Start-up shape dynamics of red blood cells in microcapillary flow. Microvasc. Res. 2011, 82, 35–41. [Google Scholar] [CrossRef] [PubMed]
- Braunmüller, S.; Schmid, L.; Sackmann, E.; Franke, T. Hydrodynamic deformation reveals two coupled modes/time scales of red blood cell relaxation. Soft Matter 2012, 8, 11240–11248. [Google Scholar] [CrossRef]
- Prado, G.; Farutin, A.; Misbah, C.; Bureau, L. Viscoelastic transient of confined red blood cells. Biophys. J. 2015, 108, 2126–2136. [Google Scholar] [CrossRef] [PubMed]
- Osaki, T.; Shin, Y.; Sivathanu, V.; Campisi, M.; Kamm, R.D. In Vitro Microfluidic Models for Neurodegenerative Disorders. Adv. Healthc. Mater. 2018, 7, 1700489. [Google Scholar] [CrossRef] [PubMed]


| Condition | Mean PLTs Volume, µm3 | % of Healthy PLTs Volume | % of Shrinkage Relative to Healthy PLTs Volume |
|---|---|---|---|
| Healthy | 3.20 ± 1.28 | - | - |
| PD | 1.48 ± 0.34 | 46.3 | 55.7% |
| ALS | 1.13 ± 0.34 | 35.3 | 64.7% |
| AD | 2.19 ± 0.55 | 68.4 | 31.6% |
| RBCs | PLTs | |||||
|---|---|---|---|---|---|---|
| Subject | Ea/Rrms | CI | Ea/Rrms | CI | ||
| r | Lower Limit | Upper Limit | r | Lower Limit | Upper Limit | |
| Healthy | −0.78 | −0.935 | −0.373 | −0.75 | −0.8794 | −0.5175 |
| PD | −0.67 | −0.875 | −0.261 | −0.78 | −0.8887 | −0.5882 |
| ALS | −0.69 | −0.879 | −0.313 | −0.84 | −0.9248 | −0.6757 |
| AD | −0.85 | −0.972 | −0.562 | −0.93 | −0.9769 | −0.7978 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Taneva, S.G.; Todinova, S.; Andreeva, T. Morphometric and Nanomechanical Screening of Peripheral Blood Cells with Atomic Force Microscopy for Label-Free Assessment of Alzheimer’s Disease, Parkinson’s Disease, and Amyotrophic Lateral Sclerosis. Int. J. Mol. Sci. 2023, 24, 14296. https://doi.org/10.3390/ijms241814296
Taneva SG, Todinova S, Andreeva T. Morphometric and Nanomechanical Screening of Peripheral Blood Cells with Atomic Force Microscopy for Label-Free Assessment of Alzheimer’s Disease, Parkinson’s Disease, and Amyotrophic Lateral Sclerosis. International Journal of Molecular Sciences. 2023; 24(18):14296. https://doi.org/10.3390/ijms241814296
Chicago/Turabian StyleTaneva, Stefka G., Svetla Todinova, and Tonya Andreeva. 2023. "Morphometric and Nanomechanical Screening of Peripheral Blood Cells with Atomic Force Microscopy for Label-Free Assessment of Alzheimer’s Disease, Parkinson’s Disease, and Amyotrophic Lateral Sclerosis" International Journal of Molecular Sciences 24, no. 18: 14296. https://doi.org/10.3390/ijms241814296
APA StyleTaneva, S. G., Todinova, S., & Andreeva, T. (2023). Morphometric and Nanomechanical Screening of Peripheral Blood Cells with Atomic Force Microscopy for Label-Free Assessment of Alzheimer’s Disease, Parkinson’s Disease, and Amyotrophic Lateral Sclerosis. International Journal of Molecular Sciences, 24(18), 14296. https://doi.org/10.3390/ijms241814296






