1. Introduction
To transmit and focus light onto the retina, the human eye lens needs to be transparent with a high refractive index of approximately 1.33 [
1]. To obtain these two key features, the microarchitecture of the lens is avascular, with a regular array of elongated fiber cells devoid of organelles and filled with high concentrations of highly stable proteins termed lens crystallins. Crystallins make up over 90% of the total lens protein content and are thought to maintain the favorable optical conditions of the lens with their compact globular structures, short-range order, and packing. Together, these result in a constant refractive index over distances approximating the wavelength of the transmitted light and hence transparency [
2,
3].
Ubiquitous crystallins include two large gene families, the α and βγ-crystallins. α-crystallins are members of the small heat shock protein family and act as chaperones [
4], while βγ-crystallins are structural proteins related to spore coat proteins [
5]. βγ-crystallins comprise two domains, each domain consisting of two Greek key motifs, and a super-secondary structural fold. Each βγ-crystallin Greek key motif consists of four β-sheets which run antiparallel to each other and form a 3,1 type of Greek key motif [
6]. Human γS-crystallin is a 178-amino-acid protein with a total of 27 aromatic residues (fourteen tyrosines Y, four tryptophans W, and nine phenylalanines F). These aromatic residues play an important role in stabilizing the Greek key fold by participating in “Greek key” and “non-Greek key” pairs or by forming “Tyrosine corners” [
7].
Five aromatic pairs are present in β-hairpin sequences in human γS-crystallin. Of these, four pairs exist in Greek key motifs and are termed Greek key pairs (Y11-F16, Y50-F55, F99-F104, and Y140-Y145; the initial methionine is counted) and one falls outside the Greek key motifs, termed a non-Greek key pair (Y21-Y33). Three of the homologous aromatic pairs (two Tyr/Tyr, one Phe/Phe) and two of the non-homologous aromatic pairs (Tyr/Phe) are present in the β-hairpins of the human γS-crystallin protein. Energetically, the optimal distance between these aromatic rings is 4.5–7 Å, either perpendicular or parallel to each other [
8]. They contribute a free energy of 0.6–1.3 kcal mol
−1 to the protein when they are perpendicular to each other [
8] or about 0.75 kcal mol
−1 stability to the protein by parallel displacement stacking [
9].
Any mutation that substitutes a non-aromatic residue for the crucial phenylalanine or tyrosine in an aromatic pair is predicted to destabilize βγ-crystallin folding severely. A dinucleotide substitution NM_017541.4:c.30_31delinsAA resulting in a NP_060011.1:p.(F10_Y11delinsLN) substitution in human γS-crystallin has been reported to be associated with autosomal dominant cortical lamellar cataracts with juvenile onset [
10]. This two-amino-acid substitution disrupts the interaction of tyrosine and phenylalanine in the first Greek key pair (Y11-F16), and in addition, the adjacent F10 is replaced by a hydrophobic leucine residue. In this study, we investigated the role of the F10_Y11delinsLN change in the destabilization of γS-crystallin and subsequent cataract formation. This change impairs the protein’s tryptophan microenvironment and increases its surface hydrophobicity under physiological conditions and decreases its stability, leading to early denaturation under both thermal and chemical stress. This eventually results in the formation of high-molecular-weight (HMW) aggregates, causing lens opacification after overcoming the main defense systems of the lens against cataract [
10] including the strong reducing environment sustained by high levels of glutathione [
11] and the chaperone activity of αA-crystallin [
12].
3. Discussion
The γ-crystallins have extremely rigid three-dimensional structures with tight packing [
17]. Five aromatic pairs Y11-F16, Y50-F55, F99-F104, Y140-Y145, and Y21-Y33 are important for maintaining their Greek key motif structures and play a critical role in providing stability and compactness to the γ-crystallin protein. In the cataract-associated mutation F10_Y11delinsLN, F10 and Y11 in the first Greek key motif of the N-terminal domain are replaced with L10 and N11. This work investigates how these substitutions affect the structure and stability of the protein and the relationship of these effects to the cortical lamellar cataracts described in family members carrying this mutation [
10].
The total accessible surface area that is occupied by the changed amino acids is decreased from 218 to 180 (Å
2) (F10 to L10) and 229 to 158 (Å
2) (Y11 to N11) [
18], suggesting that alterations in the size of these residues is not a major factor in destabilization of the Greek key motif. However, the π interactions around the site might be altered with subsequent destabilization of the protein, since the substitutions do not significantly alter the backbone conformation as estimated by CD under physiological conditions. This is consistent with molecular modeling of the wild-type and F10_Y11delinsLN mutant protein, which shows that the protein fold is essentially maintained in the carboxyl domain and only slightly distorted in the amino terminal domain (
Figure 9A). In the wild-type model, the distance between Y11 and F16 is estimated to be 4.73 Å (
Figure 9B), consistent with stabilization in a perpendicular orientation [
8]. The indel does alter the tryptophan micro-environment (
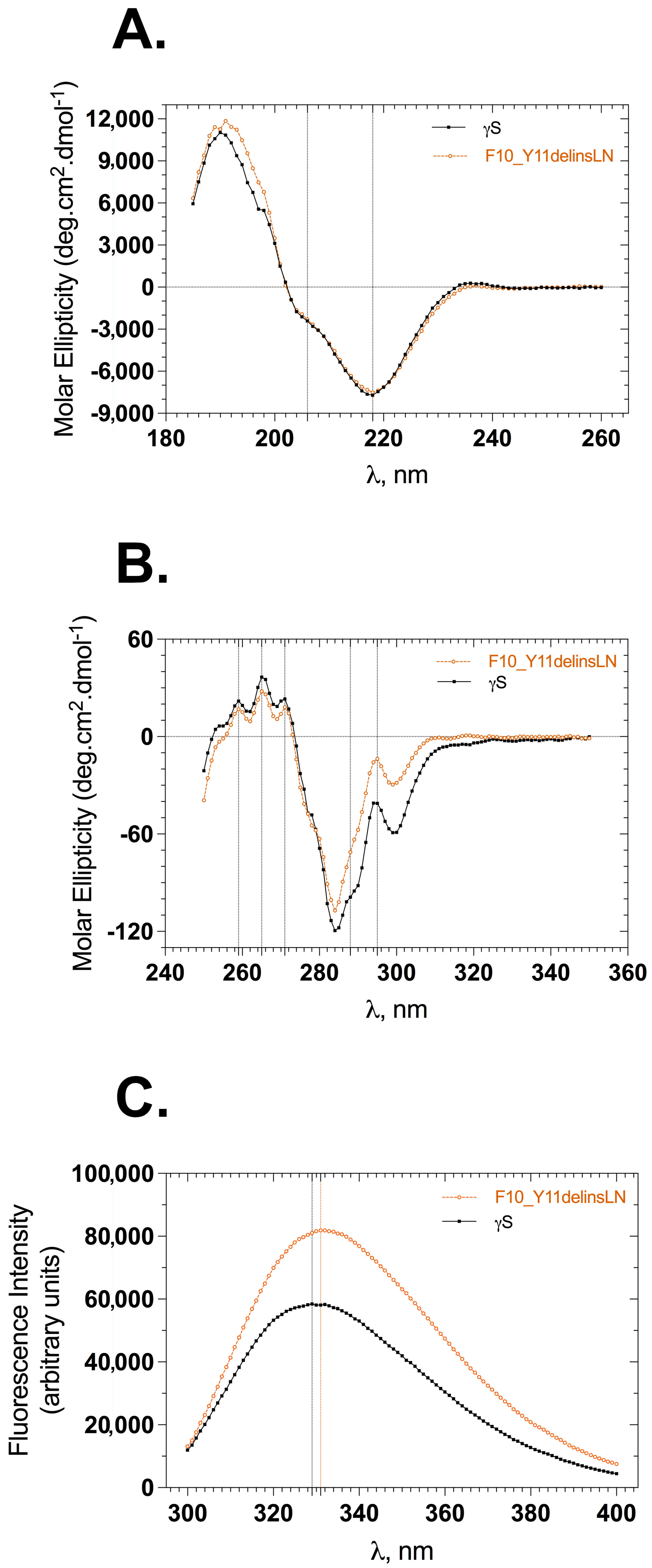
Figure 9C) consistent with the changed near-UV cd and fluorescence spectra (
Figure 1B,C). Consistent with this observation, increased exposure of hydrophobic residues to the surface of the mutant protein even under physiological conditions was confirmed via both bis-ANS and Nile Red fluorescence, similar to the results obtained with the G18V γS-crystallin and Y46D γC-crystallin mutations [
11,
13], and showing increased ANS binding to previously buried residues in the N-terminal domain and the nearby interdomain interface [
19]. However, in G18V γS-crystallin, this difference increased significantly upon exposure to 1 M GuHCl [
13] and the Y46D γC-crystallin mutant shows a greater difference in binding between bis-ANS than Nile Red at low concentrations, in contrast to the F10_Y11delinsLN γC-crystallin, which shows significant binding with both agents even at low concentrations.
While the increased exposure of hydrophobic residues to the protein surface indicates an opening of the protein fold, the circular dichroism and tryptophan fluorescence studies indicate that the mutations have only a minor effect on the overall protein structure under benign conditions approximating physiological exposures. However, the decreased stability is still present and simply becomes more obvious under stress, which accelerates the structural changes. Thus, while denaturation and aggregation would be much slower under physiological conditions, proteins in the lens fiber cells do not turn over, and even minor instability will manifest itself in structural changes over the months of prenatal development and especially the months and years of childhood and adult exposure to environmental stress. While the lens has large amounts of α-crystallins that act as chaperones to bind unstable and partially denatured proteins as well as high concentrations of reducing agents such as glutathione, these are eventually overcome, resulting in clinical cataract, either by scattering light directly or through damaging the lens cells and disrupting the lens microarchitecture [
20]. In general, one would expect an inverse correlation between the decrease in stability of a crystallin and the length of time required for denaturation and aggregation of the mutant protein under physiological conditions. In contrast, the spatial expression pattern of a mutant crystallin is better correlated with the location of resultant cataracts, although this correlation is imprecise. γS-crystallin is highly expressed in the equatorial epithelia, the cortical fiber cells and the nuclear fiber cells [
21], which correlates well with CRYGS mutations most frequently causing cortical, lamellar, sutural or nuclear cataracts [
22]. The cortical cataracts seen in this case are consistent with that pattern.
The mutations do destabilize γS-crystallin significantly under both thermal and chemical stress. In combination, the mutations decrease the overall stability to 5.0 kcal mol
−1 and lower the transition midpoint to 12.7 °C in thermal unfolding experiments monitored by CD. The energy change observed from fluorescence-based thermal unfolding data was similar, 4.3 kcal mol
−1. There is a slight difference in ΔH values and slopes in the unfolding procedures, although this is probably due to differences in the methods with regard to holding times and monitoring [
13]. The mutant appears to undergo a primarily two-state transition under thermal stress, although the shoulder seen in the tracing suggests that the N-terminal domain begins to destabilize a bit earlier when subjected to thermal stress. In contrast, the mutant shows a clear N-terminal intermediate transition under GuHCl-induced stress, during which it exhibits an intermediate around 1.25 M and the c-terminal transition is lowered to 2.59 M compared to 2.72 M in the wild type. This probably relates to the greater sensitivity of the fluorescence-based curves to small changes in the neighborhood of tryptophan and tyrosine residues while the CD measurements reflect the overall protein fold, similar to results seen with the G18V CRYGS and Y46D CRYGC mutations [
11,
13].
The role of aromatic pair interactions on the stability of the human γD-crystallin protein has been well studied. N-terminal domain substitutions affect the stability of the protein by shifting transitions of that domain to lower GuHCl concentration while the C-terminal domain transitions remained relatively unaffected. In addition, Greek key pairs tend to contribute more to the thermal and thermodynamic stability of the domain than non-Greek key pairs [
23]. The Y11-F16 aromatic pair in human γS-crystallin is homologous to the Y7-F12 pair in human γD-crystallin. When the Y7 residue of γD-crystallin is changed to alanine, γD-crystallin shows a two-state transition in thermal unfolding with an 8.9 °C decrease in its T
m [
23], similar to the present study, in which F10_Y11delinsLN γS-crystallin shows a decrease of 12.7 °C. An alanine substitution at Y6 in γD-crystallin resulted in a three-state transition under GuHCl treatment, with a concentration midpoint of 1.12 M GuHCl [
23] and a similar trend was also observed in the current study, in which the substitution caused a three-state transition with an intermediate around 1.25 M, which is consistent with Greek key pair domain substitutions showing severe effects. Phenyl rings in aromatic pairs interact optimally at a distance of 4.5–7 Å, aligning perpendicular or parallel to each other, and contributing a free energy of 0.5–1.3 kcal mol
−1 to overall protein stability [
8,
9], supporting the importance of aromatic pairs in stabilizing the protein. Substitution of two amino acids rather than a single change also contributes to the destabilization being more severe than that seen in the Y7-F12 change in γD-crystallin. Here, not only does the F10_Y11delinsLN mutation studied disturb the first Greek key aromatic pair interaction, but the adjacent F10 to L10 change also contributes to destabilization by further distorting the Greek key motif. Significant and even minor distortion of the protein fold of the Greek key motif can also lead to significant destabilization as shown by the results of Ma et al. [
13], in which even the conservative G18V CRYGS change in a critical part of the motif structure reduced the T
m by 9.5 °C, similar to the results seen here. An even greater destabilization of the second Greek key motif was seen in the Y46D γC-crystallin mutant, which showed a ΔT
m of −23 and 24.4 °C with CD and fluorescence, respectively, for the native to intermediate form and −9.9 and 9.3 °C for the intermediate to unfolded species corresponding to Δ(ΔG) values of −11 and −4.7 kcal mol
−1 for the CD curves and −9.1 and −3.5 kcal mol
−1 for the fluorescence curves. Similarly, GuHCl unfolding curves yielded ΔG values of 3.2 and 6.2 kcal mol
−1 for the intermediate and unfolded Y46D mutant species, respectively vs. 10.5 for the wild type [
11].
While both intrinsic fluorescence and ANS and Nile Red binding studies demonstrate that the surface hydrophobicity of the F10_Y11delinsLN mutant γS-crystallin is increased, size exclusion chromatography of αA-crystallin alone and mixed with wild-type and F10_Y11delinsLN mutant γS-crystallins shows that αA-crystallin does not bind either the wild-type or mutant protein to any appreciable extent under physiological conditions. However, as the F10_Y11delinsLN mutant γS-crystallin is heated at 60° both light scattering at 600 nm and dynamic light scattering show that it forms HMW aggregates, and that this process is prevented by a 1:1 molar ratio of αA-crystallin. These studies show that the F10_Y11delinsLN mutant γS-crystallin does not escape binding by αA-crystallin as some rapidly denaturing proteins do [
24,
25]. In contrast, analytical size exclusion chromatography results confirmed that αA-crystallin is not associated with F10_Y11delinsLN mutant and wild-type γS-crystallin at room temperature under benign conditions. This differs from the G18V mutant, which appears to associated with αA-crystallin by NMR analysis [
26], although the NMR technology might be more sensitive than size exclusion chromatography, which would only show stable binding. The F10_Y11delinsLN mutant aggregates and scatters light upon heating at 60 °C, and both DTT and TCEP suppress thermal aggregation by between 2- and 4-fold, differing from the Y46D γC-crystallin mutant, which does not show inhibition of aggregation by either DTT or TCEP [
11]. This probably relates to the involvement of disulfide bonds in aggregation of the F10_Y11delinsLN γS-crystallin but not the Y46D γC-crystallin protein. Few other studies have focused on the structural characterization of mutants in γS-crystallin. The D26G [
27] and G57W [
28] mutations show only minor effects on stability. In contrast, G18V [
13,
29], S39C [
7], and V42M [
30] show somewhat more severe changes, and the present work places the F10_Y11delinsLN mutation in the second group, showing a relatively normal protein fold under benign conditions, but a marked decrease in stability under thermal or chemical stress.
4. Materials and Methods
4.1. Cloning and Site-Directed Mutagenesis
Human γS-crystallin cDNA was cloned into the
NdeI
/XhoI sites of pET-20b (+) (Novagen, Burlington, MA, USA) as described earlier [
13]. To generate the F10_Y11delinsLN double mutant clone, the wild-type clone was amplified with the following primers F-5′ CAAGATTACTTTAAATGAAGACAAAAATTTTCAAGG 3′, R-5′ GAAAATTTTTGTCTTCATTTAAAGTAATCTTGGTTCC 3′. Methylated wild-type DNA was eliminated via digestion with
DpnI, and the remaining plasmid was transformed into
E. coli DH5α. Human αA-crystallin cDNA was cloned into the
NdeI
/XhoI sites of pET-21a (+) (Novagen) using a reverse primer harboring a CACCACCACCACCACCAC (6X his tag) sequence just before the stop codon. Plasmids were isolated from the transformed
E. coli colonies and bidirectionally sequenced to confirm the appropriate base pair changes and the absence of nonspecific mutations.
4.2. Protein Overexpression
Proteins were overexpressed and purified as described earlier [
13,
30]. BL21(DE3)pLysS cells were used to express the wild-type and mutant proteins. Single colonies containing the respective clones were inoculated into 12 mL of LB broth containing 50 μg/mL ampicillin and 34 μg/mL chloramphenicol and grown for 12 h at 37 °C with 225 rpm shaking. Then, 10 mL of this stationary phase culture was transferred to 1 L LB broth with the same concentration of antibiotics. The inoculated broths were grown at 37 °C with 225 rpm shaking until they reach an OD
600 of 0.6–0.8. The cultures with the F10_Y11delinsLN bacterial clone were adjusted to 0.25 mM IPTG and grown at 25 °C for an additional 6 h, whereas the cultures with wild-type γS and αA clones were induced with a final concentration of 1 mM IPTG and grown at 37 °C for an additional 4 h. Induced cells were pelleted via centrifugation at 6000×
g for 10 min at 4 °C and frozen at −80 °C until use.
4.3. γS-Crystallin Purification
Pellets containing overexpressed proteins were resuspended in 50 mM Tris-Cl pH 7.3, 100 mM NaCl, 1 mM EDTA, 1 mM DTT, 0.25 mM TCEP, and a protease inhibitor mix (Roche Diagnostics; Catalog #11836153001). A final lysozyme concentration of 0.3 mg/mL and a final DNase concentration of 7.5 µg/mL were maintained in the suspensions. Cell suspensions were freeze–thawed for 3 cycles, each cycle consisting of 5 min incubation on dry ice and 5 min incubation in a 37 °C water bath. The freeze–thawed cell suspensions were further sonicated at 4 °C and 20% amplitude with an Omni international Sonic Ruptor-400 (Kennesaw, GA, USA) using 9 cycles for the mutant, with each cycle comprising a 5 s pulse on and a 55 s pulse off, and using 6 cycles for the wild type, with each cycle comprising a 10 s pulse on 50 s pulse off. The cell lysate was centrifuged at 30,000× g for 20 min at 4 °C, and the supernatant was loaded into an 8 mL 10,000 MWCO dialysis cassette (Pierce Biotechnology, Rockford, IL, USA) and dialyzed against 2 L of buffer A (50 mM CH3COONa, 1 mM EDTA, 1 mM DTT, 50 μM TCEP, pH 5.4) for 6 h.
Soluble extracts were purified at room temperature with ion-exchange and size-exclusion chromatography using an NGC™ Chromatography System (Bio-Rad, Hercules, CA, USA). The dialyzed supernatant was centrifuged at 30,000× g for 20 min at 4 °C, and samples were loaded onto a 5 mL HiTrap SP FF (GE Healthcare, Chicago, IL, USA; Catalog #17-5157-01) column equilibrated with buffer A at a flow rate of 1.0 mL/min. The column was washed with five column volumes of buffer A. Then, a linear gradient of buffer B (50 mM CH3COONa, 1 mM EDTA, 1 mM DTT, 50 μM TCEP, pH 5.4, and 1 M NaCl) was applied and fractions were collected.
4.4. αA-Crystallin Purification
αA-Crystallin was also purified using a modification of the above-described method. The lysis buffer used in purifying αA crystallin was 50 mM KH2PO4/K2HPO4 pH 8.0, 300 mM KCl, 1 mM EDTA (buffer C) with a protease inhibitor mix (Roche Diagnostics, Basel, Switzerland; Catalog #11836153001). Sonication was carried out as described above for the wild-type protein (6 cycles, each cycle comprising a 10 s pulse on and a 50 s pulse off), and the centrifuged cell lysate was loaded onto a HisTrap HP (GE Healthcare; Catalog #17524801) column equilibrated with buffer C at a flow rate of 1.0 mL/min. Five column volumes of buffer C were passed through the column to wash away the unbound proteins completely, then, a linear gradient of buffer D (50 mM KH2PO4/K2HPO4 pH 8.0, 300 mM KCl, 1 mM EDTA, and 0.5 M imidazole) was applied and fractions were collected.
The fractions containing the proteins of interest were monitored with absorbance at 280 nm and SDS-PAGE on 12% polyacrylamide gels, and were pooled and concentrated to 10–15 mg/mL. Wild-type and mutant γS-crystallin protein pools were separately loaded onto a HiPrep™ 16/60 Sephacryl® S-100 HR column (GE Healthcare; Catalog #17116501) previously equilibrated with SEC buffer (50 mM NaH2PO4/Na2HPO4 (pH 7.3), 0.15 M NaCl, 1 mM EDTA, 1 mM DTT, and 50 μM TCEP) at a flow rate of 0.5 mL/min. Pooled wild-type αA-crystallin was injected on to a HiPrep™ 16/60 Sephacryl® S-300 HR column (GE Healthcare; Catalog #17116701) equilibrated with SEC buffer at a flow rate of 0.5 mL/min. The columns were pre-calibrated with five standards: thyroglobulin, γ-globulin, ovalbumin, myoglobin, and vitamin B12 (Bio Rad gel filtration standard; Catalog #1511901). The locations of recombinant proteins in column fractions were monitored using absorbance at 280 nm and by SDS-PAGE on 12% polyacrylamide gels. The purity of the protein was assessed and confirmed by the presence of a single band in SDS PAGE and then estimating the mass by using electrospray ionization mass spectrometry. Two independent preparations for each batch of αA, γS wild-types, and mutant proteins gave an average mass of 20,730.32 ± 0.5, 20,873.18 ± 0.5, and 20,789.76 ± 0.5 Da, respectively, in close agreement with the predicted monomer masses.
4.5. Liquid Chromatography/Mass Spectrometry (LC/MS)
Mass spectrometry data of 0.05 mg/mL proteins samples (in 50 mM Tris-Cl (pH 7.3), 150 mM NaCl, and 50 µM TCEP buffer) were acquired on an Agilent 6130 Quadrupole LC/MS System, (Agilent Technologies, Inc., Santa Clara, CA, USA) equipped with electrospray source, operated in positive-ion mode. Separation was performed on a 300SB-C3 Poroshell column (2.1 mm × 75 mm; particle size 5 μm). The analytes were eluted at a flow rate of 1 mL/min with a 5–100% organic gradient over 5 min and holding organic for 1 min. Mobile phase A contained 5% acetic acid in water, and mobile phase B was acetonitrile. Data acquisition and data analysis and deconvolution of mass spectra were performed using OpenLab ChemStation Edition software (version C.01.05).
4.6. Circular Dichroism (CD) Measurements
For determining secondary structure, 0.2 mg/mL protein samples in 10 mM NaH
2PO
4/Na
2HPO
4 (pH 7.3) were run between 260 and 185 nm at 25 °C in a 1 mm pathlength cuvette using a J-1500 (Jasco, Easton, MD, USA) circular dichroism instrument. For near-UV CD spectra, 1.0 mg/mL samples in 50 mM NaH
2PO
4/Na
2HPO
4 (pH 7.3), 150 mM NaCl, and 50 μM TCEP were analyzed between 350 and 250 nm at 25 °C in a 1 cm pathlength cuvette. A bandwidth of 1 nm, scanning speed of 20 nm/min for far-UV CD and 50 nm/min for near-UV CD and a digital integration time of 4 s at each point were maintained. At least five independent runs were measured and averaged, and spectra for the corresponding buffer for each protein were subtracted. For determining the secondary structure of wild-type and mutant proteins under GuHCl-induced stress, 0.15 mg/mL protein samples in 10 mM NaH
2PO
4/Na
2HPO
4 (pH 7.3) were incubated with varying concentrations of GuHCl (0–5 M) for 12–14 h at room temperature, and circular dichroism was measured between 260 and 185 nm at 25 °C in a 1 mm pathlength cuvette as described above. Three independent runs were measured and averaged, and blank spectra of the corresponding buffer were subtracted for each protein. Analysis of the CD spectra was performed as previously described by Fasman and coworkers [
31,
32].
Thermal denaturation experiments were performed by heating the test proteins from 25 to 95 °C with a ramping rate of 1 °C/min and collecting the 218 nm molar ellipticity using the same CD instrument as above. The protein concentration was 0.5 mg/mL in 50 mM NaH
2PO
4/Na
2HPO
4 (pH 7.3), 150 mM NaCl, and 50 μM TCEP. An equilibration time of 10 s at each temperature was maintained. Thermal denaturation curves were analyzed using GraphPad Prism 7 software according to the method of C.N. Pace [
33]. The enthalpy change, ΔH, was calculated using the Van’t Hoff equation: d(lnK)/d(1/T) = −ΔH/R. The same protein concentrations, buffer strengths, and parameters maintained in recording the far-UV CD at 25 °C were used for the far-UV CD spectra at 61 °C. Samples were pre-equilibrated at 61 °C for 200 s prior to recording the spectra and were recorded at different time points ranging from 200 to 900 s as indicated.
4.7. Fluorescence Spectroscopic Measurements
For measuring tertiary structure using tryptophan fluorescence emission, 0.05 mg/mL protein samples in 52.5 mM NaH2PO4/Na2HPO4, 150 mM NaCl, 1 mM EDTA, 5 mM DT, and 50 μM TCEP were excited with 295 nm wavelength light and emissions were collected between 300 and 400 nm using a FluouroMax-4C Spectrofluorometer (Horiba Scientific, Edison, NJ, USA).
For determining the tertiary structure of wild-type and mutant proteins under GuHCl-induced stress, 0.1 mg/mL protein samples in 50 mM NaH2PO4/Na2HPO4 (pH 7.3), 150 mM NaCl, 1 mM EDTA, 5 mM DTT, and 50 μM TCEP were incubated with varying concentrations of GuHCl (0–5 M) for 12–14 h at room temperature and tryptophan emission was collected between 300 and 400 nm at 25 °C by exciting the samples at 295 nm. Three independent runs were measured and averaged, and the values for the corresponding buffer were subtracted as blanks for each protein.
Surface hydrophobicity and aggregation propensities were assessed using bis-ANS [
14] and Nile Red [
15]. For bis-ANS experiments, protein samples (0.33 mg/mL) in 66.7 mM NaH
2PO
4/Na
2HPO
4 (pH 7.3), 150 mM NaCl, 1 mM EDTA, 5 mM DTT, and 50 μM TCEP were incubated with a final concentration of dye ranging from 0.58 to 74.31 μM. For Nile Red experiments 0.1 mg/mL protein samples in 55 mM NaH
2PO
4/Na
2HPO
4 (pH 7.3), 150 mM NaCl, 1 mM EDTA, 5 mM DTT, and 50 μM TCEP were incubated with between 0.61 and 19.63 µM of the dye in the dark at room temperature for 30 min. Spectra for bis-ANS were recorded between 400 and 600 nm by exciting the samples at 390 nm and for Nile Red samples were recorded between 570 and 700 nm after excitation at 540 nm. Fluorescence intensities at 490 nm (bis-ANS) and 650 nm (Nile Red) were plotted against dye concentration.
For GuHCl unfolding experiments [
34], samples with a final concentration of 0.1 mg/mL protein in 55 mM Tris-Cl (pH 7.3), 165 mM NaCl, 5 mM DTT, 1 mM EDTA, and 55 μM TCEP were incubated at room temperature for 14 h with stepwise increments of 1.0 M GuHCl until a final concentration of 4.8 M, sufficient to fully denature the proteins, was reached. Spectra were recorded at each step after equilibration. For refolding experiments, 1.0 mg of wild-type or mutant proteins were incubated in a final concentration of 5 M GuHCl, 64 mM Tris-Cl (pH 7.3), 191 mM NaCl, 5 mM DTT, 1 mM EDTA, and 64 μM TCEP at room temperature for 7 h, then diluted into a series of tubes containing refolding buffer (56 mM Tris-Cl (pH 7.3), 170 mM NaCl, 5 mM DTT, 1 mM EDTA, and 56 μM TCEP) until final concentrations of 0.1–4.8 M GuHCl (in 0.1 M GuHCl decrements) were reached and allowed to equilibrate at room temperature for 12–15 h. The same spectral conditions were used as for recording the tertiary structure with tryptophan emission, except for the excitation and emission slits.
GuHCl unfolding and refolding curves were analyzed by plotting the concentration of GuHCl for each sample versus the ratio of fluorescence intensities at 355 nm (maximum for the unfolded protein) and 330 nm (maximum for the native protein). The ratio of fluorescence intensities at these wavelengths was chosen for the analysis to allow simultaneous monitoring of changes in the native and unfolded maxima. Equilibria unfolding/refolding data were analyzed using GraphPad Prism 7 software, and ΔG° and m values were calculated using the method of C.N. Pace [
33].
For determining thermal aggregation, a final concentration of 2.5 µM protein in 50 mM Tris-Cl (pH 7.3) and 150 mM NaCl was heated at 60 °C for 1200 s, and static (Rayleigh) light scattering at 600 nm was monitored using a spectrofluorometer for 1200 s with a 1 cm path length quartz cuvette. An equilibration time of 2 min at 60 °C prior recording the scattering was maintained. To check the effect of DTT and TCEP on thermal aggregation 2.5 µM protein in 50 mM Tris-Cl (pH 7.3), 150 mM NaCl with varying concentrations of TCEP and DTT was heated at 60 °C for 1200 s and light scattering at 600 nm was monitored.
All fluorescence experiments were carried out at 25 °C with 2.5 nm excitation and emission slits while maintaining 1 s integration time at each point. The excitation and emission slits used in the GuHCl unfolding experiments were 5 nm, and 10 nm excitation and emission slits were maintained in the refolding experiments.
4.8. Differential Scanning Fluorimetry Measurements
Thermal denaturation was also probed with tryptophan emission using a differential scanning fluorimeter (Prometheus NT.48, Nano Temper Technologies GmbH, Munich, Germany). The same protein concentrations and buffer strengths were maintained as for the CD thermal unfolding experiments. Samples were heated from 25 to 95 °C while being excited at 285 nm, and the 350/330 nm emission ratio was collected as a function of temperature.
4.9. Dynamic Light Scattering (DLS) Measurements
Wild-type and mutant protein particle sizes and distributions were assessed with dynamic light scattering using a DynaPro NanoStar dynamic light scattering instrument (Wyatt Technology, Santa Barbara, CA, USA). Protein solutions of 25 µM in 50 mM Tris-Cl (pH 7.3) and 150 mM NaCl buffer were illuminated using a 658 nm laser, and light scattered at a 90° angle was measured from 30 to 60 °C in 10 °C increments. A minimum of 20 acquisitions at each temperature step were recorded. Each protein solution was analyzed twice, and the average of all acquisitions is presented. A 1 °C/min ramping rate and an equilibration time of 2 min at each temperature was maintained before collecting acquisitions. To obtain the hydrodynamic radii the intensity autocorrelation functions were fitted by a regularization algorithm using Dynamics software version 7.10 (Wyatt Technology, Santa Barbara, CA, USA).
4.10. Analytical Size Exclusion Chromatography
Protein at a final concentration of 37.5 µM in 50 mM Tris-Cl (pH 7.3) and 150 mM NaCl was injected into a 24 mL ENrich SEC650 (Bio-Rad #780-1650) column equilibrated with 2 column volumes of 50 mM Tris-Cl (pH 7.3) and 100 mM NaCl. A flow rate of 0.5 mL/min was maintained, and the sample volume injected was not more than 1% of the column volume (240 μL). αA-crystallin was assessed by mixing αA-crystallin and F10_Y11delinsLN mutant γS-crystallin at an 8:1 molar ratio at room temperature and stress conditions before chromatography.
4.11. Molecular Modeling
The crystal structure of wild-type human γS-crystallin was taken from 7N36 in the RCSB PDB (
https://www.rcsb.org, accessed on 30 May 2023) [
35]. The modeling of the F10_Y11delinsLN mutantγS-crystallin protein was carried out using the Expasy Swiss-Model Workspace with the fully automated option using the 7N36.1.A structure as a template and default parameters (SWISS-MODEL Workspace—SIB Swiss Institute of Bioinformatics|Expasy). This provided 99% coverage (amino acids 5–178), 98.87% sequence identity and a GMQE of 0.88. The wild-type and mutant protein structures were overlaid using the Protein Analysis and Modeling module of DNASTAR version 17.1.1.120. Backbones structures and distances were viewed using the molecular graphics program RasMol version 2.7.4.2.