Integrated Bioinformatics Analysis Confirms the Diagnostic Value of Nourin-Dependent miR-137 and miR-106b in Unstable Angina Patients
Abstract
:1. Introduction
2. Results
2.1. Homology Model of Nourin
2.2. ElliPro B-Cell Epitopes Prediction
2.3. Prediction of MHCI and MHCII Epitopes Surface Protein of COVID 19
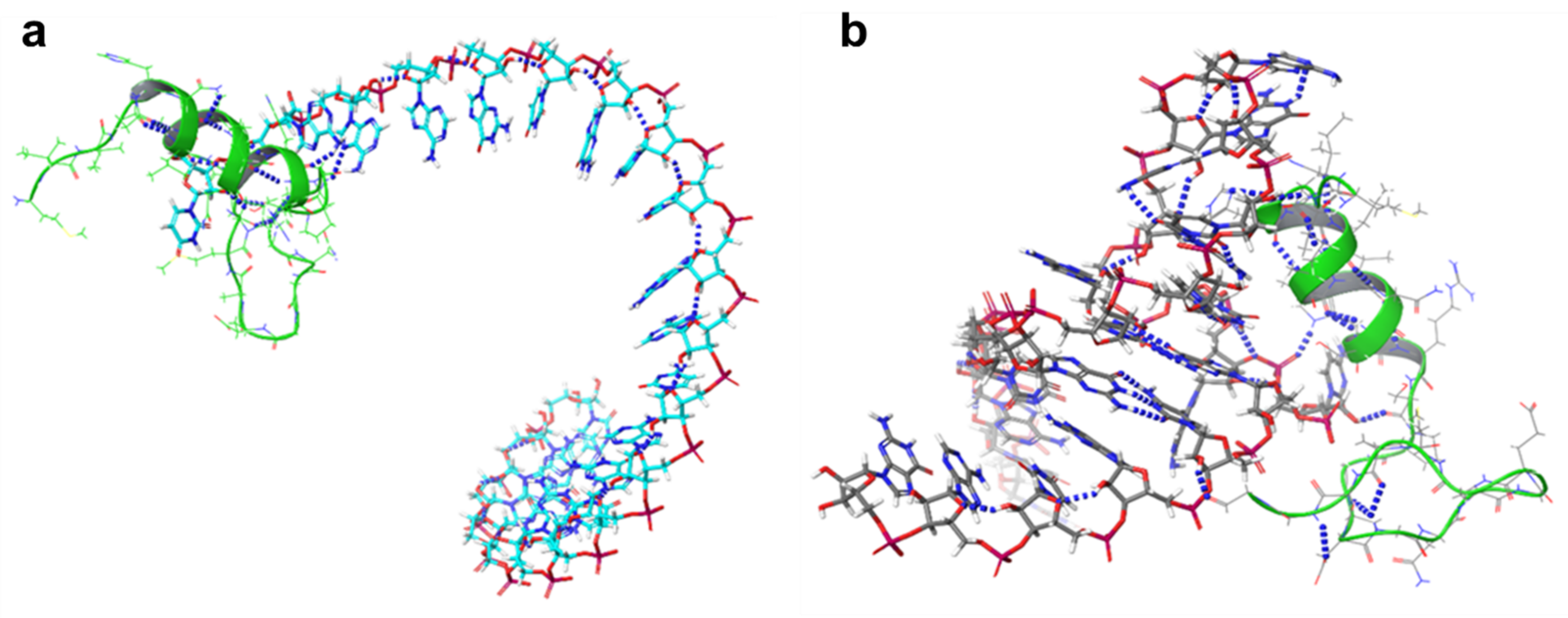
2.4. 3D Structure Prediction of miRNAs
2.5. The microRNA Interaction Network
- Protein and RNA structures were imported into the Schrödinger Maestro software.
- Protein–RNA interface was identified using the SiteMap tool.
- The binding energy of the protein–RNA complex was calculated using the Glide docking tool.
- The results of the docking were analyzed to identify the key interactions between the protein and RNA.
2.6. Association between the Nourin’s miRNAs and Hypoxia
3. Discussion
4. Methods
4.1. D Homology Model Build of Nourin Prediction
4.2. Energy Minimization and Validation
4.3. Model Optimization
4.4. Prediction of Antigenic Part
4.4.1. B-Cell Antigenic Part Prediction of Nourin
4.4.2. B-Cell Antibody Epitope Prediction Using ElliPro of Nourin
4.5. TepiTool: A Computational Prediction Pipeline for T-Cell Epitopes of Nourin
4.6. Prediction of 3D Structure of miR-3D
4.7. microRNA Interaction Network
4.8. Protein–RNA Interaction Prediction
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Roth, G.A.; Mensah, G.A.; Johnson, C.O.; Addolorato, G.; Ammirati, E.; Baddour, L.M.; Barengo, N.C.; Beaton, A.Z.; Benjamin, E.J.; Benziger, C.P.; et al. Global Burden of Cardiovascular Diseases and Risk Factors, 1990–2019: Update From the GBD 2019 Study. J. Am. Coll. Cardiol. 2020, 76, 2982–3021. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Gao, Y.; Li, G. Preclinical multi-target strategies for myocardial ischemia-reperfusion injury. Front. Cardiovasc. Med. 2022, 9, 967115. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Wang, L.; Wang, S.; Cheng, H.; Xu, L.; Pei, G.; Wang, Y.; Fu, C.; Jiang, Y.; He, C.; et al. Signaling pathways and targeted therapy for myocardial infarction. Signal Transduct. Target. Ther. 2022, 7, 78. [Google Scholar] [CrossRef] [PubMed]
- Ala, U. Competing Endogenous RNAs, Non-Coding RNAs and Diseases: An Intertwined Story. Cells 2020, 9, 1574. [Google Scholar] [CrossRef]
- Marinescu, M.C.; Lazar, A.L.; Marta, M.M.; Cozma, A.; Catana, C.S. Non-Coding RNAs: Prevention, Diagnosis, and Treatment in Myocardial Ischemia-Reperfusion Injury. Int. J. Mol. Sci. 2022, 23, 2728. [Google Scholar] [CrossRef]
- Elgebaly, S.A.; Poston, R.; Todd, R.; Helmy, T.; Almaghraby, A.M.; Elbayoumi, T.; Kreutzer, D.L. Cyclocreatine protects against ischemic injury and enhances cardiac recovery during early reperfusion. Expert Rev. Cardiovasc. Ther. 2019, 17, 683–697. [Google Scholar] [CrossRef]
- Elgebaly, S.A.; Van Buren, C.; Todd, R.; Poston, R.; Arafa, R.K.; El-Khazragy, N.; Kreutzer, D.; Rabie, M.A.; Mohamed, A.F.; Ahmed, L.A.; et al. Cyclocreatine Phosphate: A Novel Bioenergetic/Anti-Inflammatory Drug That Resuscitates Poorly Functioning Hearts and Protects against Development of Heart Failure. Pharmaceuticals 2023, 16, 453. [Google Scholar] [CrossRef]
- Elgebaly, S.A.; Todd, R.; Kreutzer, D.L.; Christenson, R.; El-Khazragy, N.; Arafa, R.K.; Rabie, M.A.; Mohamed, A.F.; Ahmed, L.A.; El Sayed, N.S. Nourin-Associated miRNAs: Novel Inflammatory Monitoring Markers for Cyclocreatine Phosphate Therapy in Heart Failure. Int. J. Mol. Sci. 2021, 22, 3575. [Google Scholar] [CrossRef]
- Elgebaly, S.A.; Christenson, R.H.; Kandil, H.; El-Khazragy, N.; Rashed, L.; Yacoub, B.; Eldeeb, H.; Ali, M.; Sharafieh, R.; Klueh, U.; et al. Nourin-Dependent miR-137 and miR-106b: Novel Early Inflammatory Diagnostic Biomarkers for Unstable Angina Patients. Biomolecules 2021, 11, 368. [Google Scholar] [CrossRef]
- Elgebaly, S.A.; Christenson, R.H.; Kandil, H.; Ibrahim, M.; Rizk, H.; El-Khazragy, N.; Rashed, L.; Yacoub, B.; Eldeeb, H.; Ali, M.M.; et al. Nourin-Dependent miR-137 and miR-106b: Novel Biomarkers for Early Diagnosis of Myocardial Ischemia in Coronary Artery Disease Patients. Diagnostics 2021, 11, 703. [Google Scholar] [CrossRef]
- Elgebaly, S.A.; Christenson, R.H.; Azrin, M.; Rizk, H.; El-Khazragy, N.; Kreutzer, D.; Van Buren, C.; Poston, R. Nourin Mirnas: Novel Blood Biomarkers for Early Identification or Exclusion of Myocardial Ischemia in Women Suspected of Having Coronary Artery Disease (CAD). Circulation 2022, 146, A9773. [Google Scholar] [CrossRef]
- Elgebaly, S.A.; Christenson, R.H.; Kandil, H.; El-Khazragy, N.; Rashed, L.; Yacoub, B.; Sharafieh, R.; Klueh, U.; Kreutzer, D.L. Nourin-dependent miRNA-106b: A Novel Early Inflammatory Diagnostic Biomarker for Cardiac Injury. Circulation 2020, 142, A13103. [Google Scholar] [CrossRef]
- Elgebaly, S.A.; Christenson, R.H.; Kandil, H.; El-Khazragy, N.; Rashed, L.; Yacoub, B.; Sharafieh, R.; Klueh, U.; Kreutzer, D.L. Nourin-dependent miRNA-137: A Novel Early Diagnostic Biomarker for Unstable Angina Patients. Circulation 2020, 142, A13051. [Google Scholar] [CrossRef]
- Shoaib, M.; Bahadur, A.; Iqbal, S.; Al-Anazy, M.M.; Laref, A.; Tahir, M.A.; Channar, P.A.; Noreen, S.; Yasir, M.; Iqbal, A.; et al. Magnesium doped mesoporous bioactive glass nanoparticles: A promising material for apatite formation and mitomycin c delivery to the MG-63 cancer cells. J. Alloys Compd. 2021, 866, 159013. [Google Scholar] [CrossRef]
- Khan, S.; Iqbal, S.; Taha, M.; Rahim, F.; Shah, M.; Ullah, H.; Bahadur, A.; Alrbyawi, H.; Dera, A.A.; Alahmdi, M.I.; et al. Synthesis, In Vitro Biological Evaluation and In Silico Molecular Docking Studies of Indole Based Thiadiazole Derivatives as Dual Inhibitor of Acetylcholinesterase and Butyrylchloinesterase. Molecules 2022, 27, 7368. [Google Scholar] [CrossRef]
- Liang, M.L.; Hsieh, T.H.; Ng, K.H.; Tsai, Y.N.; Tsai, C.F.; Chao, M.E.; Liu, D.J.; Chu, S.S.; Chen, W.; Liu, Y.R.; et al. Downregulation of miR-137 and miR-6500-3p promotes cell proliferation in pediatric high-grade gliomas. Oncotarget 2016, 7, 19723–19737. [Google Scholar] [CrossRef]
- Thomas, K.T.; Anderson, B.R.; Shah, N.; Zimmer, S.E.; Hawkins, D.; Valdez, A.N.; Gu, Q.; Bassell, G.J. Inhibition of the Schizophrenia-Associated MicroRNA miR-137 Disrupts Nrg1α Neurodevelopmental Signal Transduction. Cell Rep. 2017, 20, 1–12. [Google Scholar] [CrossRef]
- Moustafa, R.I.; Faraag, A.H.I.; El-Shenawy, R.; Agwa, M.M.; Elsayed, H. Harnessing immunoinformatics for developing a multiple-epitope peptide-based vaccination approach against SARS-CoV-2 spike protein. Saudi J. Biol. Sci. 2023, 30, 103661. [Google Scholar] [CrossRef]
- Paul, S.; Sidney, J.; Sette, A.; Peters, B. TepiTool: A Pipeline for Computational Prediction of T Cell Epitope Candidates. Curr. Protoc. Immunol. 2016, 114, 18–19. [Google Scholar] [CrossRef]
- Li, D.M.; Chen, Q.D.; Wei, G.N.; Wei, J.; Yin, J.X.; He, J.H.; Ge, X.; Shi, Z.M. Hypoxia-Induced miR-137 Inhibition Increased Glioblastoma Multiforme Growth and Chemoresistance Through LRP6. Front. Oncol. 2020, 10, 611699. [Google Scholar] [CrossRef]
- Mailloux, R.J. An Update on Mitochondrial Reactive Oxygen Species Production. Antioxidans 2020, 9, 472. [Google Scholar] [CrossRef]
- Li, H.; Zhu, Z.; Liu, J.; Wang, J.; Qu, C. MicroRNA-137 regulates hypoxia-induced retinal ganglion cell apoptosis through Notch1. Int. J. Mol. Med. 2018, 41, 1774–1782. [Google Scholar] [CrossRef]
- Ghafouri-Fard, S.; Abak, A.; Shoorei, H.; Mohaqiq, M.; Majidpoor, J.; Sayad, A.; Taheri, M. Regulatory role of microRNAs on PTEN signaling. Biomed. Pharmacother. 2021, 133, 110986. [Google Scholar] [CrossRef] [PubMed]
- Hu, Q.; Yuan, Y.; Wu, Y.; Huang, Y.; Zhao, Z.; Xiao, C. MicroRNA-137 exerts protective effects on hypoxia-induced cell injury by inhibiting autophagy/mitophagy and maintaining mitochondrial function in breast cancer stem-like cells. Oncol. Rep. 2020, 44, 1627–1637. [Google Scholar] [CrossRef] [PubMed]
- Raso, A.; Dirkx, E.; Sampaio-Pinto, V.; el Azzouzi, H.; Cubero, R.J.; Sorensen, D.W.; Ottaviani, L.; Olieslagers, S.; Huibers, M.M.; de Weger, R.; et al. A microRNA program regulates the balance between cardiomyocyte hyperplasia and hypertrophy and stimulates cardiac regeneration. Nat. Commun. 2021, 12, 4808. [Google Scholar] [CrossRef] [PubMed]
- Schulte, C.; Zeller, T. microRNA-based diagnostics and therapy in cardiovascular disease—Summing up the facts. Cardiovasc. Diagn. Ther. 2015, 5, 17–36. [Google Scholar] [CrossRef]
- Zhao, T.; Qiu, Z.; Gao, Y. MiR-137-3p exacerbates the ischemia-reperfusion injured cardiomyocyte apoptosis by targeting KLF15. Naunyn-Schmiedebergös Arch. Pharmacol. 2020, 393, 1013–1024. [Google Scholar] [CrossRef]
- Li, C.M.; Shen, S.W.; Wang, T.; Zhang, X.H. Myocardial ischemic post-conditioning attenuates ischemia reperfusion injury via PTEN/Akt signal pathway. Int. J. Clin. Exp. Med. 2015, 8, 15801–15807. [Google Scholar]
- Liang, T.; Gao, F.; Chen, J. Role of PTEN-less in cardiac injury, hypertrophy and regeneration. Cell. Regen. 2021, 10, 25. [Google Scholar] [CrossRef]
- Chen, C.-Y.; Chen, J.; He, L.; Stiles, B.L. PTEN: Tumor Suppressor and Metabolic Regulator. Front. Endocrinol. 2018, 9, 338. [Google Scholar] [CrossRef]
- El-Khazragy, N.; Gaballah, A.; Bakkar, A.; Hemida, E.H.A.; Samir, N.; Tarek, M.; Adly, H.M.; Saleh, S.A.K.; Hanna, D.H. PTEN rs701848 Polymorphism is Associated with Trastuzumab Resistance in HER2-positive Metastatic Breast Cancer and Predicts Progression-free Survival. Clin. Breast Cancer 2023, 23, e131–e139. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Q.; Zhang, Y. Small RNA modifications: Regulatory molecules and potential applications. J. Hematol. Oncol. 2023, 16, 64. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.-J.; Wu, J.; Shi, L.; Li, X.-X.; Zhu, L.; Sun, X.; Qian, J.-Y.; Wang, Y.; Wei, J.-F.; Ding, Q. PTEN expression is upregulated by a RNA-binding protein RBM38 via enhancing its mRNA stability in breast cancer. J. Exp. Clin. Cancer Res. 2017, 36, 149. [Google Scholar] [CrossRef] [PubMed]
- Mendonça, J.T. Schrödinger–Newton Model with a Background. Symmetry 2021, 13, 1007. [Google Scholar] [CrossRef]
- Suvaithenamudhan, S.; Ananth, S.; Mariappan, V.; Dhayabaran, V.V.; Parthasarathy, S.; Ganesh, P.S.; Shankar, E.M. In Silico Evaluation of Bioactive Compounds of Artemisia pallens Targeting the Efflux Protein of Multidrug-Resistant Acinetobacter baumannii (LAC-4 Strain). Molecules 2022, 27, 5188. [Google Scholar] [CrossRef] [PubMed]
- Emiliani, Y.; Muzi, G.; Sánchez, A.; Sánchez, J.; Munera, M. Prediction of molecular mimicry between proteins from Trypanosoma sp. and human antigens associated with systemic lupus erythematosus. Microb. Pathog. 2022, 172, 105760. [Google Scholar] [CrossRef] [PubMed]
- Kolaskar, A.S.; Tongaonkar, P.C. A semi-empirical method for prediction of antigenic determinants on protein antigens. FEBS Lett. 1990, 276, 172–174. [Google Scholar] [CrossRef]
- Chen, H.-Z.; Tang, L.-L.; Yu, X.-L.; Zhou, J.; Chang, Y.-F.; Wu, X. Bioinformatics analysis of epitope-based vaccine design against the novel SARS-CoV-2. Infect. Dis. Poverty 2020, 9, 88. [Google Scholar] [CrossRef]
- Agarwal, V.; Tiwari, A.; Varadwaj, P. Prediction of suitable T and B cell epitopes for eliciting immunogenic response against SARS-CoV-2 and its mutant. Netw. Model Anal. Health Inf. Bioinform. 2022, 11, 1. [Google Scholar] [CrossRef]
- Jespersen, M.C.; Peters, B.; Nielsen, M.; Marcatili, P. BepiPred-2.0: Improving sequence-based B-cell epitope prediction using conformational epitopes. Nucleic Acids Res. 2017, 45, W24–W29. [Google Scholar] [CrossRef]
- Bukhari, S.N.H.; Jain, A.; Haq, E.; Mehbodniya, A.; Webber, J. Machine Learning Techniques for the Prediction of B-Cell and T-Cell Epitopes as Potential Vaccine Targets with a Specific Focus on SARS-CoV-2 Pathogen: A Review. Pathogens 2022, 11, 146. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Duan, X.; Yang, J.; Wang, H.; Liu, F.; Xu, X.; Pan, S. Effects of high hydrostatic pressure and thermal treatment on texture properties of pickled kohlrabi. LWT 2022, 157, 113078. [Google Scholar] [CrossRef]
- Behera, S.K.; Sabarinath, T.; Mishra, P.K.K.; Deneke, Y.; Kumar, A.; Chandrasekar, S.; Senthilkumar, K.; Verma, M.R.; Ganesh, B.; Gurav, A.; et al. Immunoinformatic Study of Recombinant LigA/BCon1-5 Antigen and Evaluation of Its Diagnostic Potential in Primary and Secondary Binding Tests for Serodiagnosis of Porcine Leptospirosis. Pathogens 2021, 10, 1082. [Google Scholar] [CrossRef] [PubMed]
- Paul, S.; Weiskopf, D.; Angelo, M.A.; Sidney, J.; Peters, B.; Sette, A. HLA class I alleles are associated with peptide-binding repertoires of different size, affinity, and immunogenicity. J. Immunol. 2013, 191, 5831–5839. [Google Scholar] [CrossRef]
- Guan, Y.J.; Yu, C.Q.; Li, L.P.; You, Z.H.; Ren, Z.H.; Pan, J.; Li, Y.C. BNEMDI: A Novel MicroRNA-Drug Interaction Prediction Model Based on Multi-Source Information With a Large-Scale Biological Network. Front. Genet. 2022, 13, 919264. [Google Scholar] [CrossRef]
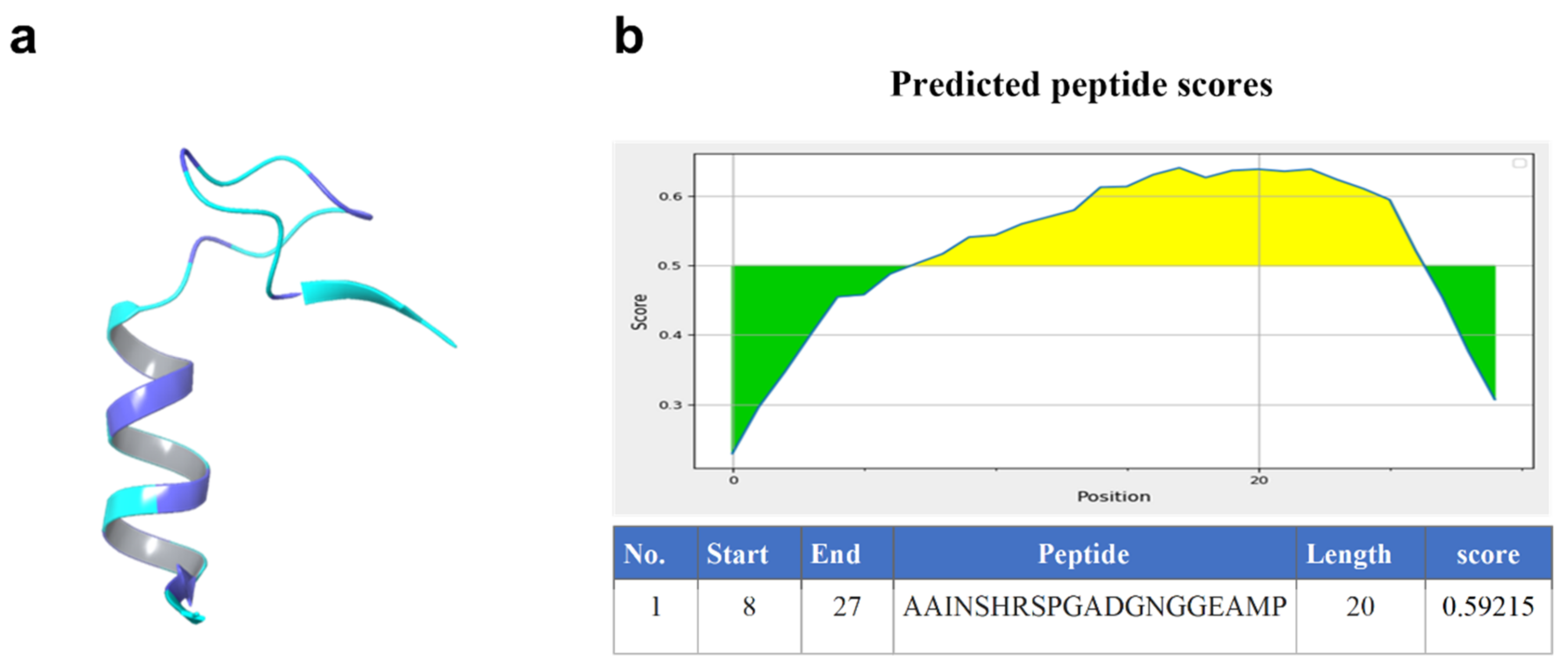
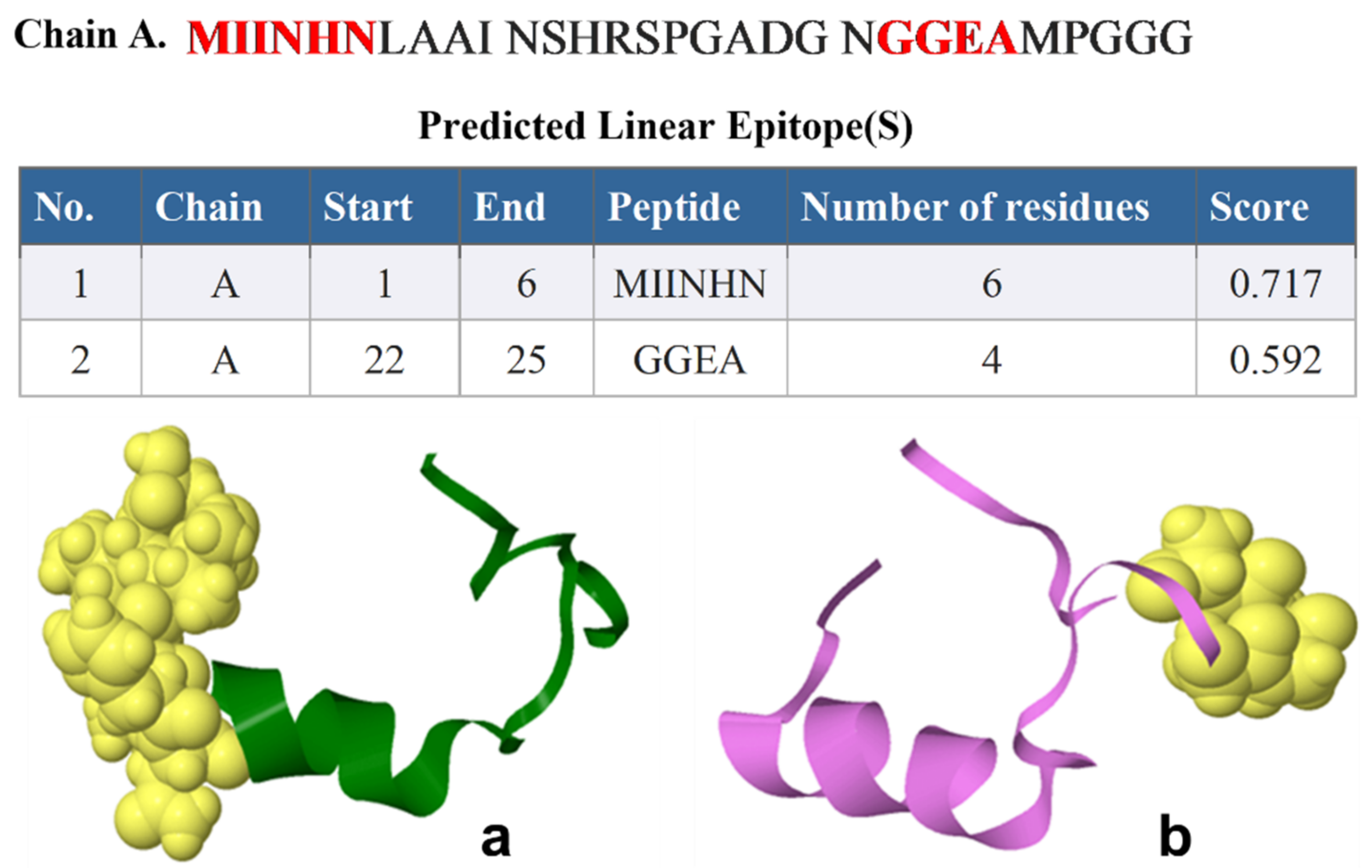
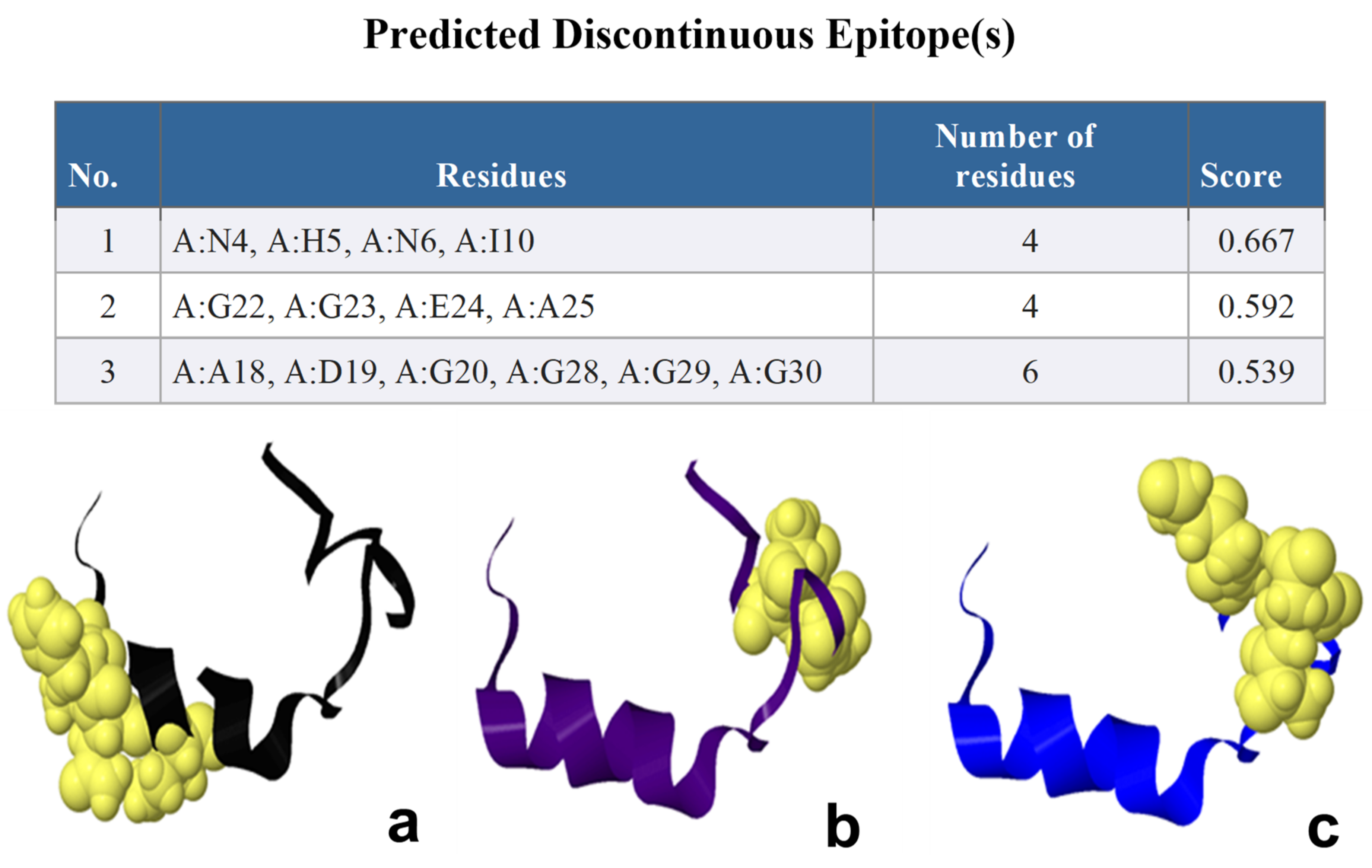
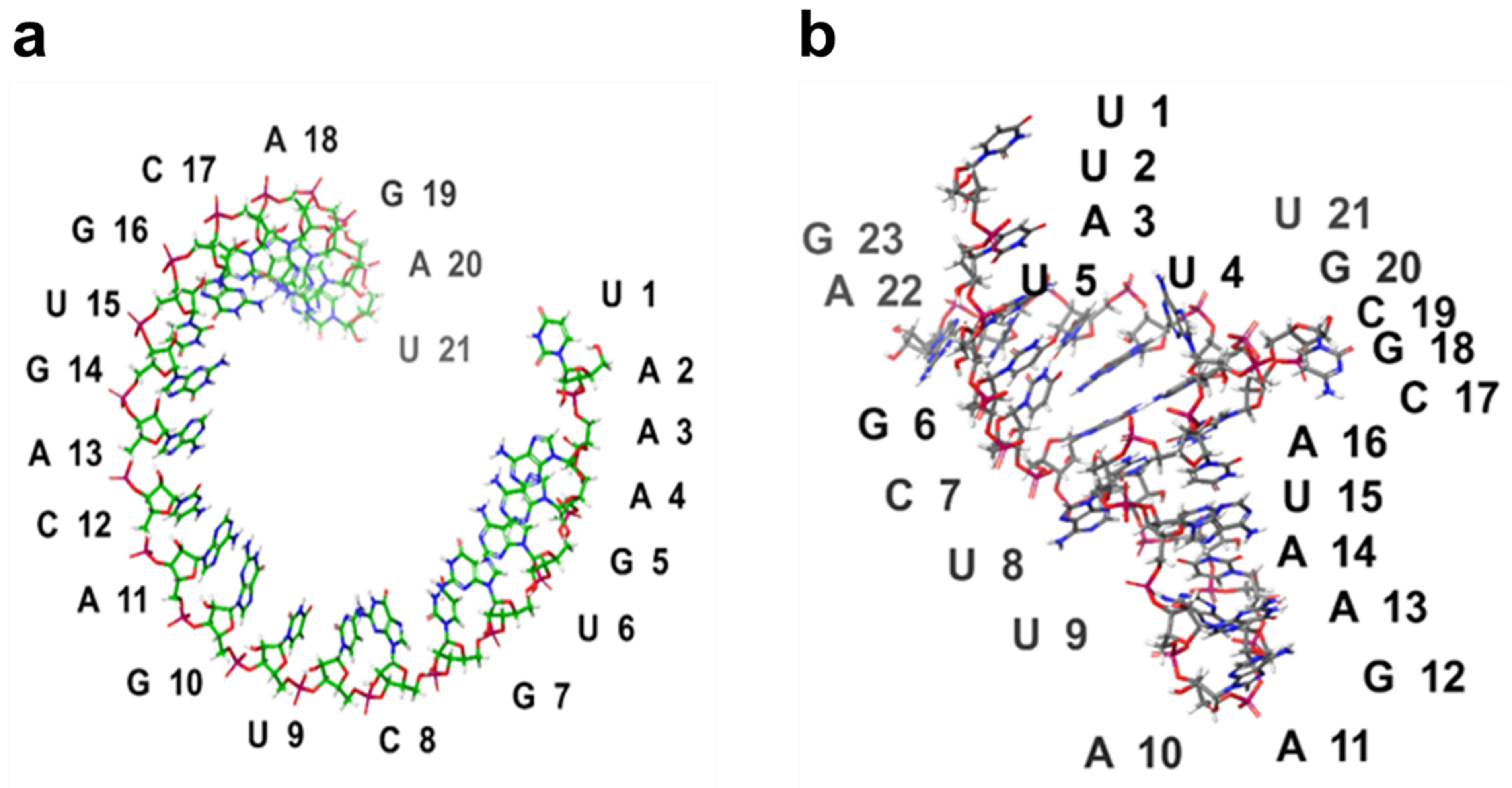


| Rank | PDB Hit | TM-Score | RMSDa | IDENa | Cov |
|---|---|---|---|---|---|
| 1 | 7m2w | 0.552 | 2.33 | 0.069 | 0.967 |
| 2 | 6xb9 | 0.543 | 2.02 | 0.107 | 0.933 |
| 3 | 4lfe | 0.533 | 2.46 | 0.069 | 0.967 |
| 4 | 2gm6 | 0.531 | 1.98 | 0.143 | 0.933 |
| 5 | 7jts | 0.530 | 2.25 | 0.034 | 0.933 |
| 6 | 5d1v | 0.528 | 2.34 | 0.172 | 0.933 |
| 7 | 1d8c | 0.526 | 2.29 | 0.100 | 0.933 |
| 8 | 5we0 | 0.524 | 2.21 | 0.034 | 0.933 |
| 9 | 6g8g | 0.523 | 1.98 | 0.071 | 0.900 |
| 10 | 6g4j | 0.521 | 2.41 | 0.069 | 0.967 |
| Name | E-Value | Score | Identity % | Positive % | Gaps % | Description |
|---|---|---|---|---|---|---|
| 6T17 | 4.92627 | 49 | 64.28571 | 85.714 | 0 | Chain A, Flagellin [Kurthia sp. 11kri321] |
| 2Q01 | 4.51369 | 49 | 58.82353 | 70.588 | 5.88235 | Crystal structure of glucuronate isomerase from Caulobacter crescentus [Caulobacter vibrioides CB15] |
| 1TTU | 9.99837 | 47 | 52.63158 | 52.631 | 0 | Crystal Structure of CSL bound to DNA [Caenorhabditis elegans] |
| 2FO1 | 9.99837 | 47 | 52.63158 | 52.631 | 0 | Crystal Structure of the CSL-Notch-Mastermind ternary complex bound to DNA [Caenorhabditis elegans] |
| 3BRD | 9.99837 | 47 | 52.63158 | 52.631 | 0 | CSL (Lag-1) bound to DNA with Lin-12 RAM peptide, P212121 [Caenorhabditis elegans] |
| 6YW5 | 3.93593 | 50 | 50 | 59.090 | 0 | Chain ZZ, 37S ribosomal protein S24, mitochondrial [Neurospora crassa OR74A] |
| 7NKG | 7.55585 | 48 | 50 | 72.222 | 0 | Chain C, Methyl-coenzyme M reductase gamma subunit [Methermicoccus shengliensis DSM 18856] |
| 1DE3 | 0.422652 | 56 | 40.74074 | 62.962 | 0 | Chain A, RIBONUCLEASE ALPHA-SARCIN [Aspergillus giganteus] |
| 1R4Y | 0.84064 | 54 | 40.74074 | 62.962 | 0 | Chain A, Ribonuclease ALPHA-SARCIN [Aspergillus giganteus] |
| Allele | # | Start | End | Length | Peptide | Core | Icore | Score | Percentile Rank |
|---|---|---|---|---|---|---|---|---|---|
| HLA A*68:01 | 1 | 6 | 14 | 9 | NLAAINSHR | NLAAINSHR | NLAAINSHR | 0.781327 | 0.22 |
| HLA-A*33:01 | 1 | 6 | 14 | 9 | NLAAINSHR | NLAAINSHR | NLAAINSHR | 0.576481 | 0.12 |
| HLA-A*68:01 | 1 | 5 | 14 | 10 | HNLAAINSHR | HLAAINSHR | HNLAAINSHR | 0.431656 | 0.77 |
| HLA-A*31:01 | 1 | 6 | 14 | 9 | NLAAINSHR | NLAAINSHR | NLAAINSHR | 0.361862 | 0.62 |
| HLA-A*02:03 | 1 | 2 | 10 | 9 | IINHNLAAI | IINHNLAAI | IINHNLAAI | 0.280018 | 0.53 |
| HLA-A*31:01 | 1 | 5 | 14 | 10 | HNLAAINSHR | HLAAINSHR | HNLAAINSHR | 0.230259 | 1.1 |
| HLA-A*33:01 | 1 | 5 | 14 | 10 | HNLAAINSHR | HLAAINSHR | HNLAAINSHR | 0.206776 | 0.69 |
| HLA-A*02:06 | 1 | 1 | 9 | 9 | MIINHNLAA | MIINHNLAA | MIINHNLAA | 0.164266 | 1.1 |
| 1 | 15 | 29 | SPGADGNGGEAMPGG | 0.0002 | HLA-DRB1*11:01 |
| 1 | 15 | 29 | SPGADGNGGEAMPGG | 0.0004 | HLA-DRB1*08:02 |
| 1 | 15 | 29 | SPGADGNGGEAMPGG | 0.0005 | HLA-DQA1*01:01/DQB1*05:01 |
| 1 | 15 | 29 | SPGADGNGGEAMPGG | 0.0005 | HLA-DRB1*04:05 |
| 1 | 9 | 23 | AINSHRSPGADGNGG | 0.0006 | HLA-DQA1*03:01/DQB1*03:02 |
| 1 | 15 | 29 | SPGADGNGGEAMPGG | 0.0007 | HLA-DPA1*02:01/DPB1*01:01 |
| 1 | 15 | 29 | SPGADGNGGEAMPGG | 0.0007 | HLA-DPA1*02:01/DPB1*05:01 |
| 1 | 15 | 29 | SPGADGNGGEAMPGG | 0.0007 | HLA-DRB1*04:01 |
| 1 | 9 | 23 | AINSHRSPGADGNGG | 0.0007 | HLA-DRB3*01:01 |
| 1 | 15 | 29 | SPGADGNGGEAMPGG | 0.0008 | HLA-DRB1*15:01 |
| miRNA | Accession Number | Sequence (5′–3′) |
|---|---|---|
| miR-137 | >hsa-miR-137 MI0000454 | UUAUUGCUUAAGAAUACGCGUAG |
| miR-106b | >hsa-miR-106b MI0000734 | UAAAGUGCUGACAGUGCAGAU |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Elgebaly, S.A.; Peacock, W.F.; Christenson, R.H.; Kreutzer, D.L.; Faraag, A.H.I.; Sarguos, A.M.M.; El-Khazragy, N. Integrated Bioinformatics Analysis Confirms the Diagnostic Value of Nourin-Dependent miR-137 and miR-106b in Unstable Angina Patients. Int. J. Mol. Sci. 2023, 24, 14783. https://doi.org/10.3390/ijms241914783
Elgebaly SA, Peacock WF, Christenson RH, Kreutzer DL, Faraag AHI, Sarguos AMM, El-Khazragy N. Integrated Bioinformatics Analysis Confirms the Diagnostic Value of Nourin-Dependent miR-137 and miR-106b in Unstable Angina Patients. International Journal of Molecular Sciences. 2023; 24(19):14783. https://doi.org/10.3390/ijms241914783
Chicago/Turabian StyleElgebaly, Salwa A., W. Frank Peacock, Robert H. Christenson, Donald L. Kreutzer, Ahmed Hassan Ibrahim Faraag, Amir Mahfouz Mokhtar Sarguos, and Nashwa El-Khazragy. 2023. "Integrated Bioinformatics Analysis Confirms the Diagnostic Value of Nourin-Dependent miR-137 and miR-106b in Unstable Angina Patients" International Journal of Molecular Sciences 24, no. 19: 14783. https://doi.org/10.3390/ijms241914783
APA StyleElgebaly, S. A., Peacock, W. F., Christenson, R. H., Kreutzer, D. L., Faraag, A. H. I., Sarguos, A. M. M., & El-Khazragy, N. (2023). Integrated Bioinformatics Analysis Confirms the Diagnostic Value of Nourin-Dependent miR-137 and miR-106b in Unstable Angina Patients. International Journal of Molecular Sciences, 24(19), 14783. https://doi.org/10.3390/ijms241914783









