How Does cGAS Avoid Sensing Self-DNA under Normal Physiological Conditions?
Abstract
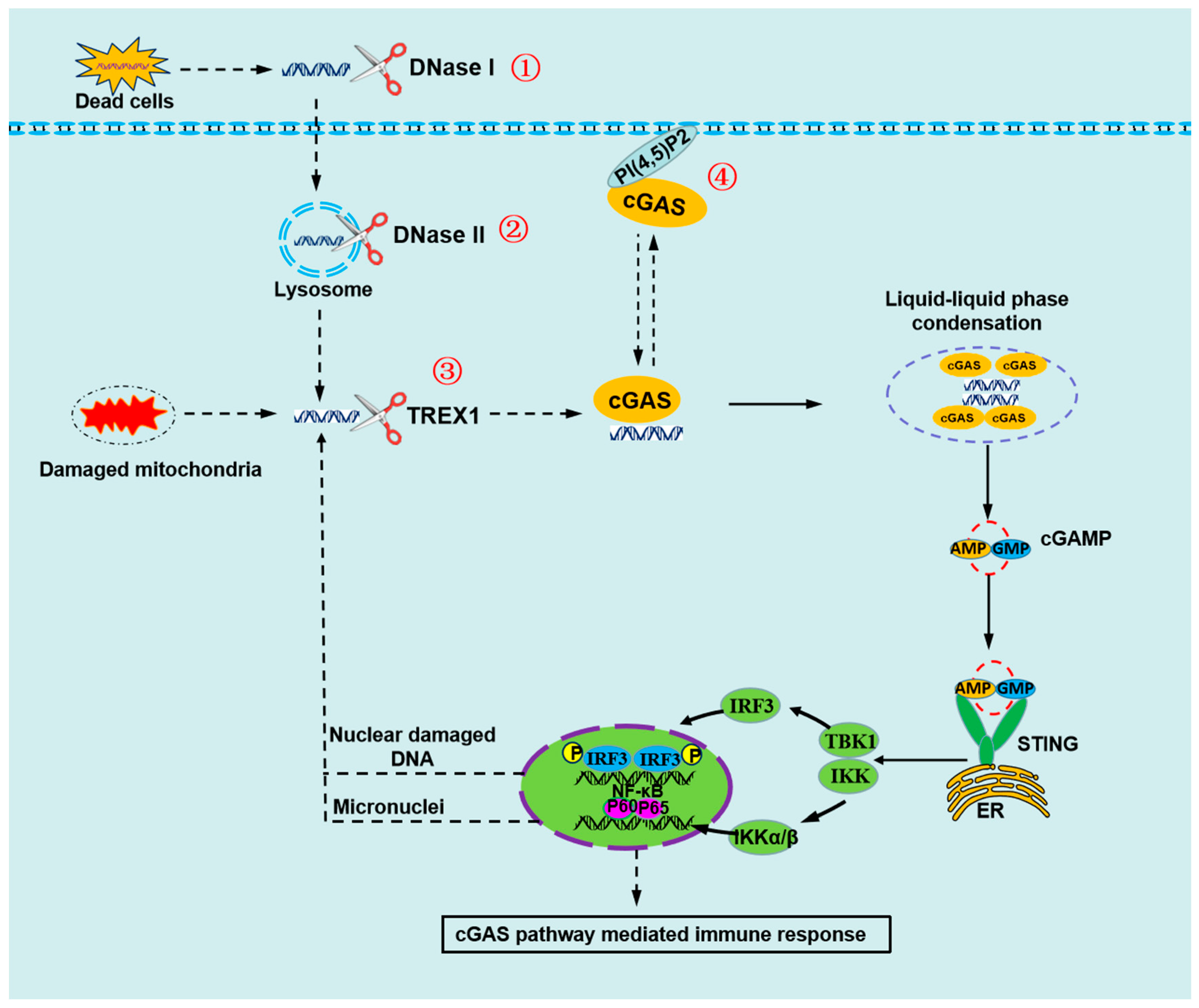
:1. Introduction
2. Structural Domains and Modification Sites of cGAS
2.1. Structural Domains of cGAS
2.2. Transcriptional and Epigenetic Regulation of cGAS
2.3. Post-Translational Modifications of cGAS
3. How Is the cGAS Activated
3.1. DNA-Induced Conformational Changes in cGAS Lead to Its Activation
3.2. Liquid-Liquid Phase Separation Can Enhance the Activation of cGAS
3.3. Divalent Cations Substantially Promote the Activity of cGAS
4. How Does cGAS Avoid Sensing Self-DNA under Normal Conditions?
4.1. Self-DNA Is Cleared by the DNases
4.2. Plasma Membrane Localization of cGAS Prevents Recognition of Self-DNA
4.3. Binding to Histones Prevents cGAS from Sensing Self-DNA during Mitosis
4.4. Barrier-to-Autointegration Factor 1 Restricts cGAS to Sense Self-DNA during Mitosis
4.5. The Activity of cGAS Is Suppressed via Phosphorylation during Mitosis
5. Consequences of Self-DNA Induced cGAS Activation
5.1. Activation of cGAS by Self-DNA Can Cause Autoimmune Diseases
5.2. Activation of cGAS by Self-DNA Is a Double-Edged Sword in Cancer
6. Conclusions and Future Perspectives
7. Methodology
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ablasser, A.; Chen, Z.J.J. cGAS in action: Expanding roles in immunity and inflammation. Science 2019, 363, aat8657. [Google Scholar] [CrossRef] [PubMed]
- Civril, F.; Deimling, T.; Mann, C.C.D.; Ablasser, A.; Moldt, M.; Witte, G.; Hornung, V.; Hopfner, K.P. Structural mechanism of cytosolic DNA sensing by cGAS. Nature 2013, 498, 332–337. [Google Scholar] [CrossRef] [PubMed]
- Zheng, W.L.; Liu, A.J.; Xia, N.W.; Chen, N.H.; Meurens, F.; Zhu, J.Z. How the Innate Immune DNA Sensing cGAS-STING Pathway Is Involved in Apoptosis. Int. J. Mol. Sci. 2023, 24, 3029. [Google Scholar] [CrossRef]
- Song, J.X.; Villagomes, D.; Zhao, H.C.; Zhu, M. cGAS in nucleus: The link between immune response and DNA damage repair. Front. Immunol. 2022, 13, 1076784. [Google Scholar] [CrossRef] [PubMed]
- Gong, T.; Liu, L.; Jiang, W.; Zhou, R.B. DAMP-sensing receptors in sterile inflammation and inflammatory diseases. Nat. Rev. Immunol. 2020, 20, 95–112. [Google Scholar] [CrossRef] [PubMed]
- Mata-Garrido, J.; Frizzi, L.; Nguyen, T.; He, X.Y.; Chang-Marchand, Y.; Xiang, Y.; Reisacher, C.; Casafont, I.; Arbibe, L. HP1 gamma Prevents Activation of the cGAS/STING Pathway by Preserving Nuclear Envelope and Genomic Integrity in Colon Adenocarcinoma Cells. Int. J. Mol. Sci. 2023, 24, 7347. [Google Scholar] [CrossRef]
- Amadio, R.; Piperno, G.M.; Benvenuti, F. Self-DNA Sensing by cGAS-STING and TLR9 in Autoimmunity: Is the Cytoskeleton in Control? Front. Immunol. 2021, 12, 657344. [Google Scholar] [CrossRef]
- Hopfner, K.P.; Hornung, V. Molecular mechanisms and cellular functions of cGAS-STING signalling. Nat. Rev. Mol. Cell Biol. 2020, 21, 501–521. [Google Scholar] [CrossRef]
- Guttinger, S.; Laurell, E.; Kutay, U. Orchestrating nuclear envelope disassembly and reassembly during mitosis. Nat. Rev. Mol. Cell Biol. 2009, 10, 178–191. [Google Scholar] [CrossRef]
- Rigby, R.E.; Leitch, A.; Jackson, A.P. Nucleic acid-mediated inflammatory diseases. Bioessays 2008, 30, 833–842. [Google Scholar] [CrossRef]
- Yu, L.; Liu, P.D. Cytosolic DNA sensing by cGAS: Regulation, function, and human diseases. Signal Transduct. Target. Ther. 2021, 6, 170. [Google Scholar] [CrossRef] [PubMed]
- Joshi, B.; Joshi, J.C.; Mehta, D. Regulation of cGAS Activity and Downstream Signaling. Cells 2022, 11, 2812. [Google Scholar] [CrossRef]
- Bai, J.L.; Liu, F. Nuclear cGAS: Sequestration and beyond. Protein Cell 2022, 13, 90–101. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.M.; Wu, F.H.; Wang, X.Q.; Wang, L.L.; Siedow, J.N.; Zhang, W.G.; Pei, Z.M. Molecular evolutionary and structural analysis of the cytosolic DNA sensor cGAS and STING. Nucleic Acids Res. 2014, 42, 8243–8257. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Sun, L.J.; Chen, Z.J.J. Regulation and function of the cGAS-STING pathway of cytosolic DNA sensing. Nat. Immunol. 2016, 17, 1142–1149. [Google Scholar] [CrossRef]
- Ma, F.; Li, B.; Liu, S.-y.; Iyer, S.S.; Yu, Y.; Wu, A.; Cheng, G. Positive feedback regulation of type I IFN production by the IFN-inducible DNA sensor cGAS. J. Immunol. 2015, 194, 1545–1554. [Google Scholar] [CrossRef]
- Chen, H.-Y.; Pang, X.-Y.; Xu, Y.-Y.; Zhou, G.-P.; Xu, H.-G. Transcriptional regulation of human cyclic GMP-AMP synthase gene. Cell. Signal. 2019, 62, 109355. [Google Scholar] [CrossRef]
- Wu, Y.K.; Li, S.T. Role of Post-Translational Modifications of cGAS in Innate Immunity. Int. J. Mol. Sci. 2020, 21, 7842. [Google Scholar] [CrossRef]
- Li, T.; Huang, T.Z.; Chen, Z.J.J. Phosphorylation and Chromatin Tethering Prevent cGAS activation During Mitosis. J. Immunol. 2021, 206, eabc5386. [Google Scholar] [CrossRef]
- Cui, Y.; Yu, H.S.; Zheng, X.; Peng, R.; Wang, Q.; Zhou, Y.; Wang, R.; Wang, J.H.; Qu, B.; Shen, N.; et al. SENP7 Potentiates cGAS Activation by Relieving SUMO-Mediated Inhibition of Cytosolic DNA Sensing. PLoS Pathog. 2017, 13, e1006156. [Google Scholar] [CrossRef]
- Wang, Q.; Huang, L.Y.; Hong, Z.; Lv, Z.S.; Mao, Z.M.; Tang, Y.J.; Kong, X.F.; Li, S.L.; Cui, Y.; Liu, H.; et al. The E3 ubiquitin ligase RNF185 facilitates the cGAS-mediated innate immune response. PLoS Pathog. 2017, 13, e1006264. [Google Scholar] [CrossRef] [PubMed]
- Ji, L.K.; Wang, Y.; Zhou, L.Y.; Lu, J.; Bao, S.W.; Shen, Q.; Wang, X.C.; Liu, Y.W.; Zhang, W. E3 Ubiquitin Ligases: The Operators of the Ubiquitin Code That Regulates the RLR and cGAS-STING Pathways. Int. J. Mol. Sci. 2022, 23, 14601. [Google Scholar] [CrossRef] [PubMed]
- Hu, M.M.; Yang, Q.; Xie, X.Q.; Liao, C.Y.; Lin, H.; Liu, T.T.; Yin, L.; Shu, H.B. Sumoylation Promotes the Stability of the DNA Sensor cGAS and the Adaptor STING to Regulate the Kinetics of Response to DNA Virus. Immunity 2016, 45, 555–569. [Google Scholar] [CrossRef] [PubMed]
- Dai, J.; Huang, Y.J.; He, X.H.; Zhao, M.; Wang, X.Z.; Liu, Z.S.; Xue, W.; Cai, H.; Zhan, X.Y.; Huang, S.Y.; et al. Acetylation Blocks cGAS Activity and Inhibits Self-DNA-Induced Autoimmunity. Cell 2019, 176, 1447–1460. [Google Scholar] [CrossRef]
- Song, Z.M.; Lin, H.; Yi, X.M.; Guo, W.; Hu, M.M.; Shu, H.B. KAT5 acetylates cGAS to promote innate immune response to DNA virus. Proc. Natl. Acad. Sci. USA 2020, 117, 21568–21575. [Google Scholar] [CrossRef]
- Dowling, Q.M.; Volkman, H.E.; Gray, E.E.; Ovchinnikov, S.; Cambier, S.; Bera, A.K.; Sankaran, B.; Johnson, M.R.; Bick, M.J.; Kang, A.L.; et al. Computational design of constitutively active cGAS. Nat. Struct. Mol. Biol. 2023, 30, 72–80. [Google Scholar] [CrossRef]
- Herzner, A.M.; Schlee, M.; Bartok, E. The many faces of cGAS: How cGAS activation is controlled in the cytosol, the nucleus, and during mitosis. Signal Transduct. Target. Ther. 2021, 6, 260. [Google Scholar] [CrossRef]
- Mann, C.D.C.; Hornung, V. Molecular mechanisms of nonself nucleic acid recognition by the innate immune system. Eur. J. Immunol. 2021, 51, 1897–1910. [Google Scholar] [CrossRef]
- Zhang, X.W.; Bai, X.C.; Chen, Z.J.J. Structures and Mechanisms in the cGAS-STING Innate Immunity Pathway. Immunity 2020, 53, 43–53. [Google Scholar] [CrossRef]
- Zheng, J.Y.; Mo, J.L.; Zhu, T.; Zhuo, W.; Yi, Y.N.; Hu, S.; Yin, J.Y.; Zhang, W.; Zhou, H.H.; Liu, Z.Q. Comprehensive elaboration of the cGAS-STING signaling axis in cancer development and immunotherapy. Mol. Cancer 2020, 19, 133. [Google Scholar] [CrossRef]
- Robb, C.G.; Dao, T.P.; Ujma, J.; Castaneda, C.; Beveridge, R. Ion Mobility Mass Spectrometry Unveils Global Protein Conformations in Response to Conditions that Promote and Reverse Liquid-Liquid Phase Separation. J. Am. Chem. Soc. 2023, 145, 12541–12549. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Q.; McAtee, C.K.; Su, X.L. Phase separation in immune signalling. Nat. Rev. Immunol. 2022, 22, 188–199. [Google Scholar] [CrossRef] [PubMed]
- Du, M.J.; Chen, Z.J.J. DNA-induced liquid phase condensation of cGAS activates innate immune signaling. Science 2018, 361, 704–709. [Google Scholar] [CrossRef] [PubMed]
- Hu, M.M.; Shu, H.B. Innate Immune Response to Cytoplasmic DNA: Mechanisms and Diseases. Annu. Rev. Immunol. 2020, 38, 79–98. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Mohr, L.; Maciejowski, J.; Kranzusch, P.J. cGAS phase separation inhibits TREX1-mediated DNA degradation and enhances cytosolic DNA sensing. Mol. Cell 2021, 81, 739–755.e7. [Google Scholar] [CrossRef]
- Li, X.; Shu, C.; Yi, G.H.; Chaton, C.T.; Shelton, C.L.; Diao, J.S.; Zuo, X.B.; Kao, C.C.; Herr, A.B.; Li, P.W. Cyclic GMP-AMP Synthase Is Activated by Double-Stranded DNA-Induced Oligomerization. Immunity 2013, 39, 1019–1031. [Google Scholar] [CrossRef]
- Mitic, N.; Miraula, M.; Selleck, C.; Hadler, K.S.; Uribe, E.; Pedroso, M.M.; Schenk, G. Catalytic Mechanisms of Metallohydrolases Containing Two Metal Ions. Adv. Protein Chem. Struct. 2014, 97, 49–81. [Google Scholar] [CrossRef]
- Zhao, Z.; Ma, Z.X.; Wang, B.; Guan, Y.K.; Su, X.D.; Jiang, Z.F. Mn2+ Directly Activates cGAS and Structural Analysis Suggests Mn2+ Induces a Noncanonical Catalytic Synthesis of 2′3′-cGAMP. Cell Rep. 2020, 32, 108053. [Google Scholar] [CrossRef]
- Rozenberg, J.M.; Kamynina, M.; Sorokin, M.; Zolotovskaia, M.; Koroleva, E.; Kremenchutckaya, K.; Gudkov, A.; Buzdin, A.; Borisov, N. The Role of the Metabolism of Zinc and Manganese Ions in Human Cancerogenesis. Biomedicines 2022, 10, 1072. [Google Scholar] [CrossRef]
- Wang, C.G.; Guan, Y.K.; Lv, M.Z.; Zhang, R.; Guo, Z.Y.; Wei, X.M.; Du, X.X.; Yang, J.; Li, T.; Wan, Y.; et al. Manganese Increases the Sensitivity of the cGAS-STING Pathway for Double-Stranded DNA and Is Required for the Host Defense against DNA Viruses. Immunity 2018, 48, 675–687.e7. [Google Scholar] [CrossRef]
- Ma, R.H.; Serrano, T.P.O.; Davis, J.; Prigge, A.D.; Ridge, K.M. The cGAS-STING pathway: The role of self-DNA sensing in inflammatory lung disease. FASEB J. 2020, 34, 13156–13170. [Google Scholar] [CrossRef] [PubMed]
- Koyama, R.; Arai, T.; Kijima, M.; Sato, S.; Miura, S.; Yuasa, M.; Kitamura, D.; Mizuta, R. DNase, DNase I and caspase-activated DNase cooperate to degrade dead cells. Genes Cells 2016, 21, 1150–1163. [Google Scholar] [CrossRef] [PubMed]
- Anindya, R. Cytoplasmic DNA in cancer cells: Several pathways that potentially limit DNase2 and TREX1 activities. BBA-Mol. Cell Res. 2022, 1869, 119278. [Google Scholar] [CrossRef] [PubMed]
- Simpson, S.R.; Hemphill, W.O.; Hudson, T.; Perrino, F.W. TREX1-Apex predator of cytosolic DNA metabolism. DNA Repair. 2020, 94, 102894. [Google Scholar] [CrossRef] [PubMed]
- Xiao, N.Y.; Wei, J.J.; Xu, S.; Du, H.K.; Huang, M.H.; Zhang, S.T.; Ye, W.W.; Sun, L.J.; Chen, Q. cGAS activation causes lupus-like autoimmune disorders in a TREX1 mutant mouse model. J. Autoimmun. 2019, 100, 84–94. [Google Scholar] [CrossRef]
- Giordano, A.M.S.; Luciani, M.; Gatto, F.; Abou Alezz, M.; Beghe, C.; Della Volpe, L.; Migliara, A.; Valsoni, S.; Genua, M.; Dzieciatkowska, M.; et al. DNA damage contributes to neurotoxic inflammation in Aicardi-Goutieres syndrome astrocytes. J. Exp. Med. 2022, 219, e20211121. [Google Scholar] [CrossRef]
- Fang, L.W.; Ying, S.C.; Xu, X.; Wu, D. TREX1 cytosolic DNA degradation correlates with autoimmune disease and cancer immunity. Clin. Exp. Immunol. 2023, 211, 193–207. [Google Scholar] [CrossRef]
- Gao, D.X.; Li, T.; Li, X.D.; Chen, X.; Li, Q.Z.; Wight-Carter, M.; Chen, Z.J. Activation of cyclic GMP-AMP synthase by self-DNA causes autoimmune diseases. Proc. Natl. Acad. Sci. USA 2015, 112, E5699–E5705. [Google Scholar] [CrossRef]
- Barnett, K.C.; Coronas-Serna, J.M.; Zhou, W.; Ernandes, M.J.; Cao, A.; Kranzusch, P.J.; Kagan, J.C. Phosphoinositide Interactions Position cGAS at the Plasma Membrane to Ensure Efficient Distinction between Self- and Viral DNA. Cell 2019, 176, 1432–1446.e11. [Google Scholar] [CrossRef]
- Ablasser, A.; Hur, S. Regulation of cGAS- and RLR-mediated immunity to nucleic acids. Nat. Immunol. 2020, 21, 17–29. [Google Scholar] [CrossRef]
- Wu, Y.K.; Song, K.; Hao, W.Z.; Li, J.; Wang, L.Y.; Li, S.T. Nuclear soluble cGAS senses double-stranded DNA virus infection. Commun. Biol. 2022, 5, 433. [Google Scholar] [CrossRef] [PubMed]
- Wischnewski, M.; Ablasser, A. Interplay of cGAS with chromatin. Trends Biochem. Sci. 2021, 46, 822–831. [Google Scholar] [CrossRef]
- Zierhut, C.; Yamaguchi, N.; Paredes, M.; Luo, J.D.; Carroll, T.; Funabiki, H. The Cytoplasmic DNA Sensor cGAS Promotes Mitotic Cell Death. Cell 2019, 178, 302–315.e23. [Google Scholar] [CrossRef] [PubMed]
- Cao, D.F.; Han, X.A.; Fan, X.Y.; Xu, R.M.; Zhang, X.Z. Structural basis for nucleosome-mediated inhibition of cGAS activity. Cell Res. 2020, 30, 1088–1097. [Google Scholar] [CrossRef] [PubMed]
- Guey, B.; Wischnewski, M.; Decout, A.; Makasheva, K.; Kaynak, M.; Sakar, M.S.; Fierz, B.; Ablasser, A. BAF restricts cGAS on nuclear DNA to prevent innate immune activation. Science 2020, 369, 823–828. [Google Scholar] [CrossRef] [PubMed]
- Jamin, A.; Wiebe, M.S. Barrier to Autointegration Factor (BANF1): Interwoven roles in nuclear structure, genome integrity, innate immunity, stress responses and progeria. Curr. Opin. Cell Biol. 2015, 34, 61–68. [Google Scholar] [CrossRef]
- Bradley, C.M.; Ronning, D.R.; Ghirlando, R.; Craigie, R.; Dyda, F. Structural basis for DNA bridging by barrier-to-autointegration factor. Nat. Struct. Mol. Biol. 2005, 12, 935–936. [Google Scholar] [CrossRef]
- Broussard, G.; Ni, G.X.; Zhang, Z.G.; Li, Q.; Cano, P.; Dittmer, D.P.; Damania, B. Barrier-to-autointegration factor 1 promotes gammaherpesvirus reactivation from latency. Nat. Commun. 2023, 14, 434. [Google Scholar] [CrossRef]
- Zhong, L.; Hu, M.M.; Bian, L.J.; Liu, Y.; Chen, Q.; Shu, H.B. Phosphorylation of cGAS by CDK1 impairs self-DNA sensing in mitosis. Cell Discov. 2020, 6, 26. [Google Scholar] [CrossRef]
- Navegantes, K.C.; Gomes, R.D.; Pereira, P.A.T.; Czaikoski, P.G.; Azevedo, C.H.M.; Monteiro, M.C. Immune modulation of some autoimmune diseases: The critical role of macrophages and neutrophils in the innate and adaptive immunity. J. Transl. Med. 2017, 15, 36. [Google Scholar] [CrossRef]
- Benmerzoug, S.; Ryffel, B.; Togbe, D.; Quesniaux, V.F.J. Self-DNA Sensing in Lung Inflammatory Diseases. Trends Immunol. 2019, 40, 719–734. [Google Scholar] [CrossRef] [PubMed]
- Tonduti, D.; Fazzi, E.; Badolato, R.; Orcesi, S. Novel and emerging treatments for Aicardi-Goutieres syndrome. Expert. Rev. Clin. Immun. 2020, 16, 189–198. [Google Scholar] [CrossRef]
- Pokatayev, V.; Hasin, N.; Chon, H.; Cerritelli, S.M.; Sakhuja, K.; Ward, J.M.; Morris, H.D.; Yan, N.; Crouch, R.J. RNase H2 catalytic core Aicardi-Goutieres syndrome-related mutant invokes cGAS-STING innate immune-sensing pathway in mice. J. Exp. Med. 2016, 213, 329–336. [Google Scholar] [CrossRef] [PubMed]
- Gray, E.E.; Treuting, P.M.; Woodward, J.J.; Stetson, D.B. Cutting Edge: cGAS Is Required for Lethal Autoimmune Disease in the Trex1-Deficient Mouse Model of Aicardi-Goutieres Syndrome. J. Immunol. 2015, 195, 1939–1943. [Google Scholar] [CrossRef]
- Crow, M.K. Pathogenesis of systemic lupus erythematosus: Risks, mechanisms and therapeutic targets. Ann. Rheum. Dis. 2023, 82, 223741. [Google Scholar] [CrossRef]
- Kato, Y.; Park, J.; Takamatsu, H.; Konaka, H.; Aoki, W.; Aburaya, S.; Ueda, M.; Nishide, M.; Koyama, S.; Hayama, Y.; et al. Apoptosis-derived membrane vesicles drive the cGAS-STING pathway and enhance type I IFN production in systemic lupus erythematosus. Ann. Rheum. Dis. 2018, 77, 1507–1515. [Google Scholar] [CrossRef]
- Duan, D.D.; Shang, M.J.; Han, Y.X.; Liu, J.Y.; Liu, J.W.; Kong, S.H.; Hou, J.Y.; Huang, B.Q.; Lu, J.; Zhang, Y. EZH2-CCF-cGAS Axis Promotes Breast Cancer Metastasis. Int. J. Mol. Sci. 2022, 23, 1788. [Google Scholar] [CrossRef]
- Huang, J.L.; Chang, Y.T.; Hong, Z.Y.; Lin, C.S. Targeting DNA Damage Response and Immune Checkpoint for Anticancer Therapy. Int. J. Mol. Sci. 2022, 23, 238. [Google Scholar] [CrossRef]
- Guscott, M.; Saha, A.; Maharaj, J.; McClelland, S.E. The multifaceted role of micronuclei in tumour progression: A whole organism perspective. Int. J. Biochem. Cell Biol. 2022, 152, 106300. [Google Scholar] [CrossRef]
- Shen, R.; Liu, D.S.; Wang, X.N.; Guo, Z.; Sun, H.A.; Song, Y.F.; Wang, D.G. DNA Damage and Activation of cGAS/ STING Pathway Induce Tumor Microenvironment Remodeling. Front. Cell Dev. Biol. 2022, 9, 828657. [Google Scholar] [CrossRef]
- Yum, S.; Li, M.H.; Chen, Z.J.J. Old dogs, new trick: Classic cancer therapies activate cGAS. Cell Res. 2020, 30, 639–648. [Google Scholar] [CrossRef] [PubMed]
- Khoo, L.T.; Chen, L.Y. Role of the cGAS-STING pathway in cancer development and oncotherapeutic approaches. EMBO Rep. 2018, 19, e46935. [Google Scholar] [CrossRef] [PubMed]
- Decout, A.; Katz, J.D.; Venkatraman, S.; Ablasser, A. The cGAS-STING pathway as a therapeutic target in inflammatory diseases. Nat. Rev. Immunol. 2021, 21, 548–569. [Google Scholar] [CrossRef] [PubMed]
- Bakhoum, S.; Ngo, B.; Bakhoum, A.; Cavallo-Fleming, J.A.; Murphy, C.W.; Powell, S.N.; Cantley, L. Chromosomal Instability Drives Metastasis through a Cytosolic DNA Response. Int. J. Radiat. Oncol. 2018, 102, S118. [Google Scholar] [CrossRef]
- Qiu, S.Q.; Zhong, X.Y.; Meng, X.; Li, S.T.; Qian, X.Y.; Lu, H.; Cai, J.; Zhang, Y.; Wang, M.J.; Ye, Z.J.; et al. Mitochondria-localized cGAS suppresses ferroptosis to promote cancer progression. Cell Res. 2023, 33, 299–311. [Google Scholar] [CrossRef]
- Chen, Q.; Boire, A.; Jin, X.; Valiente, M.; Er, E.E.; Lopez-Soto, A.; Jacob, L.S.; Patwa, R.; Shah, H.; Xu, K.; et al. Carcinoma-astrocyte gap junctions promote brain metastasis by cGAMP transfer. Nature 2017, 544, 124. [Google Scholar] [CrossRef]
- Lama, L.; Adura, C.; Xie, W.; Tomita, D.; Kamei, T.; Kuryavyi, V.; Gogakos, T.; Steinberg, J.I.; Miller, M.; Ramos-Espiritu, L. Development of human cGAS-specific small-molecule inhibitors for repression of dsDNA-triggered interferon expression. Nat. Commun. 2019, 10, 2261. [Google Scholar] [CrossRef]
- Vincent, J.; Adura, C.; Gao, P.; Luz, A.; Lama, L.; Asano, Y.; Okamoto, R.; Imaeda, T.; Aida, J.; Rothamel, K. Small molecule inhibition of cGAS reduces interferon expression in primary macrophages from autoimmune mice. Nat. Commun. 2017, 8, 750. [Google Scholar]
- Li, T.; Cheng, H.; Yuan, H.; Xu, Q.; Shu, C.; Zhang, Y.; Xu, P.; Tan, J.; Rui, Y.; Li, P. Antitumor activity of cGAMP via stimulation of cGAS-cGAMP-STING-IRF3 mediated innate immune response. Sci. Rep. 2016, 6, 19049. [Google Scholar] [CrossRef]
- Gan, Y.; Li, X.; Han, S.; Liang, Q.; Ma, X.; Rong, P.; Wang, W.; Li, W. The cGAS/STING pathway: A novel target for cancer therapy. Front. Immunol. 2022, 12, 795401. [Google Scholar] [CrossRef]
- Yum, S.; Li, M.; Frankel, A.E.; Chen, Z.J. Roles of the cGAS-STING pathway in cancer immunosurveillance and immunotherapy. Annu. Rev. Cancer Biol. 2019, 3, 323–344. [Google Scholar] [CrossRef]
- Motedayen Aval, L.; Pease, J.E.; Sharma, R.; Pinato, D.J. Challenges and opportunities in the clinical development of STING agonists for cancer immunotherapy. J. Clin. Med. 2020, 9, 3323. [Google Scholar] [CrossRef] [PubMed]
- Uhlorn, B.L.; Gamez, E.R.; Li, S.Z.; Campos, S.K. Attenuation of cGAS/STING activity during mitosis. Life Sci. Alliance 2020, 3, e201900636. [Google Scholar] [CrossRef] [PubMed]
- Cervigni, R.I.; Bonavita, R.; Barretta, M.L.; Spano, D.; Ayala, I.; Nakamura, N.; Corda, D.; Colanzi, A. JNK2 controls fragmentation of the Golgi complex and the G2/M transition through phosphorylation of GRASP65. J. Cell Sci. 2015, 128, 2249–2260. [Google Scholar] [CrossRef]
- Liu, H.P.; Wang, F.; Cao, Y.J.; Dang, Y.F.; Ge, B.X. The multifaceted functions of cGAS. J. Mol. Cell Biol. 2022, 14, mjac031. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Zou, Y.; Zhou, X.; Zhou, J. Inhibitory targeting cGAS-STING-TBK1 axis: Emerging strategies for autoimmune diseases therapy. Front. Immunol. 2022, 13, 954129. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zheng, W.; Chen, N.; Meurens, F.; Zheng, W.; Zhu, J. How Does cGAS Avoid Sensing Self-DNA under Normal Physiological Conditions? Int. J. Mol. Sci. 2023, 24, 14738. https://doi.org/10.3390/ijms241914738
Zheng W, Chen N, Meurens F, Zheng W, Zhu J. How Does cGAS Avoid Sensing Self-DNA under Normal Physiological Conditions? International Journal of Molecular Sciences. 2023; 24(19):14738. https://doi.org/10.3390/ijms241914738
Chicago/Turabian StyleZheng, Wangli, Nanhua Chen, François Meurens, Wanglong Zheng, and Jianzhong Zhu. 2023. "How Does cGAS Avoid Sensing Self-DNA under Normal Physiological Conditions?" International Journal of Molecular Sciences 24, no. 19: 14738. https://doi.org/10.3390/ijms241914738
APA StyleZheng, W., Chen, N., Meurens, F., Zheng, W., & Zhu, J. (2023). How Does cGAS Avoid Sensing Self-DNA under Normal Physiological Conditions? International Journal of Molecular Sciences, 24(19), 14738. https://doi.org/10.3390/ijms241914738








