The Pro-Oncogenic Protein IF1 Promotes Proliferation of Anoxic Cancer Cells during Re-Oxygenation
Abstract
:1. Introduction
2. Results
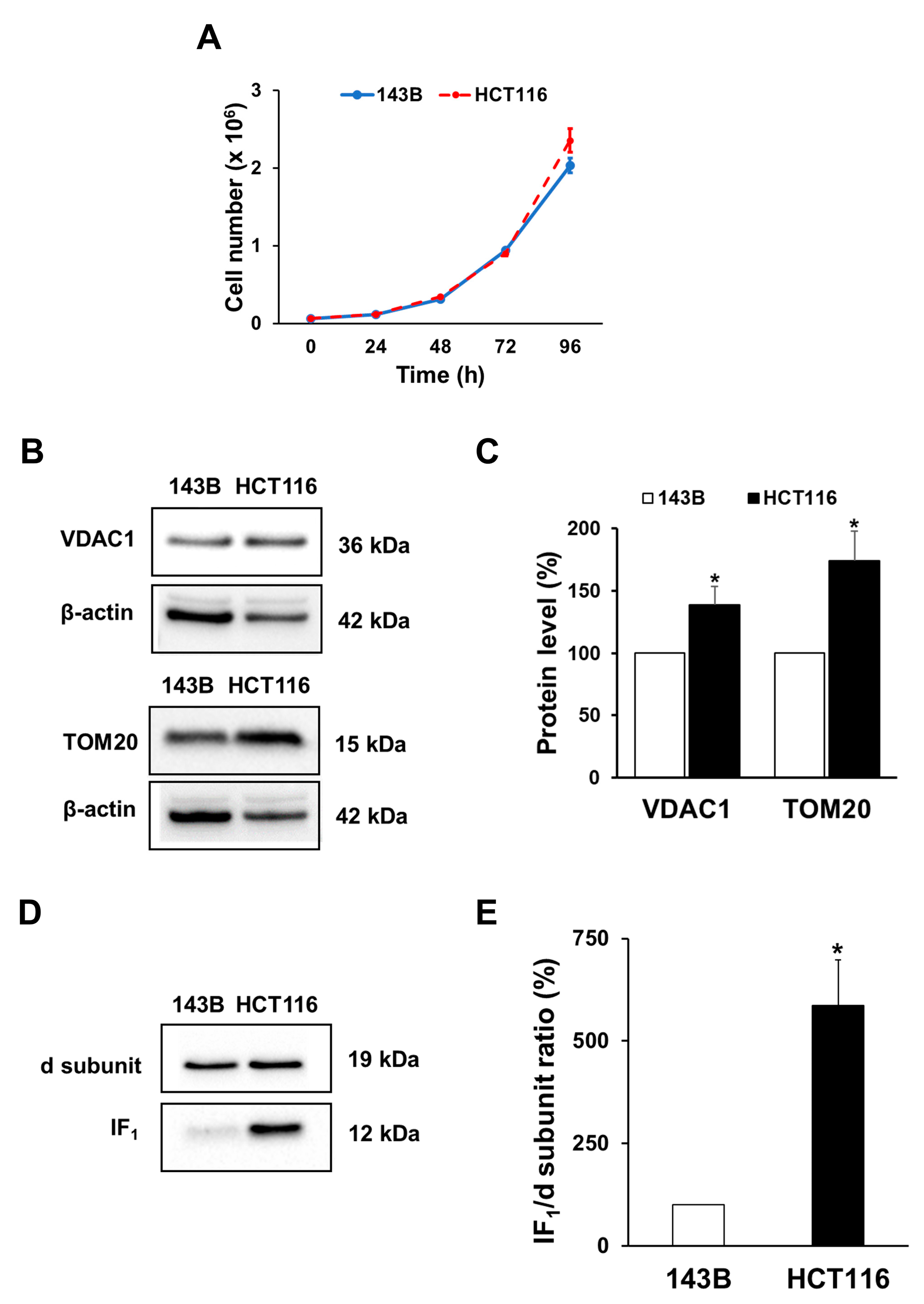
2.1. Cell Growth and Mitochondrial Mass of Both Osteosarcoma 143B and Colon Carcinoma HCT116 Cell Lines
2.2. IF1 Preserves the Energy Charge of Cancer Cells under Uncoupling Conditions
2.3. Uncoupling Promotes Mitophagy in Cancer Cells
2.4. IF1 Does Not Affect the Level of Mitochondrial Mass in Uncoupled Cancer Cells
2.5. IF1 Expression Favors the Proliferation of Uncoupled Cancer Cells When Re-Oxygenated
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Cell Clones
4.3. Proliferation of Parental Cells
4.4. Cell Growth after Exposure to Uncoupling Conditions
4.5. Cell Viability
4.6. SDS-PAGE and Western Blot Analysis
4.7. Flow Cytometry Assay of Mitochondrial Membrane Potential
4.8. Fluorescence Microscopy Evaluation of Mitochondrial Membrane Potential
4.9. ADP/ATP Ratio Assay
4.10. Fluorescence Microscopy Evaluation of Mitophagy Activation
4.11. Reagents
4.12. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Grasso, D.; Zampieri, L.X.; Capelôa, T.; Van de Velde, J.A.; Sonveaux, P. Mitochondria in Cancer. Cell Stress 2020, 4, 114–146. [Google Scholar] [CrossRef] [PubMed]
- Porporato, P.E.; Filigheddu, N.; Pedro, J.M.B.-S.; Kroemer, G.; Galluzzi, L. Mitochondrial Metabolism and Cancer. Cell Res. 2018, 28, 265–280. [Google Scholar] [CrossRef] [PubMed]
- Moreno-Sánchez, R.; Marín-Hernández, A.; Saavedra, E.; Pardo, J.P.; Ralph, S.J.; Rodríguez-Enríquez, S. Who Controls the ATP Supply in Cancer Cells? Biochemistry Lessons to Understand Cancer Energy Metabolism. Int. J. Biochem. Cell Biol. 2014, 50, 10–23. [Google Scholar] [CrossRef] [PubMed]
- Boland, M.L.; Chourasia, A.H.; Macleod, K.F. Mitochondrial Dysfunction in Cancer. Front. Oncol. 2013, 3, 292. [Google Scholar] [CrossRef] [PubMed]
- Solaini, G.; Sgarbi, G.; Baracca, A. Oxidative Phosphorylation in Cancer Cells. Biochim. Biophys. Acta Bioenerg. 2011, 1807, 534–542. [Google Scholar] [CrossRef] [PubMed]
- Gundamaraju, R.; Lu, W.; Manikam, R. Revisiting Mitochondria Scored Cancer Progression and Metastasis. Cancers 2021, 13, 432. [Google Scholar] [CrossRef] [PubMed]
- Fiorillo, M.; Ózsvári, B.; Sotgia, F.; Lisanti, M.P. High ATP Production Fuels Cancer Drug Resistance and Metastasis: Implications for Mitochondrial ATP Depletion Therapy. Front. Oncol. 2021, 11, 740720. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. The Hallmarks of Cancer. Cell 2000, 100, 57–70. [Google Scholar] [CrossRef]
- Zong, W.-X.; Rabinowitz, J.D.; White, E. Mitochondria and Cancer. Mol. Cell 2016, 61, 667–676. [Google Scholar] [CrossRef]
- Izyumov, D.S.; Avetisyan, A.V.; Pletjushkina, O.Y.; Sakharov, D.V.; Wirtz, K.W.; Chernyak, B.V.; Skulachev, V.P. “Wages of Fear”: Transient Threefold Decrease in Intracellular ATP Level Imposes Apoptosis. Biochim. Et Biophys. Acta Bioenerg. 2004, 1658, 141–147. [Google Scholar] [CrossRef]
- Faccenda, D.; Tan, C.H.; Seraphim, A.; Duchen, M.R.; Campanella, M. IF1 Limits the Apoptotic-Signalling Cascade by Preventing Mitochondrial Remodelling. Cell Death Differ. 2013, 20, 686–697. [Google Scholar] [CrossRef] [PubMed]
- Gerencser, A.A.; Chinopoulos, C.; Birket, M.J.; Jastroch, M.; Vitelli, C.; Nicholls, D.G.; Brand, M.D. Quantitative Measurement of Mitochondrial Membrane Potential in Cultured Cells: Calcium-Induced de- and Hyperpolarization of Neuronal Mitochondria: Absolute Mitochondrial Membrane Potential in Cultured Cells. J. Physiol. 2012, 590, 2845–2871. [Google Scholar] [CrossRef] [PubMed]
- Nicholls, D.G. Fluorescence Measurement of Mitochondrial Membrane Potential Changes in Cultured Cells. In Mitochondrial Bioenergetics; Palmeira, C.M., Moreno, A.J., Eds.; Methods in Molecular Biology; Springer: New York, NY, USA, 2018; Volume 1782, pp. 121–135. ISBN 978-1-4939-7830-4. [Google Scholar]
- Solaini, G.; Baracca, A.; Lenaz, G.; Sgarbi, G. Hypoxia and Mitochondrial Oxidative Metabolism. Biochim. Biophys. Acta Bioenerg. 2010, 1797, 1171–1177. [Google Scholar] [CrossRef] [PubMed]
- Rouslin, W.; Broge, C.W. Mechanisms of ATP Conservation during Ischemia in Slow and Fast Heart Rate Hearts. Am. J. Physiol.-Cell Physiol. 1993, 264, C209–C216. [Google Scholar] [CrossRef]
- Rouslin, W.; Broge, C.W. IF1 Function in Situ in Uncoupler-Challenged Ischemic Rabbit, Rat, and Pigeon Hearts. J. Biol. Chem. 1996, 271, 23638–23641. [Google Scholar] [CrossRef]
- Cabezón, E.; Montgomery, M.G.; Leslie, A.G.W.; Walker, J.E. The Structure of Bovine F1-ATPase in Complex with Its Regulatory Protein IF1. Nat. Struct. Biol. 2003, 10, 744–750. [Google Scholar] [CrossRef] [PubMed]
- Rieger, B.; Arroum, T.; Borowski, M.; Villalta, J.; Busch, K.B. Mitochondrial F 1 F O ATP Synthase Determines the Local Proton Motive Force at Cristae Rims. EMBO Rep. 2021, 22, e52727. [Google Scholar] [CrossRef]
- Zorova, L.D.; Popkov, V.A.; Plotnikov, E.Y.; Silachev, D.N.; Pevzner, I.B.; Jankauskas, S.S.; Babenko, V.A.; Zorov, S.D.; Balakireva, A.V.; Juhaszova, M.; et al. Mitochondrial Membrane Potential. Anal. Biochem. 2018, 552, 50–59. [Google Scholar] [CrossRef]
- Stotland, A.; Gottlieb, R.A. Mitochondrial Quality Control: Easy Come, Easy Go. Biochim. Biophys. Acta Mol. Cell Res. 2015, 1853, 2802–2811. [Google Scholar] [CrossRef]
- Ng, M.Y.W.; Wai, T.; Simonsen, A. Quality Control of the Mitochondrion. Dev. Cell 2021, 56, 881–905. [Google Scholar] [CrossRef]
- Aman, Y.; Schmauck-Medina, T.; Hansen, M.; Morimoto, R.I.; Simon, A.K.; Bjedov, I.; Palikaras, K.; Simonsen, A.; Johansen, T.; Tavernarakis, N.; et al. Autophagy in Healthy Aging and Disease. Nat. Aging 2021, 1, 634–650. [Google Scholar] [CrossRef] [PubMed]
- Dikic, I.; Elazar, Z. Mechanism and Medical Implications of Mammalian Autophagy. Nat. Rev. Mol. Cell Biol. 2018, 19, 349–364. [Google Scholar] [CrossRef] [PubMed]
- Vara-Perez, M.; Felipe-Abrio, B.; Agostinis, P. Mitophagy in Cancer: A Tale of Adaptation. Cells 2019, 8, 493. [Google Scholar] [CrossRef]
- Lazarou, M.; Sliter, D.A.; Kane, L.A.; Sarraf, S.A.; Wang, C.; Burman, J.L.; Sideris, D.P.; Fogel, A.I.; Youle, R.J. The Ubiquitin Kinase PINK1 Recruits Autophagy Receptors to Induce Mitophagy. Nature 2015, 524, 309–314. [Google Scholar] [CrossRef] [PubMed]
- Hanna, R.A.; Quinsay, M.N.; Orogo, A.M.; Giang, K.; Rikka, S.; Gustafsson, Å.B. Microtubule-Associated Protein 1 Light Chain 3 (LC3) Interacts with Bnip3 Protein to Selectively Remove Endoplasmic Reticulum and Mitochondria via Autophagy. J. Biol. Chem. 2012, 287, 19094–19104. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Chen, Z.; Wang, Y.; Tan, Z.; Zhu, C.; Li, Y.; Han, Z.; Chen, L.; Gao, R.; Liu, L.; et al. Mitophagy Receptor FUNDC1 Regulates Mitochondrial Dynamics and Mitophagy. Autophagy 2016, 12, 689–702. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Feng, D.; Chen, G.; Chen, M.; Zheng, Q.; Song, P.; Ma, Q.; Zhu, C.; Wang, R.; Qi, W.; et al. Mitochondrial Outer-Membrane Protein FUNDC1 Mediates Hypoxia-Induced Mitophagy in Mammalian Cells. Nat. Cell Biol. 2012, 14, 177–185. [Google Scholar] [CrossRef]
- Klionsky, D.J.; Abdel-Aziz, A.K.; Abdelfatah, S.; Abdellatif, M.; Abdoli, A.; Abel, S.; Abeliovich, H.; Abildgaard, M.H.; Abudu, Y.P.; Acevedo-Arozena, A.; et al. Guidelines for the Use and Interpretation of Assays for Monitoring Autophagy (4th Edition). Autophagy 2021, 17, 1–382. [Google Scholar] [CrossRef]
- Su, M.; Mei, Y.; Sinha, S. Role of the Crosstalk between Autophagy and Apoptosis in Cancer. J. Oncol. 2013, 2013, 102735. [Google Scholar] [CrossRef]
- Poole, L.P.; Macleod, K.F. Mitophagy in Tumorigenesis and Metastasis. Cell. Mol. Life Sci. 2021, 78, 3817–3851. [Google Scholar] [CrossRef]
- Panigrahi, D.P.; Praharaj, P.P.; Bhol, C.S.; Mahapatra, K.K.; Patra, S.; Behera, B.P.; Mishra, S.R.; Bhutia, S.K. The Emerging, Multifaceted Role of Mitophagy in Cancer and Cancer Therapeutics. Semin. Cancer Biol. 2020, 66, 45–58. [Google Scholar] [CrossRef]
- Ferro, F.; Servais, S.; Besson, P.; Roger, S.; Dumas, J.-F.; Brisson, L. Autophagy and Mitophagy in Cancer Metabolic Remodelling. Semin. Cell Dev. Biol. 2020, 98, 129–138. [Google Scholar] [CrossRef]
- Galber, C.; Fabbian, S.; Gatto, C.; Grandi, M.; Carissimi, S.; Acosta, M.J.; Sgarbi, G.; Tiso, N.; Argenton, F.; Solaini, G.; et al. The Mitochondrial Inhibitor IF1 Binds to the ATP Synthase OSCP Subunit and Protects Cancer Cells from Apoptosis. Cell Death Dis. 2023, 14, 54. [Google Scholar] [CrossRef]
- Campanella, M.; Seraphim, A.; Abeti, R.; Casswell, E.; Echave, P.; Duchen, M.R. IF1, the Endogenous Regulator of the F1Fo-ATPsynthase, Defines Mitochondrial Volume Fraction in HeLa Cells by Regulating Autophagy. Biochim. Et Biophys. Acta-Bioenerg. 2009, 1787, 393–401. [Google Scholar] [CrossRef]
- Faccenda, D.; Gorini, G.; Jones, A.; Thornton, C.; Baracca, A.; Solaini, G.; Campanella, M. The ATPase Inhibitory Factor 1 (IF1) Regulates the Expression of the Mitochondrial Ca2+ Uniporter (MCU) via the AMPK/CREB Pathway. Biochim. Et Biophys. Acta-Mol. Cell Res. 2021, 1868, 118860. [Google Scholar] [CrossRef] [PubMed]
- Sgarbi, G.; Barbato, S.; Costanzini, A.; Solaini, G.; Baracca, A. The Role of the ATPase Inhibitor Factor 1 (IF1) in Cancer Cells Adaptation to Hypoxia and Anoxia. Biochim. Biophys. Acta-Bioenerg. 2018, 1859, 99–109. [Google Scholar] [CrossRef] [PubMed]
- Fujikawa, M.; Imamura, H.; Nakamura, J.; Yoshida, M. Assessing Actual Contribution of IF1, Inhibitor of Mitochondrial FoF1, to ATP Homeostasis, Cell Growth, Mitochondrial Morphology, and Cell Viability. J. Biol. Chem. 2012, 287, 18781–18787. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Cenizo, L.; Formentini, L.; Aldea, M.; Ortega, Á.D.; García-Huerta, P.; Sánchez-Aragó, M.; Cuezva, J.M. Up-Regulation of the ATPase Inhibitory Factor 1 (IF1) of the Mitochondrial H+-ATP Synthase in Human Tumors Mediates the Metabolic Shift of Cancer Cells to a Warburg Phenotype. J. Biol. Chem. 2010, 285, 25308–25313. [Google Scholar] [CrossRef] [PubMed]
- Solaini, G.; Sgarbi, G.; Baracca, A. The F1Fo-ATPase Inhibitor, IF1, Is a Critical Regulator of Energy Metabolism in Cancer Cells. Biochem. Soc. Trans. 2021, 49, 815–827. [Google Scholar] [CrossRef] [PubMed]
- Xie, H.; Simon, M.C. Oxygen Availability and Metabolic Reprogramming in Cancer. J. Biol. Chem. 2017, 292, 16825–16832. [Google Scholar] [CrossRef]
- Sgarbi, G.; Gorini, G.; Liuzzi, F.; Solaini, G.; Baracca, A. Hypoxia and IF₁ Expression Promote ROS Decrease in Cancer Cells. Cells 2018, 7, 64. [Google Scholar] [CrossRef] [PubMed]
- Andersson, B.S.; Aw, T.Y.; Jones, D.P. Mitochondrial Transmembrane Potential and PH Gradient during Anoxia. Am. J. Physiol.-Cell Physiol. 1987, 252, C349–C355. [Google Scholar] [CrossRef] [PubMed]
- Hawrysh, P.J.; Buck, L.T. Mitochondrial Matrix pH Acidifies during Anoxia and Is Maintained by the F1 Fo—ATP Ase in Anoxia-tolerant Painted Turtle Cortical Neurons. FEBS Open Bio. 2019, 9, 571–581. [Google Scholar] [CrossRef] [PubMed]
- Jin, S.M.; Lazarou, M.; Wang, C.; Kane, L.A.; Narendra, D.P.; Youle, R.J. Mitochondrial Membrane Potential Regulates PINK1 Import and Proteolytic Destabilization by PARL. J. Cell Biol. 2010, 191, 933–942. [Google Scholar] [CrossRef] [PubMed]
- Narendra, D.P.; Jin, S.M.; Tanaka, A.; Suen, D.-F.; Gautier, C.A.; Shen, J.; Cookson, M.R.; Youle, R.J. PINK1 Is Selectively Stabilized on Impaired Mitochondria to Activate Parkin. PLoS Biol. 2010, 8, e1000298. [Google Scholar] [CrossRef]
- Baracca, A.; Sgarbi, G.; Padula, A.; Solaini, G. Glucose Plays a Main Role in Human Fibroblasts Adaptation to Hypoxia. Int. J. Biochem. Cell Biol. 2013, 45, 1356–1365. [Google Scholar] [CrossRef]
- Coppi, L.; Ligorio, S.; Mitro, N.; Caruso, D.; De Fabiani, E.; Crestani, M. PGC1s and Beyond: Disentangling the Complex Regulation of Mitochondrial and Cellular Metabolism. Int. J. Mol. Sci. 2021, 22, 6913. [Google Scholar] [CrossRef]
- Bost, F.; Kaminski, L. The Metabolic Modulator PGC-1α in Cancer. Am. J. Cancer Res. 2019, 9, 198–211. [Google Scholar]
- Lee, D.H. Sirt1 as a New Therapeutic Target in Metabolic and Age-Related Diseases. Chonnam Med. J. 2010, 46, 67. [Google Scholar] [CrossRef]
- Cantó, C.; Auwerx, J. PGC-1α, SIRT1 and AMPK, an Energy Sensing Network That Controls Energy Expenditure. Curr. Opin. Lipidol. 2009, 20, 98–105. [Google Scholar] [CrossRef]
- Li, Y.; Xu, S.; Li, J.; Zheng, L.; Feng, M.; Wang, X.; Han, K.; Pi, H.; Li, M.; Huang, X.; et al. SIRT1 Facilitates Hepatocellular Carcinoma Metastasis by Promoting PGC-1α-Mediated Mitochondrial Biogenesis. Oncotarget 2016, 7, 29255–29274. [Google Scholar] [CrossRef]
- Thomas, L.W.; Ashcroft, M. Exploring the Molecular Interface between Hypoxia-Inducible Factor Signalling and Mitochondria. Cell. Mol. Life Sci. 2019, 76, 1759–1777. [Google Scholar] [CrossRef]
- Bosetti, F.; Yu, G.; Zucchi, R.; Ronca-Testoni, S.; Solaini, G. Myocardial Ischemic Preconditioning and Mitochondrial F1F0-ATPase Activity. Mol. Cell Biochem. 2000, 215, 31–37. [Google Scholar] [CrossRef]
- Soares, R.O.S.; Losada, D.M.; Jordani, M.C.; Évora, P.; Castro-e-Silva, O. Ischemia/Reperfusion Injury Revisited: An Overview of the Latest Pharmacological Strategies. Int. J. Mol. Sci. 2019, 20, 5034. [Google Scholar] [CrossRef] [PubMed]
- Kalogeris, T.; Baines, C.P.; Krenz, M.; Korthuis, R.J. Cell Biology of Ischemia/Reperfusion Injury. In International Review of Cell and Molecular Biology; Elsevier: Amsterdam, The Netherlands, 2012; Volume 298, pp. 229–317. ISBN 978-0-12-394309-5. [Google Scholar] [CrossRef]
- Saemann, L.; Naujoks, P.; Hartrumpf, L.; Pohl, S.; Simm, A.; Szabó, G. Sex-Specific Protection of Endothelial Function after Vascular Ischemia/Reperfusion Injury by the Senomorphic Agent Ruxolitinib. Int. J. Mol. Sci. 2023, 24, 11727. [Google Scholar] [CrossRef] [PubMed]
- Gruszczyk, A.V.; Casey, A.M.; James, A.M.; Prag, H.A.; Burger, N.; Bates, G.R.; Hall, A.R.; Allen, F.M.; Krieg, T.; Saeb-Parsy, K.; et al. Mitochondrial Metabolism and Bioenergetic Function in an Anoxic Isolated Adult Mouse Cardiomyocyte Model of in Vivo Cardiac Ischemia-Reperfusion Injury. Redox Biol. 2022, 54, 102368. [Google Scholar] [CrossRef] [PubMed]
- Ploumi, C.; Daskalaki, I.; Tavernarakis, N. Mitochondrial Biogenesis and Clearance: A Balancing Act. FEBS J. 2017, 284, 183–195. [Google Scholar] [CrossRef]
- Chourasia, A.H.; Boland, M.L.; Macleod, K.F. Mitophagy and Cancer. Cancer Metab. 2015, 3, 4. [Google Scholar] [CrossRef] [PubMed]
- Gao, F.; Zhang, Y.; Hou, X.; Tao, Z.; Ren, H.; Wang, G. Dependence of PINK1 Accumulation on Mitochondrial Redox System. Aging Cell 2020, 19, e13211. [Google Scholar] [CrossRef]
- Lefebvre, V.; Du, Q.; Baird, S.; Ng, A.C.-H.; Nascimento, M.; Campanella, M.; McBride, H.M.; Screaton, R.A. Genome-Wide RNAi Screen Identifies ATPase Inhibitory Factor 1 (ATPIF1) as Essential for PARK2 Recruitment and Mitophagy. Autophagy 2013, 9, 1770–1779. [Google Scholar] [CrossRef]
- Malena, A.; Pantic, B.; Borgia, D.; Sgarbi, G.; Solaini, G.; Holt, I.J.; Spinazzola, A.; Perissinotto, E.; Sandri, M.; Baracca, A.; et al. Mitochondrial Quality Control: Cell-Type-Dependent Responses to Pathological Mutant Mitochondrial DNA. Autophagy 2016, 12, 2098–2112. [Google Scholar] [CrossRef]
- Matsuda, N.; Sato, S.; Shiba, K.; Okatsu, K.; Saisho, K.; Gautier, C.A.; Sou, Y.; Saiki, S.; Kawajiri, S.; Sato, F.; et al. PINK1 Stabilized by Mitochondrial Depolarization Recruits Parkin to Damaged Mitochondria and Activates Latent Parkin for Mitophagy. J. Cell Biol. 2010, 189, 211–221. [Google Scholar] [CrossRef] [PubMed]
- Barbato, S.; Sgarbi, G.; Gorini, G.; Baracca, A.; Solaini, G. The Inhibitor Protein (IF1) of the F1F0-ATPase Modulates Human Osteosarcoma Cell Bioenergetics. J. Biol. Chem. 2015, 290, 6338–6348. [Google Scholar] [CrossRef] [PubMed]
- Costanzini, A.; Sgarbi, G.; Maresca, A.; Del Dotto, V.; Solaini, G.; Baracca, A. Mitochondrial Mass Assessment in a Selected Cell Line under Different Metabolic Conditions. Cells 2019, 8, 1454. [Google Scholar] [CrossRef] [PubMed]
- Waterborg, J.H.; Matthews, H.R. The Lowry Method for Protein Quantitation. Methods Mol. Biol. 1984, 1, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Lowry, O.H.; Rosebrough, N.J.; Farr, A.L.; Randall, R.J. Protein Measurement with the Folin Phenol Reagent. J. Biol. Chem. 1951, 193, 265–275. [Google Scholar] [CrossRef] [PubMed]
- Sgarbi, G.; Gorini, G.; Costanzini, A.; Barbato, S.; Solaini, G.; Baracca, A. Hypoxia Decreases ROS Level in Human Fibroblasts. Int. J. Biochem. Cell Biol. 2017, 88, 133–144. [Google Scholar] [CrossRef] [PubMed]
- Creed, S.; McKenzie, M. Measurement of Mitochondrial Membrane Potential with the Fluorescent Dye Tetramethylrhodamine Methyl Ester (TMRM). In Cancer Metabolism; Haznadar, M., Ed.; Methods in Molecular Biology; Springer: New York, NY, USA, 2019; Volume 1928, pp. 69–76. ISBN 978-1-4939-9026-9. [Google Scholar]











Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Righetti, R.; Grillini, S.; Del Dotto, V.; Costanzini, A.; Liuzzi, F.; Zanna, C.; Sgarbi, G.; Solaini, G.; Baracca, A. The Pro-Oncogenic Protein IF1 Promotes Proliferation of Anoxic Cancer Cells during Re-Oxygenation. Int. J. Mol. Sci. 2023, 24, 14624. https://doi.org/10.3390/ijms241914624
Righetti R, Grillini S, Del Dotto V, Costanzini A, Liuzzi F, Zanna C, Sgarbi G, Solaini G, Baracca A. The Pro-Oncogenic Protein IF1 Promotes Proliferation of Anoxic Cancer Cells during Re-Oxygenation. International Journal of Molecular Sciences. 2023; 24(19):14624. https://doi.org/10.3390/ijms241914624
Chicago/Turabian StyleRighetti, Riccardo, Silvia Grillini, Valentina Del Dotto, Anna Costanzini, Francesca Liuzzi, Claudia Zanna, Gianluca Sgarbi, Giancarlo Solaini, and Alessandra Baracca. 2023. "The Pro-Oncogenic Protein IF1 Promotes Proliferation of Anoxic Cancer Cells during Re-Oxygenation" International Journal of Molecular Sciences 24, no. 19: 14624. https://doi.org/10.3390/ijms241914624
APA StyleRighetti, R., Grillini, S., Del Dotto, V., Costanzini, A., Liuzzi, F., Zanna, C., Sgarbi, G., Solaini, G., & Baracca, A. (2023). The Pro-Oncogenic Protein IF1 Promotes Proliferation of Anoxic Cancer Cells during Re-Oxygenation. International Journal of Molecular Sciences, 24(19), 14624. https://doi.org/10.3390/ijms241914624









