Insulin-like Androgenic Gland Hormone Induced Sex Reversal and Molecular Pathways in Macrobrachium nipponense: Insights into Reproduction, Growth, and Sex Differentiation
Abstract
:1. Introduction
2. Results
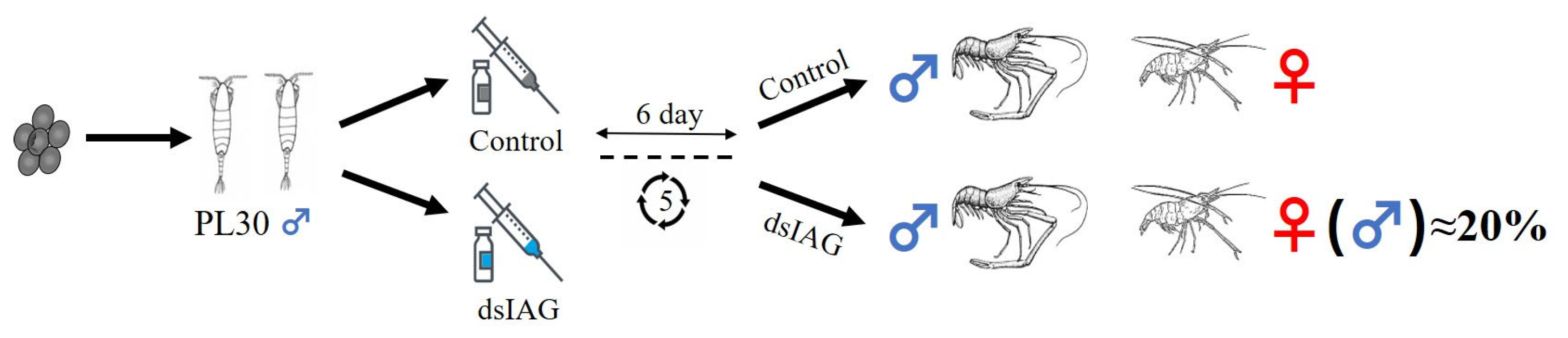
2.1. Effects of dsIAG on Male PL30 Juvenile Prawn
2.1.1. Interference Efficiency and Sex Ratio
2.1.2. Histological Observations of the Gonad
2.1.3. Growth Traits
2.2. The Comparative Transcriptomic Analysis
2.2.1. Overview of Transcriptome Sequencing
2.2.2. Identification and Functional Analysis of DEGs
2.2.3. GO and COG Enrichment Analysis of DEGs
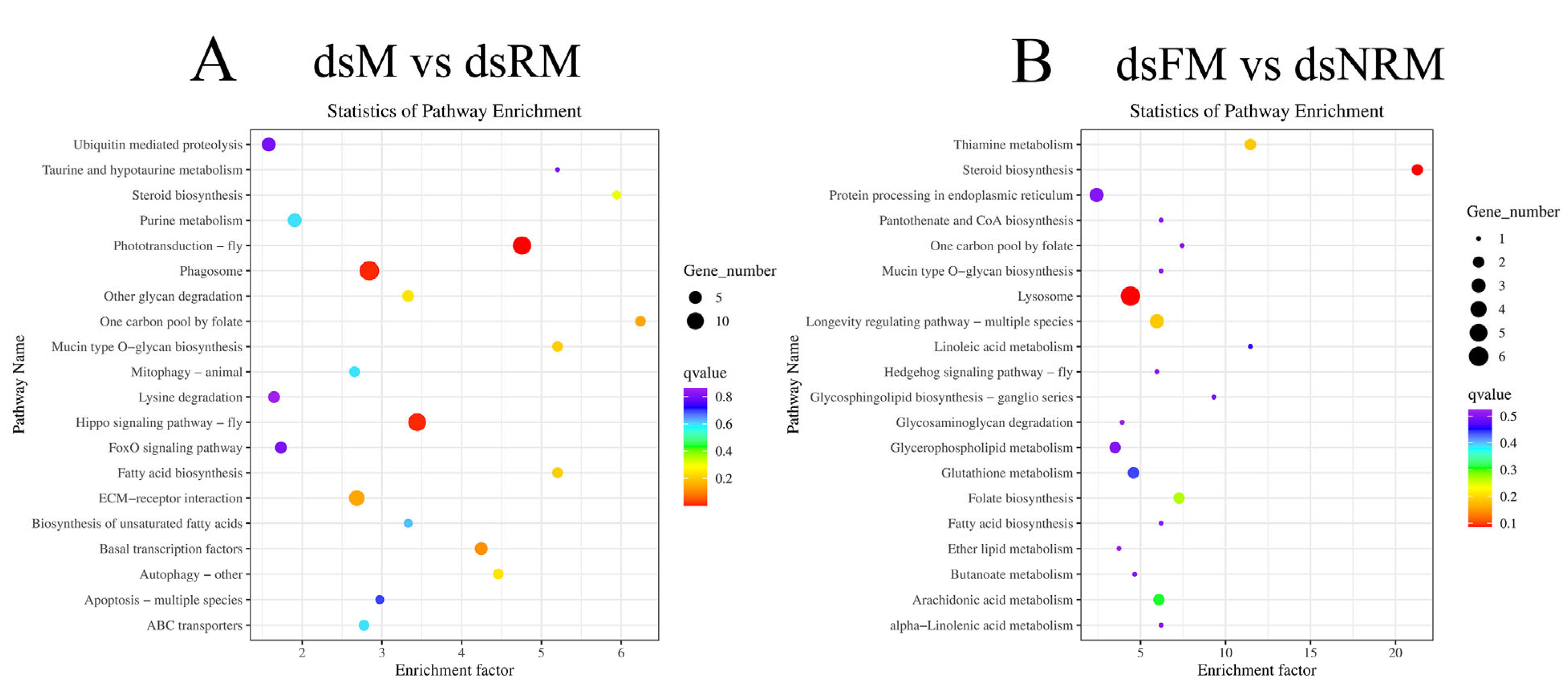
2.2.4. KEGG Analysis and Important Differentially Expressed Pathways
2.2.5. Key Pathways and Genes for Reproduction in M. nipponense
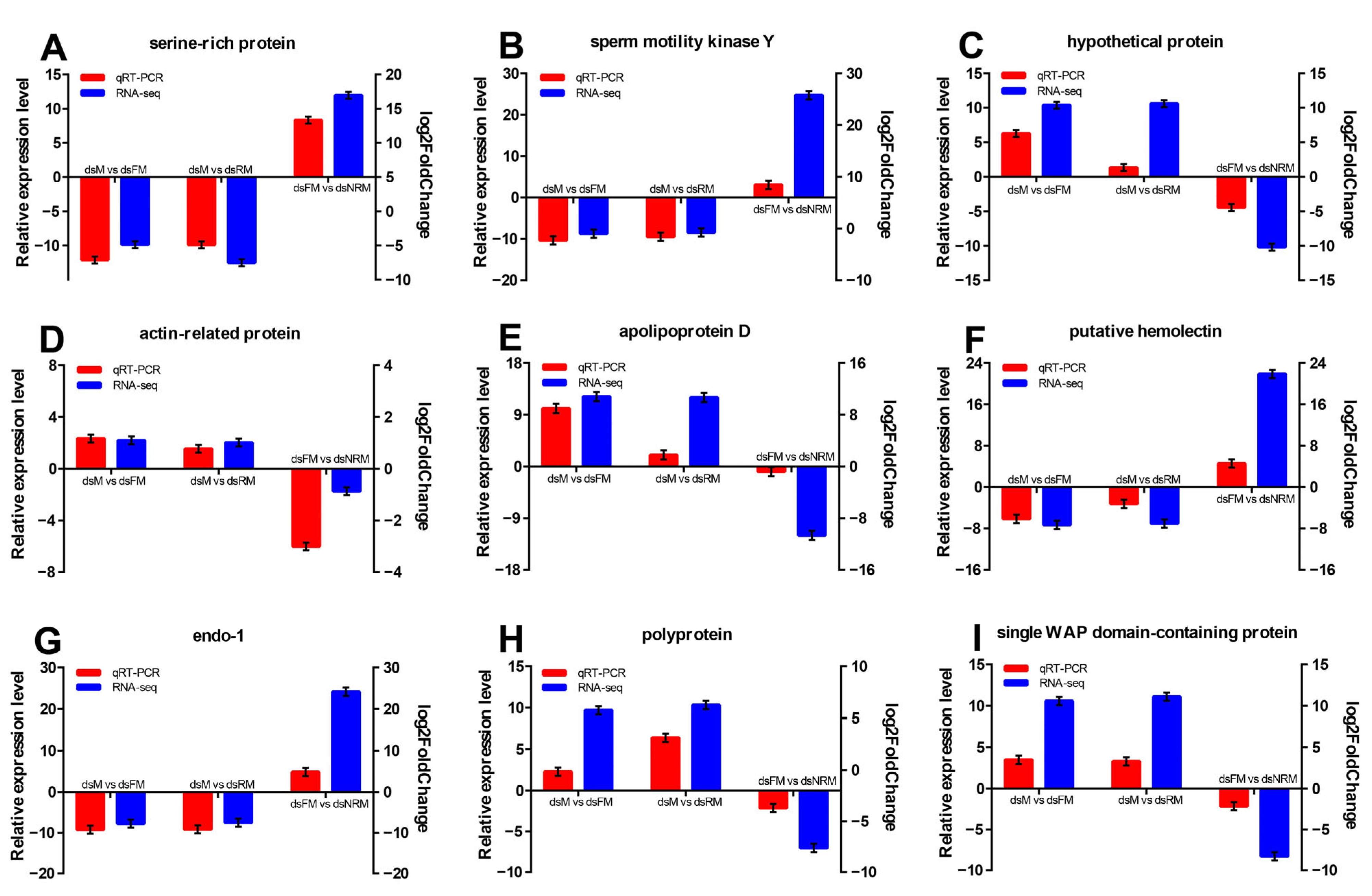
2.2.6. Validation of DEGs by qRT-PCR
3. Discussion
4. Materials and Methods
4.1. Experimental Design
4.2. dsIAG Synthesis and Injection
4.3. Histological Observations
4.4. Growth Traits
4.5. Sex Ratio and Feminization Rate Statistics
4.6. RNA Isolation and Quantitative Real-Time PCR Analysis
4.7. Transcriptomic Sequencing
4.8. Quantification and Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sun, R.; Li, Y. A sex-reversing factor: Insulin-like androgenic gland hormone in decapods. Rev. Aquac. 2021, 13, 1352–1366. [Google Scholar] [CrossRef]
- Sagi, A. Sexual differentiation in decapod crustaceans: Role of the androgenic gland. Int. J. Invertebr. Reprod. 1997, 31, 55–61. [Google Scholar] [CrossRef]
- Ventura, T.; Fitzgibbon, Q.; Battaglene, S.; Sagi, A.; Elizur, A. Identification and characterization of androgenic gland specific insulin-like peptide-encoding transcripts in two spiny lobster species: Sagmariasus verreauxi and Jasus edwardsii. Gen. Comp. Endocrinol. 2015, 214, 126–133. [Google Scholar] [CrossRef] [PubMed]
- Manor, R.; Weil, S.; Oren, S.; Glazer, L.; Aflalo, E.D.; Ventura, T.; Chalifa-Caspi, V.; Lapidot, M.; Sagi, A. Insulin and gender: An insulin-like gene expressed exclusively in the androgenic gland of the male crayfish. Gen. Comp. Endocrinol. 2004, 150, 326–336. [Google Scholar] [CrossRef]
- Rosen, O.; Manor, R.; Weil, S.; Gafni, O.; Linial, A.; Aflalo, E.D.; Ventura, T.; Sagi, A. A sexual shift induced by silencing of a single insulin-like gene in Crayfish: Ovarian Upregulation and Testicular Degeneration. PLoS ONE 2010, 5, e15281. [Google Scholar] [CrossRef]
- Parnes, S.; Khalaila, I.; Hulata, G.; Sagi, A. Sex determination in crayfish: Are intersex Cherax quadricarinatus (Decapoda, Parastacidae) genetically females? Genet. Res. 2003, 82, 107–116. [Google Scholar] [CrossRef]
- Vázquez-Islas, G.; Garza-Torres, R.; Guerrero-Tortolero, D.; Campos-Ramos, R. Histology of the androgenic gland and expression of the insulin-like androgenic gland hormone precursor gene in the genital organ of pacific white shrimp Litopenaeus vannamei. J. Crustac. Biol. 2014, 34, 293–299. [Google Scholar] [CrossRef]
- Tinikul, Y.; Poljaroen, J.; Kornthong, N.; Chotwiwatthanakun, C.; Anuracpreeda, P.; Poomtong, T.; Hanna, P.J.; Sobhon, P. Distribution and changes of serotonin and dopamine levels in the central nervous system and ovary of the Pacific white shrimp, Litopenaeus vannamei, during ovarian maturation cycle. Cell Tissue Res. 2011, 345, 103–124. [Google Scholar] [CrossRef]
- Ma, M.; Gard, A.; Xiang, F.; Wang, J.; Davoodian, N.; Lenz, P.; Malecha, S.; Christie, A.; Li, L. Combining in silico transcriptome mining and biological mass spectrometry for neuropeptide discovery in the Pacific white shrimp Litopenaeus vannamei. Peptides 2010, 31, 27–43. [Google Scholar] [CrossRef]
- Zhang, D.; Sun, M.; Liu, X. Phase-specific expression of an insulin-like androgenic gland factor in a marine shrimp Lysmata wurdemanni: Implication for maintaining protandric simultaneous hermaphroditism. PLoS ONE 2017, 12, e0172782. [Google Scholar] [CrossRef]
- Sroyraya, M.; Chotwiwatthanakun, C.; Stewart, M.; Soonklang, N.; Kornthong, N.; Phoungpetchara, I.; Hanna, P.; Sobhon, P. Bilateral eyestalk ablation of the blue swimmer crab, Portunus pelagicus, produces hypertrophy of the androgenic gland and an increase of cells producing insulin-like androgenic gland hormone. Tissue Cell 2010, 42, 293–300. [Google Scholar] [CrossRef] [PubMed]
- Chung, J. An insulin-like growth factor found in hepatopancreas implicates carbohydrate metabolism of the blue crab Callinectes sapidus. Gen. Comp. Endocrinol. 2014, 199, 56–64. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Ye, H.; Chung, J. The presence of an insulin-like androgenic gland factor (IAG) and insulin-like peptide binding protein (ILPBP) in the ovary of the blue crab, Callinectes sapidus and their roles in ovarian development. Gen. Comp. Endocrinol. 2017, 249, 64–70. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Ye, H.; Huang, H.; Yang, Y.; Gong, J. An insulin-like androgenic gland hormone gene in the mud crab, Scylla paramamosain, extensively expressed and involved in the processes of growth and female reproduction. Gen. Comp. Endocrinol. 2014, 204, 229–238. [Google Scholar] [CrossRef] [PubMed]
- Lawrence, A.; Green, S.; Chung, J. Isolation and tissue distribution of an insulin-like androgenic gland hormone (IAG) of the male red deep-sea crab, Chaceon quinquedens. Mar. Drugs 2017, 15, 241. [Google Scholar] [CrossRef]
- Ventura, T.; Sagi, A. The insulin-like androgenic gland hormone in crustaceans: From a single gene silencing to a wide array of sexual manipulation-based biotechnologies. Biotechnol. Adv. 2012, 30, 1543–1550. [Google Scholar] [CrossRef]
- Aflalo, E.; Hoang, T.; Nguyen, V.; Lam, Q.; Nguyen, D.; Trinh, Q.; Raviv, S.; Sagi, A. A novel two-step procedure for mass production of all-male populations of the giant freshwater prawn Macrobrachium rosenbergii. Aquaculture 2006, 256, 468–478. [Google Scholar] [CrossRef]
- Ventura, T.; Manor, R.; Aflalo, E.; Weil, S.; Raviv, S.; Glazer, L.; Sagi, A. Temporal silencing of an androgenic gland-specific insulin-like gene affecting phenotypical gender differences and spermatogenesis. Endocrinology 2009, 150, 1278–1286. [Google Scholar] [CrossRef]
- Cai, Y.; Ng, P. The freshwater palaemonid prawns (Crustacea: Decapoda: Caridea) of Myanmar. Hydrobiologia 2002, 487, 59–83. [Google Scholar] [CrossRef]
- De Grave, S.; Ghane, A. The establishment of the oriental river prawn, Macrobrachium nipponense (de Haan, 1849) in Anzali Lagoon, Iran. Aquat. Invasions 2006, 1, 204–208. [Google Scholar] [CrossRef]
- Hongtuo, F.; Sufei, J.; Yiwei, X. Current status and prospects of farming the giant river prawn (Macrobrachium rosenbergii) and the oriental river prawn (Macrobrachium nipponense) in China. Aquac. Res. 2012, 43, 993–998. [Google Scholar] [CrossRef]
- Cai, P.; Yuan, H.; Gao, Z.; Qiao, H.; Zhang, W.; Jiang, S.; Xiong, Y.; Gong, Y.; Wu, Y.; Jin, S.; et al. 17β-estradiol induced sex reversal and gonadal transcriptome analysis in the oriental river prawn (Macrobrachium nipponense): Mechanisms, pathways, and potential Harm. Int. J. Mol. Sci. 2023, 24, 8481. [Google Scholar] [CrossRef] [PubMed]
- Jin, S.; Yue, D.; Fu, H.; Jiang, S.; Xiong, Y.; Qiao, H.; Zhang, W.; Gong, Y.; Wu, Y. Effects of dietary supplementation with 17β-estradiol and 17α-methyltestosterone on growth performance and gonadal development of the juvenile oriental river prawn (Macrobrachium nipponense). Aquac. Rep. 2022, 23, 101042. [Google Scholar] [CrossRef]
- Cai, P.; Yuan, H.; Gao, Z.; Daka, P.; Qiao, H.; Zhang, W.; Jiang, S.; Xiong, Y.; Gong, Y.; Wu, Y.; et al. Sex reversal induced by dietary supplementation with 17α-methyltestosterone during the critical period of sex differentiation in oriental river prawn (Macrobrachium nipponense). Animals 2023, 13, 1369. [Google Scholar] [CrossRef]
- Qiao, H.; Xiong, Y.; Zhang, W.; Fu, H.; Jiang, S.; Sun, S.; Bai, H.; Jin, S.; Gong, Y. Characterization, expression, and function analysis of gonad-inhibiting hormone in Oriental River prawn, Macrobrachium nipponense and its induced expression by temperature. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2015, 185, 1–8. [Google Scholar] [CrossRef]
- Poljaroen, J.; Tinikul, Y.; Phoungpetchara, I.; Kankoun, W.; Suwansa-ard, S.; Siangcham, T.; Meeratana, P.; Cummins, S.; Sretarugsa, P.; Hanna, P. The effects of biogenic amines, gonadotropin-releasing hormones and corazonin on spermatogenesis in sexually mature small giant freshwater prawns, Macrobrachium rosenbergii (De Man, 1879). Aquaculture 2011, 321, 121–129. [Google Scholar] [CrossRef]
- Vázquez-Islas, G.; Guerrero-Tortolero, D.; Garza-Torres, R.; Álvarez-Ruiz, P.; Mejía-Ruiz, H.; Campos-Ramos, R. Quantitative analysis of hypertrophy and hyperactivity in the androgenic gland of eyestalk-ablated male Pacific white shrimp Litopenaeus vannamei during molt stages. Aquaculture 2015, 439, 7–13. [Google Scholar] [CrossRef]
- Senthilan, P.; Grebler, R.; Reinhard, N.; Rieger, D.; Helfrich-Förster, C. Role of rhodopsins as circadian photoreceptors in the Drosophila melanogaster. Biology 2019, 8, 6. [Google Scholar] [CrossRef]
- Hanon, E.; Lincoln, G.; Fustin, J.; Dardente, H.; Masson-Pévet, M.; Morgan, P.; Hazlerigg, D. Ancestral TSH mechanism signals summer in a photoperiodic mammal. Curr. Biol. 2008, 18, 1147–1152. [Google Scholar] [CrossRef]
- García-Fernández, J.; Cernuda-Cernuda, R.; Davies, W.; Rodgers, J.; Turton, M.; Peirson, S.; Follett, B.; Halford, S.; Hughes, S.; Hankins, M. The hypothalamic photoreceptors regulating seasonal reproduction in birds: A prime role for VA opsin. Front. Neuroendocrinol. 2015, 37, 13–28. [Google Scholar] [CrossRef]
- Karplus, I.; Sagi, A.; Khalaila, I.; Barki, A. The influence of androgenic gland implantation on the agonistic behavior of female crayfish (Cherax quadricarinatus) in Interactions with Males. Behaviour 2003, 140, 649–663. [Google Scholar] [CrossRef]
- Qiao, H.; Fu, H.; Xiong, Y.; Jiang, S.; Zhang, W.; Sun, S.; Jin, S.; Gong, Y.; Wang, Y.; Shan, D. Molecular insights into reproduction regulation of female Oriental River prawns Macrobrachium nipponense through comparative transcriptomic analysis. Sci. Rep. 2017, 7, 12161. [Google Scholar] [CrossRef] [PubMed]
- Fu, C.; Li, F.; Wang, L.; Li, T. Molecular insights into ovary degeneration induced by environmental factors in female oriental river prawns Macrobrachium nipponense. Environ. Pollut. 2019, 253, 882–888. [Google Scholar] [CrossRef]
- Ono, K.; Sandell, L.; Trainor, P.; Wu, D. Retinoic acid synthesis and autoregulation mediate zonal patterning of vestibular organs and inner ear morphogenesis. Development 2020, 147, dev192070. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Qiao, H.; Fu, H.; Sun, S.; Zhang, W.; Jin, S.; Jiang, S.; Gong, Y.; Xiong, Y.; Wu, Y. Identification and characterization of opsin gene and its role in ovarian maturation in the oriental river prawn Macrobrachium nipponense. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2018, 218, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Watson, C.; Gray, S.; Hoffmann, M.; Lubieniecki, K.; Joy, J.; Sandkam, B.; Weigel, D.; Loew, E.; Dreyer, C.; Davidson, W. Gene duplication and divergence of long wavelength-sensitive opsin genes in the guppy, Poecilia reticulata. J. Mol. Evol. 2011, 72, 240–252. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Wang, X. Lysosome biogenesis: Regulation and functions. J. Cell Biol. 2021, 220, e202102001. [Google Scholar] [CrossRef]
- Schiffer, L.; Barnard, L.; Baranowski, E.; Gilligan, L.; Taylor, A.; Arlt, W.; Shackleton, C.; Storbeck, K. Human steroid biosynthesis, metabolism and excretion are differentially reflected by serum and urine steroid metabolomes: A comprehensive review. J. Steroid Biochem. Mol. Biol. 2019, 194, 105439. [Google Scholar] [CrossRef]
- Yang, L.; Zhang, X.; Liu, S.; Zhao, C.; Miao, Y.; Jin, L.; Wang, D.; Zhou, L. Cyp17a1 is required for female sex determination and male fertility by regulating sex steroid biosynthesis in fish. Endocrinology 2021, 162, bqab205. [Google Scholar] [CrossRef]
- Calderón-Ospina, C.; Nava-Mesa, M. B Vitamins in the nervous system: Current knowledge of the biochemical modes of action and synergies of thiamine, pyridoxine, and cobalamin. CNS Neurosci. Ther. 2020, 26, 5–13. [Google Scholar] [CrossRef]
- Henrique Mazucanti, C.; Victor Cabral-Costa, J.; Rodrigues Vasconcelos, A.; Zukas Andreotti, D.; Scavone, C.; Mitiko Kawamoto, E. Longevity pathways (mTOR, SIRT, Insulin/IGF-1) as key modulatory targets on aging and neurodegeneration. Curr. Top. Med. Chem. 2015, 15, 2116–2138. [Google Scholar] [CrossRef] [PubMed]
- Brownsey, R.; Boone, A.; Elliott, J.; Kulpa, J.; Lee, W. Regulation of acetyl-CoA carboxylase. Biochem. Soc. Trans. 2006, 34, 223–227. [Google Scholar] [CrossRef] [PubMed]
- Smith, S.; Witkowski, A.; Joshi, A. Structural and functional organization of the animal fatty acid synthase. Prog. Lipid Res. 2003, 42, 289–317. [Google Scholar] [CrossRef]
- Strange, R.; Spiteri, M.; Ramachandran, S.; Fryer, A. Glutathione-S-transferase family of enzymes. Mutat. Res./Fundam. Mol. Mech. Mutagen. 2001, 482, 21–26. [Google Scholar] [CrossRef] [PubMed]
- Zerai, D.; Fitzsimmons, K.; Collier, R. Transcriptional response of delta-9-desaturase gene to acute and chronic cold stress in Nile tilapia, Oreochromis niloticus. J. World Aquac. Soc. 2010, 41, 800–806. [Google Scholar] [CrossRef]
- Yao, W.; Jiang, P.; Bai, J. Effects of 17α-methyltestosterone on gonadal development and hormone levels in grass carp (Ctenopharyngodon idella). J. Fish. China 2019, 43, 801–809. [Google Scholar] [CrossRef]
- Tsutsui, S.; Sato, M.; Miyashita, M.; Amano, H.; Maeda, T.; Nakamura, O. Vitellogenin-derived fragment in embryos of Japanese flounder Paralichthys olivaceus with binding and bactericidal activities against an infectious bacterium via an interaction with saccharides. Mol. Immunol. 2022, 142, 76–82. [Google Scholar] [CrossRef]
- Bai, H.; Qiao, H.; Li, F.; Fu, H.; Jiang, S.; Zhang, W.; Yan, Y.; Xiong, Y.; Sun, S.; Jin, S. Molecular and functional characterization of the vitellogenin receptor in oriental river prawn, Macrobrachium nipponense. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2016, 194, 45–55. [Google Scholar] [CrossRef]
- Du, Y.; Ma, K.; Qiu, G. Discovery of the genes in putative GnRH signaling pathway with focus on characterization of GnRH-like receptor transcripts in the brain and ovary of the oriental river prawn Macrobrachium nipponense. Aquaculture 2015, 442, 1–11. [Google Scholar] [CrossRef]
- Sun, S.; Zheng, C.; Shi, X. Effect of paternal exposure to microcystin-LR on testicular dysfunction, reproduction, and offspring immune response in the oriental river prawn (Macrobrachium nipponense). Aquaculture 2021, 534, 736332. [Google Scholar] [CrossRef]
- Maya, F.; Debby, I.; Haim, B. Regulation of sperm motility by PIP2(4,5) and actin polymerization. Dev. Biol. 2013, 381, 62–72. [Google Scholar] [CrossRef]
- Breitbart, H.; Etkovitz, N. Role and regulation of EGFR in actin remodeling in sperm capacitation and the acrosome reaction. Asian J. Androl. 2011, 13, 106–110. [Google Scholar] [CrossRef] [PubMed]
- Inoue, N.; Hamada, D.; Kamikubo, H.; Hirata, K.; Kataoka, M.; Yamamoto, M.; Ikawa, M.; Okabe, M.; Hagihara, Y. Molecular dissection of IZUMO1, a sperm protein essential for sperm-egg fusion. Development 2013, 140, 3221–3229. [Google Scholar] [CrossRef]
- Ahn, S.; Bak, H.; Park, J.; Kim, S.; Kim, N.; Lee, J.; Sung, J.; Jeon, S.; Chung, J.; Lee, H. Olive flounder (Paralichthys olivaceus) cystatin B: Cloning, tissue distribution, expression and inhibitory profile of piscine cystatin B. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2013, 165, 211–218. [Google Scholar] [CrossRef] [PubMed]
- Teal, C.; Schill, D.; Fogelson, S.; Roberts, C.; Fitzsimmons, K.; Bauder, J.; Stewart, W.; Bonar, S. The effects of estradiol-17β on the sex reversal, survival, and growth of green sunfish Lepomis cyanellus. Aquaculture 2022, 562, 0044–8486. [Google Scholar] [CrossRef]
- Nagahama, Y.; Chakraborty, T.; Paul-Prasanth, B.; Ohta, K.; Nakamura, M. Sex determination, gonadal sex differentiation, and plasticity in vertebrate species. Physiol. Rev. 2021, 101, 1237–1308. [Google Scholar] [CrossRef] [PubMed]
- Faeed, M.; Kasra, R.; PourKazemi, M.; Darboee, M.; Haghighi, S. Study on effect feedings with probiotics in increasing resistance to Aeromonas hydrophila and Changes in gut bacterial communities Sander lucioperca. Biol. J. Microorg. 2018, 7, 1–12. [Google Scholar] [CrossRef]
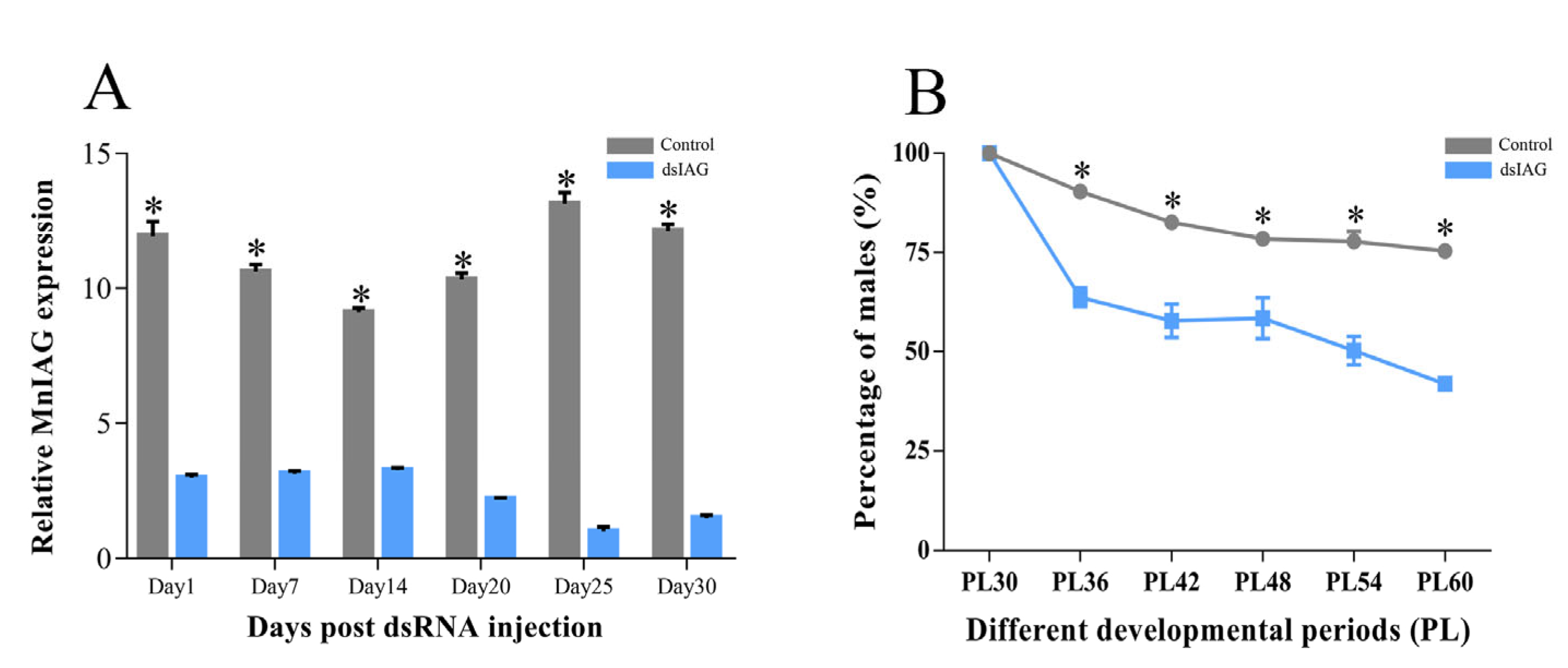

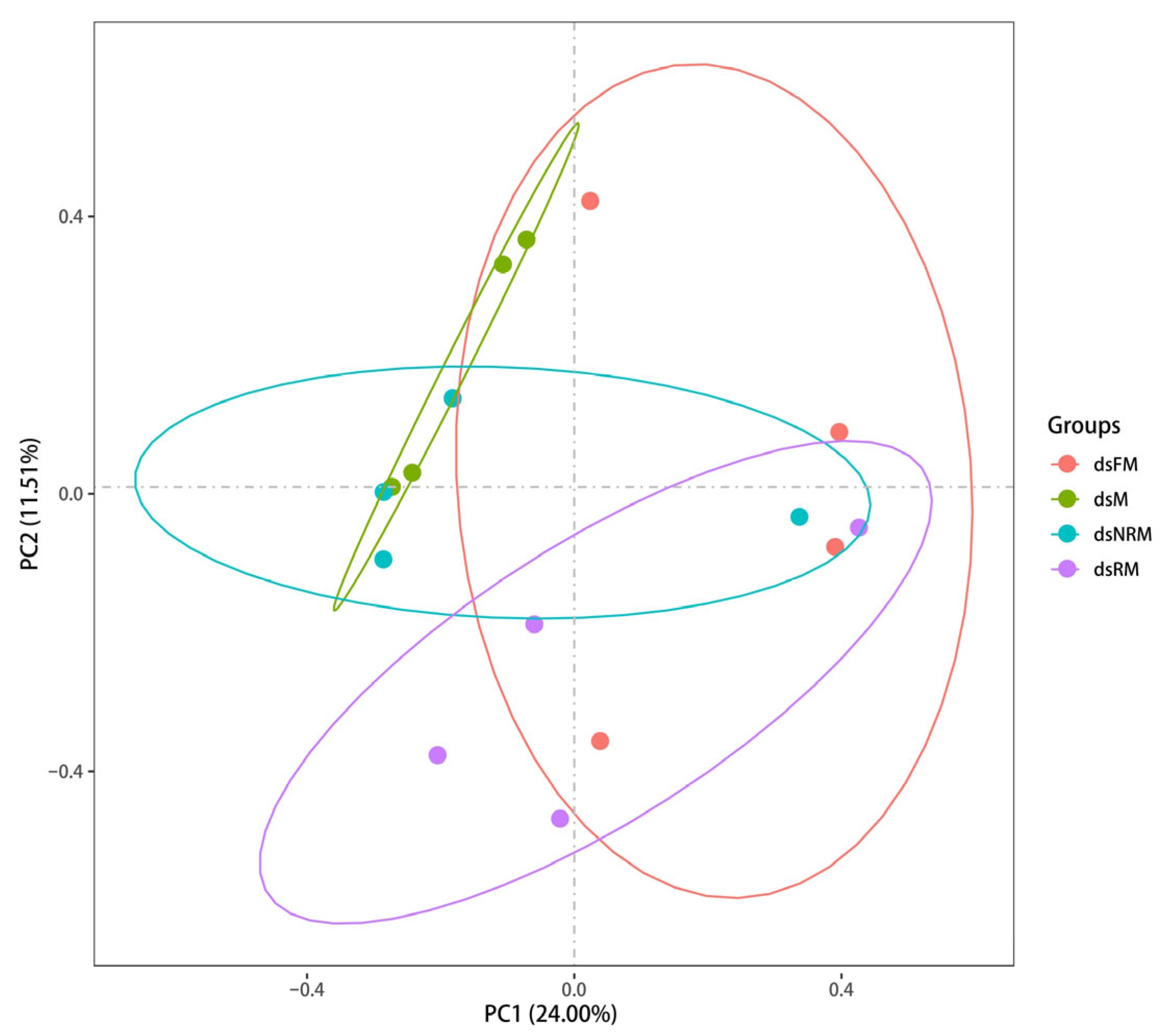

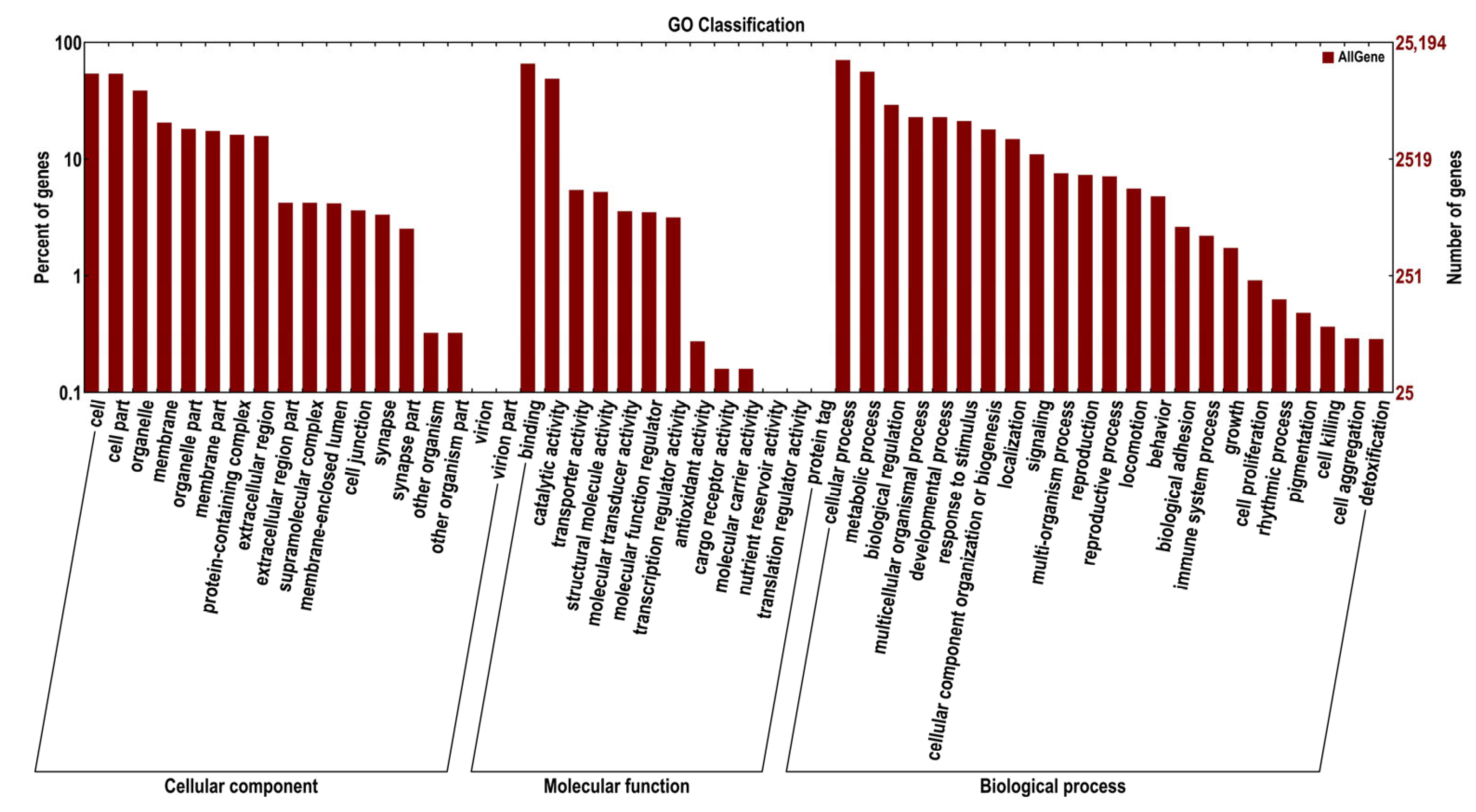
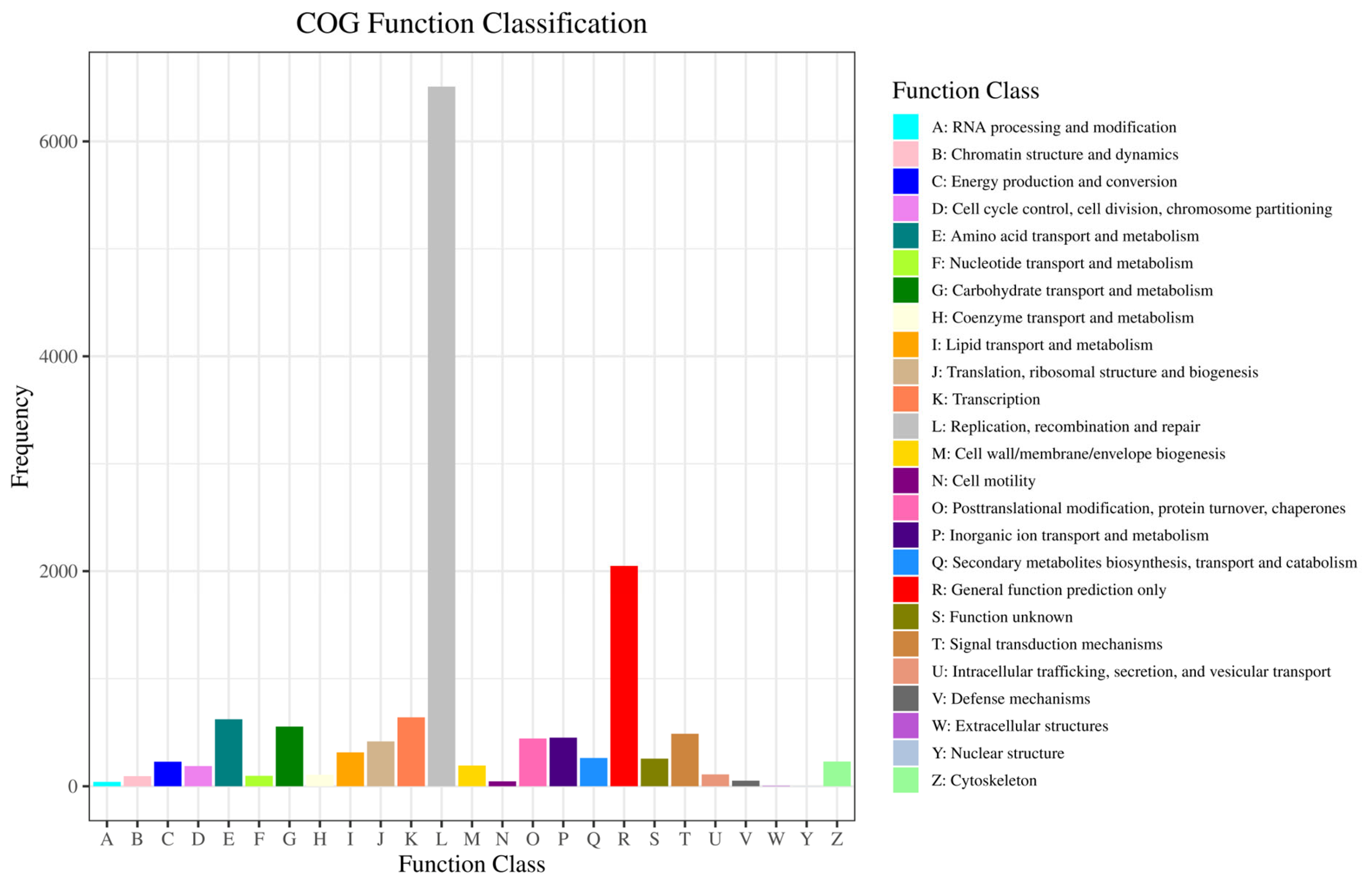
| Group (μg/g) | Sex | Index | |||
|---|---|---|---|---|---|
| IMW (mg) | FMW (mg) | WGR (%/g) | SGR (%/d) | ||
| Control | Male | 129.85 ± 1.90 a | 262.57 ± 9.49 a | 50.52 ± 1.82 a | 2.34 ± 0.12 a |
| Female | 141.41 ± 1.98 A | 353.37 ± 19.52 B | 60.02 ± 2.16 A | 3.06 ± 0.18 A | |
| DsIAG | Neo-female | 112.32 ± 2.64 A | 265.40 ± 17.28 A | 57.56 ± 2.86 A | 2.86 ± 0.22 A |
| Unsex-reversed male | 112.32 ± 2.64 a | 305.93 ± 6.82 b | 63.28 ± 0.83 b | 3.34 ± 0.07 b | |
| Sample | ReadSum | BaseSum | GC(%) | Q20(%) | Q30(%) |
|---|---|---|---|---|---|
| dsM1 | 22,290,216 | 6,687,064,800 | 44.04 | 96.48 | 90.96 |
| dsM2 | 21,213,797 | 6,364,139,100 | 44.67 | 96.00 | 90.02 |
| dsM3 | 27,395,675 | 8,218,702,500 | 43.89 | 96.51 | 90.99 |
| dsM4 | 19,865,327 | 5,959,598,100 | 44.42 | 96.62 | 91.16 |
| dsFM1 | 21,757,005 | 6,527,101,500 | 43.57 | 96.34 | 90.61 |
| dsFM2 | 24,507,624 | 7,352,287,200 | 43.38 | 96.09 | 90.19 |
| dsFM3 | 24,153,175 | 7,245,952,500 | 42.94 | 95.97 | 89.94 |
| dsFM4 | 19,776,829 | 5,933,048,700 | 43.49 | 96.19 | 90.31 |
| dsRM1 | 18,570,225 | 5,571,067,500 | 43.99 | 96.47 | 90.90 |
| dsRM2 | 21,045,097 | 6,313,529,100 | 43.33 | 96.10 | 90.21 |
| dsRM3 | 21,327,753 | 6,398,325,900 | 44.41 | 96.43 | 90.78 |
| dsRM4 | 25,840,035 | 7,752,010,500 | 43.60 | 96.04 | 90.07 |
| dsNRM1 | 21,015,809 | 6,304,742,700 | 44.41 | 96.15 | 90.24 |
| dsNRM2 | 21,060,098 | 6,318,029,400 | 45.14 | 96.50 | 91.01 |
| dsNRM3 | 21,433,329 | 6,429,998,700 | 43.24 | 96.54 | 91.05 |
| dsNRM4 | 20,540,615 | 6,162,184,500 | 44.69 | 96.35 | 90.69 |
| No. | Pathway | Pathway ID | dsM vs. dsFM | dsM vs. dsRM | ||
|---|---|---|---|---|---|---|
| q-Value | DEGs Number | q-Value | DEGs Number | |||
| 1 | Phototransduction-fly | map04745 | 0.989 | 4 | 0.000 | 12 |
| 2 | Hippo signaling pathway-fly | map04391 | 1.000 | 4 | 0.012 | 11 |
| 3 | Phagosome | map04145 | 0.905 | 10 | 0.012 | 14 |
| 4 | ECM-receptor interaction | map04512 | 0.905 | 6 | 0.145 | 8 |
| No. | Pathway | Pathway ID | dsM vs. dsFM | dsFM vs. dsNRM | ||
|---|---|---|---|---|---|---|
| q-Value | DEGs Number | q-Value | DEGs Number | |||
| 1 | Lysosome | map04142 | 0.989 | 8 | 0.086 | 6 |
| 2 | Steroid biosynthesis | map00100 | 0.940 | 1 | 0.086 | 2 |
| 3 | Thiamine metabolism | map00730 | 0.535 | 3 | 0.188 | 2 |
| 4 | Longevity regulating pathway-multiple species | map04213 | 0.905 | 4 | 0.188 | 3 |
| No. | Name | Accession Number | Up or Down | ||
|---|---|---|---|---|---|
| dsM/dsFM | dsM/dsRM | dsFM/dsNRM | |||
| Sex-related genes | |||||
| 1 | vitellogenin | AJP60219.1 | up | up | down |
| 2 | vitellogenin receptor | AJP60220.1 | up | up | |
| 3 | VASA-like | AEQ19569.1 | up | up | |
| 4 | cyclin B | ADB44902.1 | up | ||
| 5 | Fem1b | ANN47504.1 | up | ||
| 6 | ferritin | QDA69873.1 | up | up | |
| 7 | gonadotropin-releasing hormone receptor | AHB33640.1 | up | up | |
| 8 | cystatin | AXS76129.1 | up | down | |
| 9 | doublesex and mab-3 related transcription factor | QDE10512.1 | down | down | up |
| 10 | heat shock protein | QCC72758.1 | down | down | up |
| 11 | sperm gelatinase | AFM38794.1 | down | down | |
| 12 | Kazal-type protease inhibitor | AEW24505.1 | down | ||
| 13 | male reproductive-related protein | ABQ41234.1 | down | ||
| Growth-related genes | |||||
| 1 | fatty acid synthase | QDK64693.1 | up | up | |
| 2 | acetyl-CoA carboxylas | ALK82309.1 | up | ||
| 3 | delta-9 desaturase | AMQ48727.1 | up | ||
| 4 | long wavelength sensitive opsin | ASS36969.1 | up | ||
| 5 | glutathione S-transferase | AGJ70295.1 | up | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cai, P.; Zhang, W.; Jiang, S.; Xiong, Y.; Yuan, H.; Gao, Z.; Gao, X.; Ma, C.; Zhou, Y.; Gong, Y.; et al. Insulin-like Androgenic Gland Hormone Induced Sex Reversal and Molecular Pathways in Macrobrachium nipponense: Insights into Reproduction, Growth, and Sex Differentiation. Int. J. Mol. Sci. 2023, 24, 14306. https://doi.org/10.3390/ijms241814306
Cai P, Zhang W, Jiang S, Xiong Y, Yuan H, Gao Z, Gao X, Ma C, Zhou Y, Gong Y, et al. Insulin-like Androgenic Gland Hormone Induced Sex Reversal and Molecular Pathways in Macrobrachium nipponense: Insights into Reproduction, Growth, and Sex Differentiation. International Journal of Molecular Sciences. 2023; 24(18):14306. https://doi.org/10.3390/ijms241814306
Chicago/Turabian StyleCai, Pengfei, Wenyi Zhang, Sufei Jiang, Yiwei Xiong, Huwei Yuan, Zijian Gao, Xuanbing Gao, Cheng Ma, Yongkang Zhou, Yongsheng Gong, and et al. 2023. "Insulin-like Androgenic Gland Hormone Induced Sex Reversal and Molecular Pathways in Macrobrachium nipponense: Insights into Reproduction, Growth, and Sex Differentiation" International Journal of Molecular Sciences 24, no. 18: 14306. https://doi.org/10.3390/ijms241814306
APA StyleCai, P., Zhang, W., Jiang, S., Xiong, Y., Yuan, H., Gao, Z., Gao, X., Ma, C., Zhou, Y., Gong, Y., Qiao, H., Jin, S., & Fu, H. (2023). Insulin-like Androgenic Gland Hormone Induced Sex Reversal and Molecular Pathways in Macrobrachium nipponense: Insights into Reproduction, Growth, and Sex Differentiation. International Journal of Molecular Sciences, 24(18), 14306. https://doi.org/10.3390/ijms241814306








