Development of Engineered-U1 snRNA Therapies: Current Status
Abstract
1. Introduction
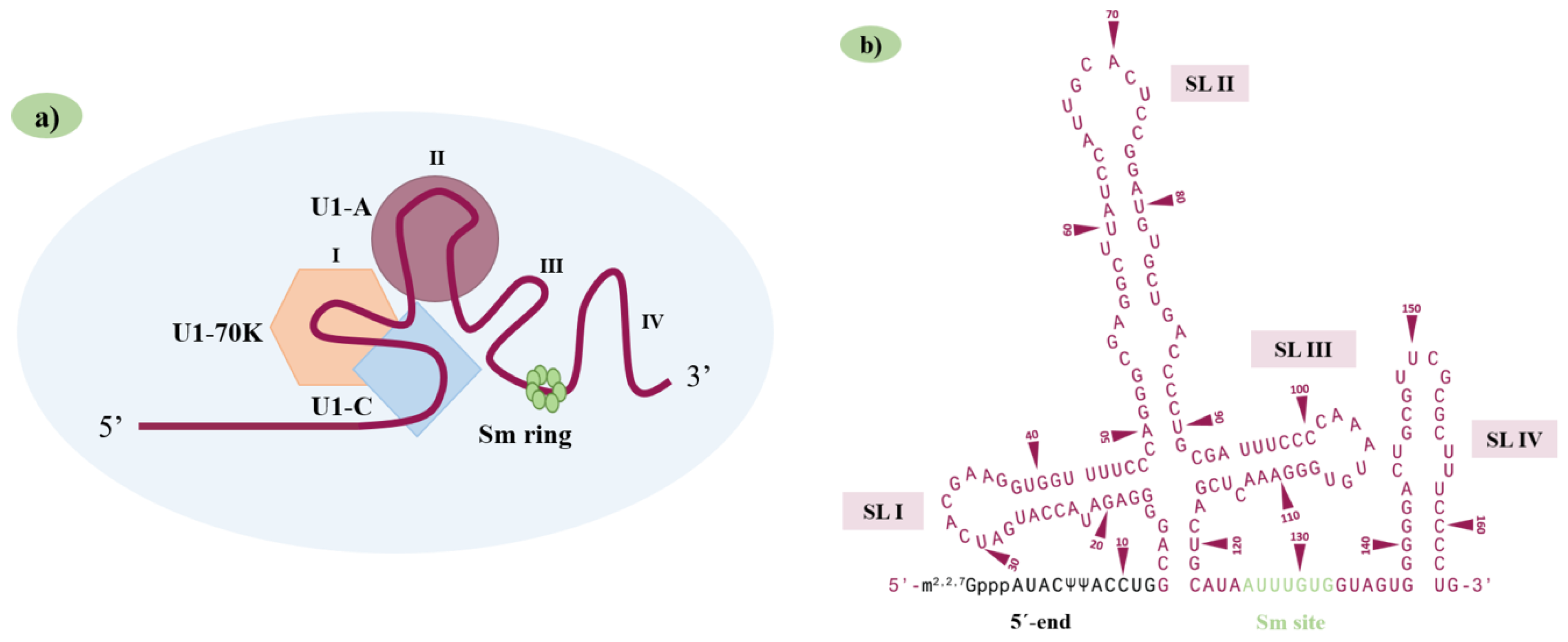
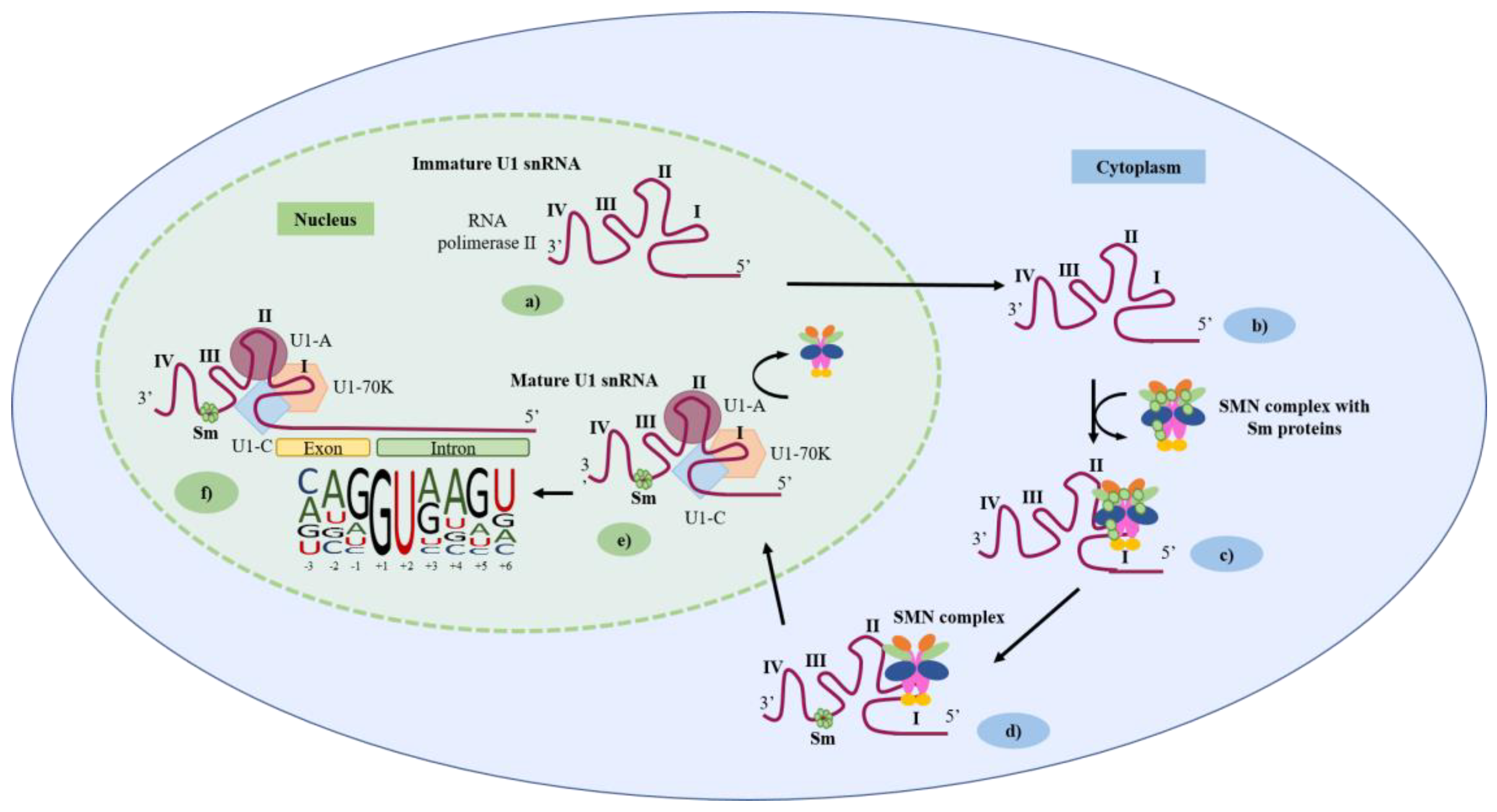
2. The U1 snRNA Structure/U1 snRNP Complex and 5′ Splice-Site Features
3. The U1 snRNA Molecule as a Promising Therapeutic Tool for Splicing Correction
3.1. First Generation of Synthetic U1 snRNAs: The Modified U1 snRNAs
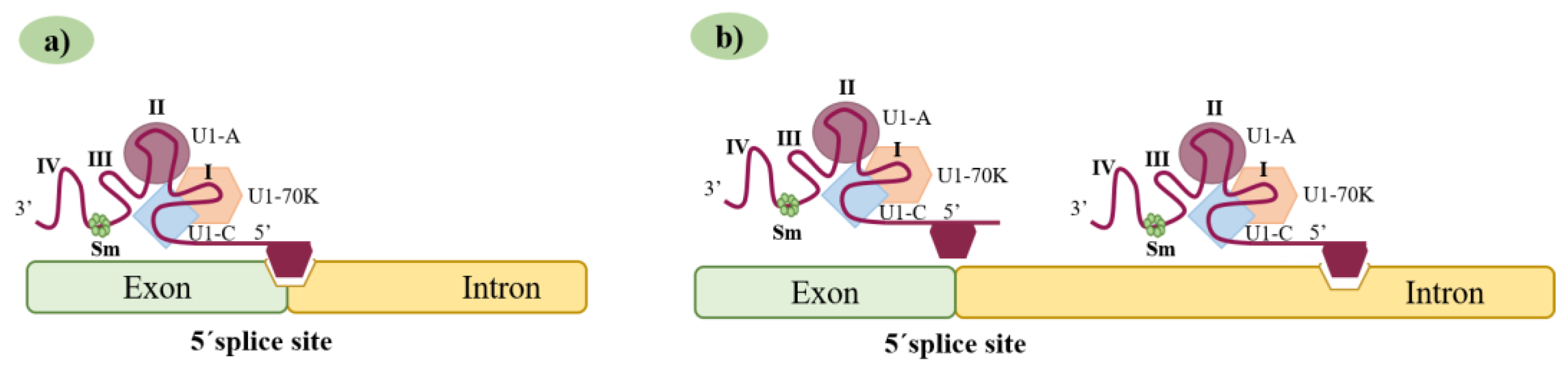
3.2. A New Generation of Synthetic U1 snRNAs: The Exon-Specific U1 snRNAs
3.3. When Modified U1 snRNAs Meet Other Therapeutic Molecules: Combined Therapeutic Strategies
4. Factors to Consider in the Effectiveness and Use of Engineered U1 snRNA Vectors
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Singh, R.N.; Singh, N.N. A Novel Role of U1 SnRNP: Splice Site Selection from a Distance. Biochim. Biophys. Acta Gene Regul. Mech. 2019, 1862, 634–642. [Google Scholar] [CrossRef]
- Shi, Y. The Spliceosome: A Protein-Directed Metalloribozyme. J. Mol. Biol. 2017, 429, 2640–2653. [Google Scholar] [CrossRef]
- Schmidt, V.; Kirschner, K.M. Alternative Pre-MRNA Splicing. Acta Physiol. 2018, 222, e13053. [Google Scholar] [CrossRef] [PubMed]
- Yan, C.; Wan, R.; Shi, Y. Molecular Mechanisms of Pre-MRNA Splicing through Structural Biology of the Spliceosome. Cold Spring Harb. Perspect. Biol. 2019, 11, a032409. [Google Scholar] [CrossRef] [PubMed]
- Wan, R.; Bai, R.; Shi, Y. Molecular Choreography of Pre-MRNA Splicing by the Spliceosome. Curr. Opin. Struct. Biol. 2019, 59, 124–133. [Google Scholar] [CrossRef] [PubMed]
- Suñé-Pou, M.; Limeres, M.J.; Moreno-Castro, C.; Hernández-Munain, C.; Suñé-Negre, J.M.; Cuestas, M.L.; Suñé, C. Innovative Therapeutic and Delivery Approaches Using Nanotechnology to Correct Splicing Defects Underlying Disease. Front. Genet. 2020, 11, 731. [Google Scholar] [CrossRef]
- Baralle, D.; Buratti, E. RNA Splicing in Human Disease and in the Clinic. Clin. Sci. 2017, 131, 355–368. [Google Scholar] [CrossRef]
- Anna, A.; Monika, G. Splicing Mutations in Human Genetic Disorders: Examples, Detection, and Confirmation. J. Appl. Genet. 2018, 59, 253–268. [Google Scholar] [CrossRef]
- Montes, M.; Sanford, B.L.; Comiskey, D.F.; Chandler, D.S. RNA Splicing and Disease: Animal Models to Therapies. Trends Genet. 2019, 35, 68–87. [Google Scholar] [CrossRef]
- van der Woerd, W.L.; Mulder, J.; Pagani, F.; Beuers, U.; Houwen, R.H.J.; van de Graaf, S.F.J. Analysis of Aberrant Pre-Messenger RNA Splicing Resulting from Mutations in ATP8B1 and Efficient in Vitro Rescue by Adapted U1 Small Nuclear RNA. Hepatology 2015, 61, 1382–1391. [Google Scholar] [CrossRef]
- Matos, L.; Canals, I.; Dridi, L.; Choi, Y.; Prata, M.J.; Jordan, P.; Desviat, L.R.; Pérez, B.; Pshezhetsky, A.V.; Grinberg, D.; et al. Therapeutic Strategies Based on Modified U1 Snrnas and Chaperones for Sanfilippo c Splicing Mutations. Orphanet J. Rare Dis. 2014, 9, 180. [Google Scholar] [CrossRef]
- Tajnik, M.; Rogalska, M.E.; Bussani, E.; Barbon, E.; Balestra, D.; Pinotti, M.; Pagani, F. Molecular Basis and Therapeutic Strategies to Rescue Factor IX Variants That Affect Splicing and Protein Function. PLoS Genet. 2016, 12, e1006082. [Google Scholar] [CrossRef]
- Balestra, D.; Scalet, D.; Ferrarese, M.; Lombardi, S.; Ziliotto, N.; Croes, C.C.; Petersen, N.; Bosma, P.; Riccardi, F.; Pagani, F.; et al. A Compensatory U1snRNA Partially Rescues FAH Splicing and Protein Expression in a Splicing-Defective Mouse Model of Tyrosinemia Type I. Int. J. Mol. Sci. 2020, 21, 2136. [Google Scholar] [CrossRef] [PubMed]
- Buratti, E.; Baralle, D. Novel Roles of U1 SnRNP in Alternative Splicing Regulation. RNA Biol. 2010, 7, 412–419. [Google Scholar] [CrossRef] [PubMed]
- Matos, L.; Santos, J.I.; Coutinho, M.F.; Alves, S. How to Design U1 SnRNA Molecules for Splicing Rescue. In Methods in Molecular Biology; Humana Press Inc.: New York, NY, USA, 2022; Volume 2434, pp. 89–102. [Google Scholar]
- Lee, N.-C.; Lee, Y.-M.; Chen, P.-W.; Byrne, B.J.; Hwu, W.-L. Mutation-Adapted U1 SnRNA Corrects a Splicing Error of the Dopa Decarboxylase Gene. Hum. Mol. Genet. 2016, 25, ddw323. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Scalet, D.; Maestri, I.; Branchini, A.; Bernardi, F.; Pinotti, M.; Balestra, D. Disease-Causing Variants of the Conserved +2T of 5′ Splice Sites Can Be Rescued by Engineered U1snRNAs. Hum. Mutat. 2019, 40, 48–52. [Google Scholar] [CrossRef]
- Coutinho, M.F.; Matos, L.; Santos, J.I.; Alves, S. RNA Therapeutics: How Far Have We Gone? Cell 2019, 46, 133–177. [Google Scholar]
- Naylor, S.L.; Zabel, B.U.; Manser, T.; Gesteland, R.; Sakaguchi, A.Y. Localization of Human U1 Small Nuclear RNA Genes to Band P36.3 of Chromosome 1 by in Situ Hybridization. Somat. Cell Mol. Genet. 1984, 10, 307–313. [Google Scholar] [CrossRef]
- Lindgren, V.; Bernstein, L.B.; Weiner, A.M.; Francke1, U. Human Ul Small Nuclear RNA Pseudogenes Do Not Map to the Site of the Ul Genes in Lp36 but Are Clustered in Lql2–q22. Mol. Cell. Biol. 1985, 5, 2172–2180. [Google Scholar]
- Lund, E.; Dahlberg, J.E. True Genes for Human U1 Small Nuclear RNA. Copy Number, Polymorphism, and Methylation. J. Biol. Chem. 1984, 259, 2013–2021. [Google Scholar] [CrossRef]
- Lund, E.; Bostock, C.J.; Dahlberg, J.E. The Transcription of Xenopus Laevis Embryonic U1 SnRNA Genes Changes When Oocytes Mature into Eggs. Genes Dev. 1987, 1, 47–56. [Google Scholar] [CrossRef]
- O’Reilly, D.; Dienstbier, M.; Cowley, S.A.; Vazquez, P.; Drozdz, M.; Taylor, S.; James, W.S.; Murphy, S. Differentially Expressed, Variant U1 SnRNAs Regulate Gene Expression in Human Cells. Genome Res. 2013, 23, 281–291. [Google Scholar] [CrossRef] [PubMed]
- Vazquez-Arango, P.; Vowles, J.; Browne, C.; Hartfield, E.; Fernandes, H.J.R.; Mandefro, B.; Sareen, D.; James, W.; Wade-Martins, R.; Cowley, S.A.; et al. Variant U1 SnRNAs Are Implicated in Human Pluripotent Stem Cell Maintenance and Neuromuscular Disease. Nucleic Acids Res. 2016, 44, 10960–10973. [Google Scholar] [CrossRef]
- Guiro, J.; O’Reilly, D. Insights into the U1 Small Nuclear Ribonucleoprotein Complex Superfamily. Wiley Interdiscip. Rev. RNA 2015, 6, 79–92. [Google Scholar] [CrossRef]
- Pomeranz Krummel, D.A.; Oubridge, C.; Leung, A.K.W.; Li, J.; Nagai, K. Crystal Structure of Human Spliceosomal U1 SnRNP at 5.5 Resolution. Nature 2009, 458, 475–480. [Google Scholar] [CrossRef] [PubMed]
- Blazquez, L.; Fortes, P. U1 SnRNP Control of 3′-End Processing and the Therapeutic Application of U1 Inhibition Combined with RNA Interference. Curr. Mol. Med. 2013, 13, 1203–1216. [Google Scholar] [CrossRef] [PubMed]
- Kondo, Y.; Oubridge, C.; van Roon, A.M.M.; Nagai, K. Crystal Structure of Human U1 SnRNP, a Small Nuclear Ribonucleoprotein Particle, Reveals the Mechanism of 5’ Splice Site Recognition. Elife 2015, 4, e04986. [Google Scholar] [CrossRef]
- Roca, X.; Krainer, A.R.; Eperon, I.C. Pick One, but Be Quick: 5′ Splice Sites and the Problems of Too Many Choices. Genes Dev. 2013, 27, 129–144. [Google Scholar] [CrossRef] [PubMed]
- Freund, M.; Hicks, M.J.; Konermann, C.; Otte, M.; Hertel, K.J.; Schaal, H. Extended Base Pair Complementarity between U1 SnRNA and the 5′ Splice Site Does Not Inhibit Splicing in Higher Eukaryotes, but Rather Increases 5′ Splice Site Recognition. Nucleic Acids Res. 2005, 33, 5112–5119. [Google Scholar] [CrossRef]
- Tan, J.; Ho, J.X.J.; Zhong, Z.; Luo, S.; Chen, G.; Roca, X. Noncanonical Registers and Base Pairs in Human 5′ Splice-Site Selection. Nucleic Acids Res. 2016, 44, 3908–3921. [Google Scholar] [CrossRef][Green Version]
- Carmel, I.; Tal, S.; Vig, I.; Ast, G. Comparative Analysis Detects Dependencies among the 5′ Splice-Site Positions. RNA 2004, 10, 828–840. [Google Scholar] [CrossRef] [PubMed]
- Malard, F.; Mackereth, C.D.; Campagne, S. Principles and Correction of 5’-Splice Site Selection. RNA Biol. 2022, 19, 943–960. [Google Scholar] [CrossRef] [PubMed]
- Fredericks, A.; Cygan, K.; Brown, B.; Fairbrother, W. RNA-Binding Proteins: Splicing Factors and Disease. Biomolecules 2015, 5, 893–909. [Google Scholar] [CrossRef] [PubMed]
- Hwu, W.L.; Lee, Y.M.; Lee, N.C. Gene Therapy with Modified U1 Small Nuclear RNA. Expert. Rev. Endocrinol. Metab. 2017, 12, 171–175. [Google Scholar] [CrossRef] [PubMed]
- Baralle, M. Identification of a Mutation That Perturbs NF1 Agene Splicing Using Genomic DNA Samples and a Minigene Assay. J. Med. Genet. 2003, 40, 220–222. [Google Scholar] [CrossRef]
- Pinotti, M.; Balestra, D.; Rizzotto, L.; Maestri, I.; Pagani, F.; Bernardi, F. Rescue of Coagulation Factor VII Function by the U1+5A SnRNA. Blood 2009, 113, 6461–6464. [Google Scholar] [CrossRef]
- Pinotti, M.; Rizzotto, L.; Balestra, D.; Lewandowska, M.A.; Cavallari, N.; Marchetti, G.; Bernardi, F.; Pagani, F. U1-SnRNA–Mediated Rescue of MRNA Processing in Severe Factor VII Deficiency. Blood 2008, 111, 2681–2684. [Google Scholar] [CrossRef]
- Balestra, D.; Faella, A.; Margaritis, P.; Cavallari, N.; Pagani, F.; Bernardi, F.; Arruda, V.R.; Pinotti, M. An Engineered U1 Small Nuclear RNA Rescues Splicing- Defective Coagulation F7 Gene Expression in Mice. J. Thromb. Haemost. 2014, 12, 177–185. [Google Scholar] [CrossRef]
- Tanner, G.; Glaus, E.; Barthelmes, D.; Ader, M.; Fleischhauer, J.; Pagani, F.; Berger, W.; Neidhardt, J. Therapeutic Strategy to Rescue Mutation-Induced Exon Skipping in Rhodopsin by Adaptation of U1 SnRNA. Hum. Mutat. 2009, 30, 255–263. [Google Scholar] [CrossRef]
- Glaus, E.; Schmid, F.; Da Costa, R.; Berger, W.; Neidhardt, J. Gene Therapeutic Approach Using Mutation-Adapted U1 SnRNA to Correct a RPGR Splice Defect in Patient-Derived Cells. Mol. Ther. 2011, 19, 936–941. [Google Scholar] [CrossRef]
- Schmid, F.; Glaus, E.; Barthelmes, D.; Fliegauf, M.; Gaspar, H.; Nürnberg, G.; Nürnberg, P.; Omran, H.; Berger, W.; Neidhardt, J. U1 SnRNA-Mediated Gene Therapeutic Correction of Splice Defects Caused by an Exceptionally Mild BBS Mutation. Hum. Mutat. 2011, 32, 815–824. [Google Scholar] [CrossRef]
- Sánchez-Alcudia, R.; Pérez, B.; Pérez-Cerdá, C.; Ugarte, M.; Desviat, L.R. Overexpression of Adapted U1snRNA in Patients’ Cells to Correct a 5′ Splice Site Mutation in Propionic Acidemia. Mol. Genet. Metab. 2011, 102, 134–138. [Google Scholar] [CrossRef] [PubMed]
- Scalet, D.; Sacchetto, C.; Bernardi, F.; Pinotti, M.; van de Graaf, S.F.J.; Balestra, D. The somatic FAH C. 1061C>A change counteracts the frequent FAH c. 1062+ 5G>A mutation and permits U1snRNA-based splicing correction. J. Hum. Genet. 2018, 63, 683–686. [Google Scholar] [CrossRef] [PubMed]
- Hartmann, L.; Neveling, K.; Borkens, S.; Schneider, H.; Freund, M.; Grassman, E.; Theiss, S.; Wawer, A.; Burdach, S.; Auerbach, A.D.; et al. Correct MRNA Processing at a Mutant TT Splice Donor in FANCC Ameliorates the Clinical Phenotype in Patients and Is Enhanced by Delivery of Suppressor U1 SnRNAs. Am. J. Hum. Genet. 2010, 87, 480–493. [Google Scholar] [CrossRef]
- Peretto, L.; Tonetto, E.; Maestri, I.; Bezzerri, V.; Valli, R.; Cipolli, M.; Pinotti, M.; Balestra, D. Counteracting the Common Shwachman–Diamond Syndrome-Causing SBDS c.258+2T>C Mutation by RNA Therapeutics and Base/Prime Editing. Int. J. Mol. Sci. 2023, 24, 4024. [Google Scholar] [CrossRef] [PubMed]
- Balestra, D.; Giorgio, D.; Bizzotto, M.; Fazzari, M.; Ben Zeev, B.; Pinotti, M.; Landsberger, N.; Frasca, A. Splicing Mutations Impairing CDKL5 Expression and Activity Can Be Efficiently Rescued by U1snRNA-Based Therapy. Int. J. Mol. Sci. 2019, 20, 4130. [Google Scholar] [CrossRef]
- Fernandez Alanis, E.; Pinotti, M.; Dal Mas, A.; Balestra, D.; Cavallari, N.; Rogalska, M.E.; Bernardi, F.; Pagani, F. An Exon-Specific U1 Small Nuclear RNA (SnRNA) Strategy to Correct Splicing Defects. Hum. Mol. Genet. 2012, 21, 2389–2398. [Google Scholar] [CrossRef]
- Barbon, E.; Ferrarese, M.; van Wittenberghe, L.; Sanatine, P.; Ronzitti, G.; Collaud, F.; Colella, P.; Pinotti, M.; Mingozzi, F. Transposon-Mediated Generation of Cellular and Mouse Models of Splicing Mutations to Assess the Efficacy of SnRNA-Based Therapeutics. Mol. Ther. Nucleic Acids 2016, 5, e392. [Google Scholar] [CrossRef]
- Donegà, S.; Rogalska, M.E.; Pianigiani, G.; Igreja, S.; Amaral, M.D.; Pagani, F. Rescue of Common Exon-Skipping Mutations in Cystic Fibrosis with Modified U1 SnRNAs. Hum. Mutat. 2020, 41, 2143–2154. [Google Scholar] [CrossRef] [PubMed]
- Rogalska, M.E.; Tajnik, M.; Licastro, D.; Bussani, E.; Camparini, L.; Mattioli, C.; Pagani, F. Therapeutic Activity of Modified U1 Core Spliceosomal Particles. Nat. Commun. 2016, 7, 11168. [Google Scholar] [CrossRef]
- Donadon, I.; Bussani, E.; Riccardi, F.; Licastro, D.; Romano, G.; Pianigiani, G.; Pinotti, M.; Konstantinova, P.; Evers, M.; Lin, S.; et al. Rescue of Spinal Muscular Atrophy Mouse Models with AAV9-Exon-Specific U1 SnRNA. Nucleic Acids Res. 2019, 47, 7618–7632. [Google Scholar] [CrossRef] [PubMed]
- Dal Mas, A.; Rogalska, M.E.; Bussani, E.; Pagani, F. Improvement of SMN2 Pre-MRNA Processing Mediated by Exon-Specific U1 Small Nuclear RNA. Am. J. Hum. Genet. 2015, 96, 93–103. [Google Scholar] [CrossRef] [PubMed]
- Balestra, D.; Maestri, I.; Branchini, A.; Ferrarese, M.; Bernardi, F.; Pinotti, M. An Altered Splicing Registry Explains the Differential ExSpeU1-Mediated Rescue of Splicing Mutations Causing Haemophilia A. Front. Genet. 2019, 10, 974. [Google Scholar] [CrossRef]
- Lombardi, S.; Leo, G.; Merlin, S.; Follenzi, A.; McVey, J.H.; Maestri, I.; Bernardi, F.; Pinotti, M.; Balestra, D. Dissection of Pleiotropic Effects of Variants in and Adjacent to F8 Exon 19 and Rescue of MRNA Splicing and Protein Function. Am. J. Hum. Genet. 2021, 108, 1512–1525. [Google Scholar] [CrossRef]
- Dal Mas, A.; Fortugno, P.; Donadon, I.; Levati, L.; Castiglia, D.; Pagani, F. Exon-Specific U1s Correct SPINK 5 Exon 11 Skipping Caused by a Synonymous Substitution That Affects a Bifunctional Splicing Regulatory Element. Hum. Mutat. 2015, 36, 504–512. [Google Scholar] [CrossRef] [PubMed]
- Mattioli, C.; Pianigiani, G.; De Rocco, D.; Bianco, A.M.R.; Cappelli, E.; Savoia, A.; Pagani, F. Unusual Splice Site Mutations Disrupt FANCA Exon 8 Definition. Biochim. Biophys. Acta (BBA)-Mol. Basis Dis. 2014, 1842, 1052–1058. [Google Scholar] [CrossRef]
- Donadon, I.; Pinotti, M.; Rajkowska, K.; Pianigiani, G.; Barbon, E.; Morini, E.; Motaln, H.; Rogelj, B.; Mingozzi, F.; Slaugenhaupt, S.A.; et al. Exon-Specific U1 SnRNAs Improve ELP1 Exon 20 Definition and Rescue ELP1 Protein Expression in a Familial Dysautonomia Mouse Model. Hum. Mol. Genet. 2018, 27, 2466–2476. [Google Scholar] [CrossRef] [PubMed]
- Romano, G.; Riccardi, F.; Bussani, E.; Vodret, S.; Licastro, D.; Ragone, I.; Ronzitti, G.; Morini, E.; Slaugenhaupt, S.A.; Pagani, F. Rescue of a Familial Dysautonomia Mouse Model by AAV9-Exon-Specific U1 SnRNA. Am. J. Hum. Genet. 2022, 109, 1534–1548. [Google Scholar] [CrossRef]
- Jüschke, C.; Klopstock, T.; Catarino, C.B.; Owczarek-Lipska, M.; Wissinger, B.; Neidhardt, J. Autosomal Dominant Optic Atrophy: A Novel Treatment for OPA1 Splice Defects Using U1 SnRNA Adaption. Mol. Ther. Nucleic Acids 2021, 26, 1186–1197. [Google Scholar] [CrossRef]
- Martínez-Pizarro, A.; Dembic, M.; Pérez, B.; Andresen, B.S.; Desviat, L.R. Intronic PAH Gene Mutations Cause a Splicing Defect by a Novel Mechanism Involving U1snRNP Binding Downstream of the 5’ Splice Site. PLoS Genet. 2018, 14, e1007360. [Google Scholar] [CrossRef]
- Balestra, D.; Ferrarese, M.; Lombardi, S.; Ziliotto, N.; Branchini, A.; Petersen, N.; Bosma, P.; Pinotti, M.; van de Graaf, S.F.J. An Exon-Specific Small Nuclear U1 Rna (Exspeu1) Improves Hepatic Otc Expression in a Splicing-Defective Spf/Ash Mouse Model of Ornithine Transcarbamylase Deficiency. Int. J. Mol. Sci. 2020, 21, 8735. [Google Scholar] [CrossRef] [PubMed]
- Sacchetto, C.; Peretto, L.; Baralle, F.; Maestri, I.; Tassi, F.; Bernardi, F.; van de Graaf, S.F.J.; Pagani, F.; Pinotti, M.; Balestra, D. OTC Intron 4 Variations Mediate Pathogenic Splicing Patterns Caused by the c.386G>A Mutation in Humans and Spfash Mice, and Govern Susceptibility to RNA-Based Therapies. Mol. Med. 2021, 27, 157. [Google Scholar] [CrossRef]
- Breuel, S.; Vorm, M.; Bräuer, A.U.; Owczarek-Lipska, M.; Neidhardt, J. Combining Engineered U1 SnRNA and Antisense Oligonucleotides to Improve the Treatment of a BBS1 Splice Site Mutation. Mol. Ther. Nucleic Acids 2019, 18, 123–130. [Google Scholar] [CrossRef] [PubMed]
- Schmid, F.; Hiller, T.; Korner, G.; Glaus, E.; Berger, W.; Neidhardt, J. A Gene Therapeutic Approach to Correct Splice Defects with Modified U1 and U6 SnRNPs. Hum. Gene Ther. 2013, 24, 97–104. [Google Scholar] [CrossRef]
- Swirski, S.; May, O.; Ahlers, M.; Wissinger, B.; Greschner, M.; Jüschke, C.; Neidhardt, J. In Vivo Efficacy and Safety Evaluations of Therapeutic Splicing Correction Using U1 SnRNA in the Mouse Retina. Cells 2023, 12, 955. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, Y.; Weiner, A.M. A Compensatory Base Change in U1 SnRNA Suppresses a 5′ Splice Site Mutation. Cell 1986, 46, 827–835. [Google Scholar] [CrossRef]
- Boocock, G.R.B.; Morrison, J.A.; Popovic, M.; Richards, N.; Ellis, L.; Durie, P.R.; Rommens, J.M. Mutations in SBDS Are Associated with Shwachman-Diamond Syndrome. Nat. Genet. 2003, 33, 97–101. [Google Scholar] [CrossRef]
- Bezzerri, V.; Cipolli, M. Shwachman-Diamond Syndrome: Molecular Mechanisms and Current Perspectives. Mol. Diagn. Ther. 2019, 23, 281–290. [Google Scholar] [CrossRef]
- Blázquez, L.; Fortes, P. U1 Interference (U1i) for Antiviral Approaches. In Advances in Experimental Medicine and Biology; Springer: New York, NY, USA, 2015; Volume 848, pp. 51–69. [Google Scholar]
- Imbert, M.; Dias-Florencio, G.; Goyenvalle, A. Viral Vector-Mediated Antisense Therapy for Genetic Diseases. Genes 2017, 8, 51. [Google Scholar] [CrossRef]
- Balestra, D.; Scalet, D.; Pagani, F.; Rogalska, M.E.; Mari, R.; Bernardi, F.; Pinotti, M. An Exon-Specific U1snRNA Induces a Robust Factor IX Activity in Mice Expressing Multiple Human FIX Splicing Mutants. Mol. Ther. Nucleic Acids 2016, 5, e370. [Google Scholar] [CrossRef][Green Version]
- Manfredsson, F.P.; Rising, A.C.; Mandel, R.J. AAV9: A Potential Blood-Brain Barrier Buster. Mol. Ther. 2009, 17, 403–405. [Google Scholar] [CrossRef]
- Kido, J.; Sugawara, K.; Nakamura, K. Gene Therapy for Lysosomal Storage Diseases: Current Clinical Trial Prospects. Front. Genet. 2023, 14, 1064924. [Google Scholar] [CrossRef] [PubMed]
- Parenti, G.; Pignata, C.; Vajro, P.; Salerno, M. New Strategies for the Treatment of Lysosomal Storage Diseases (Review). Int. J. Mol. Med. 2013, 31, 11–20. [Google Scholar] [CrossRef] [PubMed]
- De Angelis, F.G.; Sthandier, O.; Berarducci, B.; Toso, S.; Galluzzi, G.; Ricci, E.; Cossu, G.; Bozzoni, I. Chimeric SnRNA Molecules Carrying Antisense Sequences against the Splice Junctions of Exon 51 of the Dystrophin Pre-MRNA Induce Exon Skipping and Restoration of a Dystrophin Synthesis in Δ48–50 DMD Cells. Proc. Natl. Acad. Sci. USA 2002, 99, 9456–9461. [Google Scholar] [CrossRef]
- Hatch, S.T.; Smargon, A.A.; Yeo, G.W. Engineered U1 SnRNAs to Modulate Alternatively Spliced Exons. Methods 2022, 205, 140–148. [Google Scholar] [CrossRef]
- Happi Mbakam, C.; Lamothe, G.; Tremblay, J.P. Therapeutic Strategies for Dystrophin Replacement in Duchenne Muscular Dystrophy. Front. Med. 2022, 9, 859930. [Google Scholar] [CrossRef] [PubMed]
- Denti, M.A.; Rosa, A.; D’Antona, G.; Sthandier, O.; De Angelis, F.G.; Nicoletti, C.; Allocca, M.; Pansarasa, O.; Parente, V.; Musarò, A.; et al. Body-Wide Gene Therapy of Duchenne Muscular Dystrophy in the Mdx Mouse Model. Proc. Natl. Acad. Sci. USA 2006, 103, 3758–3763. [Google Scholar] [CrossRef]
- Denti, M.A.; Rosa, A.; D’Antona, G.; Sthandier, O.; De Angelis, F.G.; Nicoletti, C.; Allocca, M.; Pansarasa, O.; Parente, V.; Musarò, A.; et al. Chimeric Adeno-Associated Virus/Antisense U1 Small Nuclear RNA Effectively Rescues Dystrophin Synthesis and Muscle Function by Local Treatment of Mdx Mice. Hum. Gene Ther. 2006, 17, 565–574. [Google Scholar] [CrossRef] [PubMed]
- Incitti, T.; De Angelis, F.G.; Cazzella, V.; Sthandier, O.; Pinnarò, C.; Legnini, I.; Bozzoni, I. Exon Skipping and Duchenne Muscular Dystrophy Therapy: Selection of the Most Active U1 SnRNA Antisense Able to Induce Dystrophin Exon 51 Skipping. Mol. Ther. 2010, 18, 1675–1682. [Google Scholar] [CrossRef]
- Cazzella, V.; Martone, J.; Pinnarò, C.; Santini, T.; Twayana, S.S.; Sthandier, O.; D’amico, A.; Ricotti, V.; Bertini, E.; Muntoni, F.; et al. Exon 45 Skipping through U1-SnRNA Antisense Molecules Recovers the Dys-NNOS Pathway and Muscle Differentiation in Human DMD Myoblasts. Mol. Ther. 2012, 20, 2134–2142. [Google Scholar] [CrossRef]
- Brun, C.; Suter, D.; Pauli, C.; Dunant, P.; Lochmüller, H.; Burgunder, J.-M.; Schümperli, D.; Weis, J. U7 SnRNAs Induce Correction of Mutated Dystrophin Pre-MRNA by Exon Skipping. Cell. Mol. Life Sci. 2003, 60, 557–566. [Google Scholar] [CrossRef]
- Goyenvalle, A.; Vulin, A.; Fougerousse, F.; Leturcq, F.; Kaplan, J.-C.; Garcia, L.; Danos, O. Rescue of Dystrophic Muscle Through U7 SnRNA-Mediated Exon Skipping. Science 2004, 306, 1796–1799. [Google Scholar] [CrossRef]
- Goyenvalle, A.; Wright, J.; Babbs, A.; Wilkins, V.; Garcia, L.; Davies, K.E. Engineering Multiple U7snRNA Constructs to Induce Single and Multiexon-Skipping for Duchenne Muscular Dystrophy. Mol. Ther. 2012, 20, 1212–1221. [Google Scholar] [CrossRef]
- Goyenvalle, A.; Babbs, A.; van Ommen, G.J.B.; Garcia, L.; Davies, K.E. Enhanced Exon-Skipping Induced by U7 SnRNA Carrying a Splicing Silencer Sequence: Promising Tool for DMD Therapy. Mol. Ther. 2009, 17, 1234–1240. [Google Scholar] [CrossRef] [PubMed]
- Benchaouir, R.; Meregalli, M.; Farini, A.; D’Antona, G.; Belicchi, M.; Goyenvalle, A.; Battistelli, M.; Bresolin, N.; Bottinelli, R.; Garcia, L.; et al. Restoration of Human Dystrophin Following Transplantation of Exon-Skipping-Engineered DMD Patient Stem Cells into Dystrophic Mice. Cell Stem Cell 2007, 1, 646–657. [Google Scholar] [CrossRef] [PubMed]
- Denti, M.A.; Incitti, T.; Sthandier, O.; Nicoletti, C.; De Angelis, F.G.; Rizzuto, E.; Auricchio, A.; Musarò, A.; Bozzoni, I. Long-Term Benefit of Adeno-Associated Virus/Antisense-Mediated Exon Skipping in Dystrophic Mice. Hum. Gene Ther. 2008, 19, 601–608. [Google Scholar] [CrossRef] [PubMed]
- Martone, J.; De Angelis, F.G.; Bozzoni, I. U1 SnRNA as an Effective Vector for Stable Expression of Antisense Molecules and for the Inhibition of the Splicing Reaction. Methods Mol. Biol. 2012, 867, 239–257. [Google Scholar] [CrossRef]
| Genetic Disease | Gene | Type of Study | Splicing Variants * | Reference | |
|---|---|---|---|---|---|
| Modified U1 snRNAs | Neurofibromatosis type 1 | NF1 | in vitro | c.288+5G>C | [36] |
| Coagulation factor VII deficiency | F7 | in vitro | 9726+5G>A (c.805+5G>A) | [37,38] | |
| in vivo | 859+5G>A (c.805+5G>A) | [39] | |||
| Retinitis pigmentosa | RHO | in vitro | c.936G>A | [40] | |
| RPGR | in vitro | c.1245+3A>T | [41] | ||
| Bardet-Biedl syndrome | BBS1 | in vitro | c.479G>A | [42] | |
| Propionyl-CoA carboxylase deficiency | PCCA | in vitro | c.1209+3A>G | [43] | |
| Tyrosinaemia type 1 | FAH | in vitro | c.1062+5G>A | [44] | |
| Fanconi anemia | FANCC | in vitro | c.165+1G>T | [45] | |
| Mucopolysaccharidosis type IIIC | HGSNAT | in vitro | c.234+1G>A; c.633+1G>A; c.1542+4dupA | [11] | |
| Shwachman-Diamond syndrome | SBDS | in vitro | c.258+2T>C | [46] | |
| Aromatic L-Amino Acid Decarboxylase deficiency | AADC | in vitro | IVS6+4A>T | [16] | |
| in vivo | (c.714+4A>T) | ||||
| ATP8B1 deficiency | ATP8B1 | in vitro | c.2932-3C>A; c.2418+5G>A; c.279G>A; c.625_627+5delinsACAGTAAT | [10] | |
| CDKL5-deficiency | CDKL5 | in vitro | c.99+1G>T | [47] | |
| c.99+5G>A | |||||
| c.463+1G>A | |||||
| c.744+1G>C | |||||
| c.2376+5G>A | |||||
| Tyrosinaemia type 1 | Fah | in vivo | c.706G>A | [13] | |
| Exon-Specific U1 snRNAs | Hemophilia B | F9 | in vitro | c.392-9T>G; c.392-8T>G; | [48] |
| c.519A>C; c.519A>G; c.519A>T | |||||
| c.520G>T; c.520+1G>A; c.520+2T>C | |||||
| p.V107V (c.459G>A); | [12] | ||||
| p.R116R (c.484C>A); | |||||
| p.A118V (c.491C>T); | |||||
| p. Q121H (c.501G>T) | |||||
| c.277+1G>A; c.277+1G>T, | [17] | ||||
| c.277+2T>C; c.277+2T>G; | |||||
| c.277+4A>G | |||||
| in vitro | c.519A>C | [49] | |||
| in vivo | |||||
| Cystic fibrosis | CFTR | in vitro | c.1766+3A>G; c.1766+3A>C; c.1766+5G>A; c.1766G>T; c.1766G>A; p.A566T (c.1696G>A); p.Y577Y (c.1731C>T) | [48] | |
| 711+3A>C (c.579+3A>C); | [50] | ||||
| 711+3A>G (c.579+3A>G); | |||||
| 711+5G>A (c.579+5G>A); | |||||
| 1863C>T (c.1731C>T); | |||||
| 1898+3A>G (c.1766+3A>G); | |||||
| 2789+5G>A (c.2657+5G>A); | |||||
| 3120G>A (c.2988G>A); | |||||
| TG13T3, TG13T5; TG12T5 | |||||
| Spinal muscular atrophy | SMN2 | in vitro | c.840C>T | [48] | |
| in vitro | c.840C>T | [51,52,53] | |||
| in vivo | |||||
| ATP8B1 deficiency | ATP8B1 | in vitro | c.625_627+5delinsACAGTAAT | [10] | |
| CDKL5-deficiency | CDKL5 | in vitro | c.99+1G>T | [47] | |
| c.99+5G>A | |||||
| c.463+1G>A | |||||
| c.744+1G>C | |||||
| c.2376+5G>A | |||||
| Hemophilia A | F8 | in vitro | c.602-32A>G; c.602-10T>G; c.669A>G; c.669A>T; c.670G>T; c.670+1G>T; c.670+1G>A; 670+2T>G; c.670+5G>A; c.670+6T>C | [54] | |
| in vitro | p.G2000A (c.5999G>C); | [55] | |||
| p.Y2036Y (c.6108C>T); | |||||
| p.N2038S (c.6113A>G); | |||||
| c.6115+1G>A; c.6115+2T>C; | |||||
| c.6115+3G>T; c.6115+4A>G; | |||||
| c.6115+5G>A; c.6115+6T>A | |||||
| Netherton syndrome | SPINK5 | in vitro | c.891C>T | [56] | |
| Fanconi anemia | FANCA | in vitro | c.790C>T | [57] | |
| Familial dysautonomia | ELP1 | in vitro | c.2204+6T>C | [58,59] | |
| in vivo | |||||
| Autosomal dominant optic atrophy | OPA1 | in vitro | c.1065+5G>A | [60] | |
| Phenylketonuria | PAH | in vitro | c.1199+17G>A; c. 1199+20G>C | [61] | |
| Shwachman-Diamond syndrome | SBDS | in vitro | c.258+2T>C | [46] | |
| Ornithine transcarbamylase deficiency | Otc | in vivo | c.386G>A | [62] | |
| in vitro | |||||
| OTC | in vitro | c.386G>A | [63] | ||
| Combined therapies | Bardet-Biedl syndrome | BBS1 | in vitro | c.479G>A | [64] |
| Exon 5 5’ss (position +5) # | [65] | ||||
| Autosomal dominant optic atrophy | Opa1 | in vivo | c.1065+5G>A | [66] | |
| Hemophilia A | F8 | in vitro | c.6115+5G>A | [55] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gonçalves, M.; Santos, J.I.; Coutinho, M.F.; Matos, L.; Alves, S. Development of Engineered-U1 snRNA Therapies: Current Status. Int. J. Mol. Sci. 2023, 24, 14617. https://doi.org/10.3390/ijms241914617
Gonçalves M, Santos JI, Coutinho MF, Matos L, Alves S. Development of Engineered-U1 snRNA Therapies: Current Status. International Journal of Molecular Sciences. 2023; 24(19):14617. https://doi.org/10.3390/ijms241914617
Chicago/Turabian StyleGonçalves, Mariana, Juliana Inês Santos, Maria Francisca Coutinho, Liliana Matos, and Sandra Alves. 2023. "Development of Engineered-U1 snRNA Therapies: Current Status" International Journal of Molecular Sciences 24, no. 19: 14617. https://doi.org/10.3390/ijms241914617
APA StyleGonçalves, M., Santos, J. I., Coutinho, M. F., Matos, L., & Alves, S. (2023). Development of Engineered-U1 snRNA Therapies: Current Status. International Journal of Molecular Sciences, 24(19), 14617. https://doi.org/10.3390/ijms241914617











