Ecdysteroid Biosynthesis Halloween Gene Spook Plays an Important Role in the Oviposition Process of Spider Mite, Tetranychus urticae
Abstract
:1. Introduction
2. Results
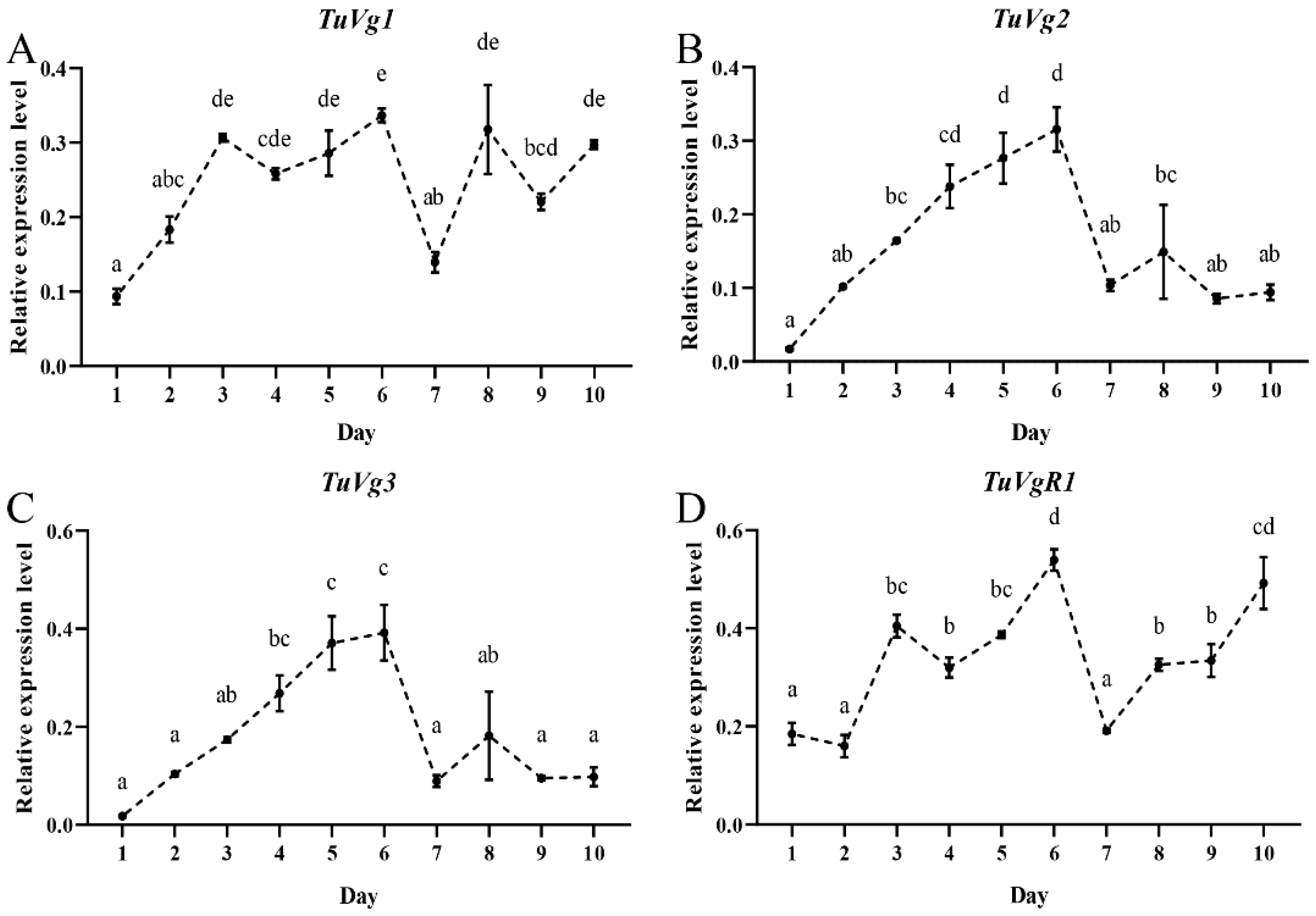
2.1. Expression Dynamics of the Halloween Genes in the Oviposition Period
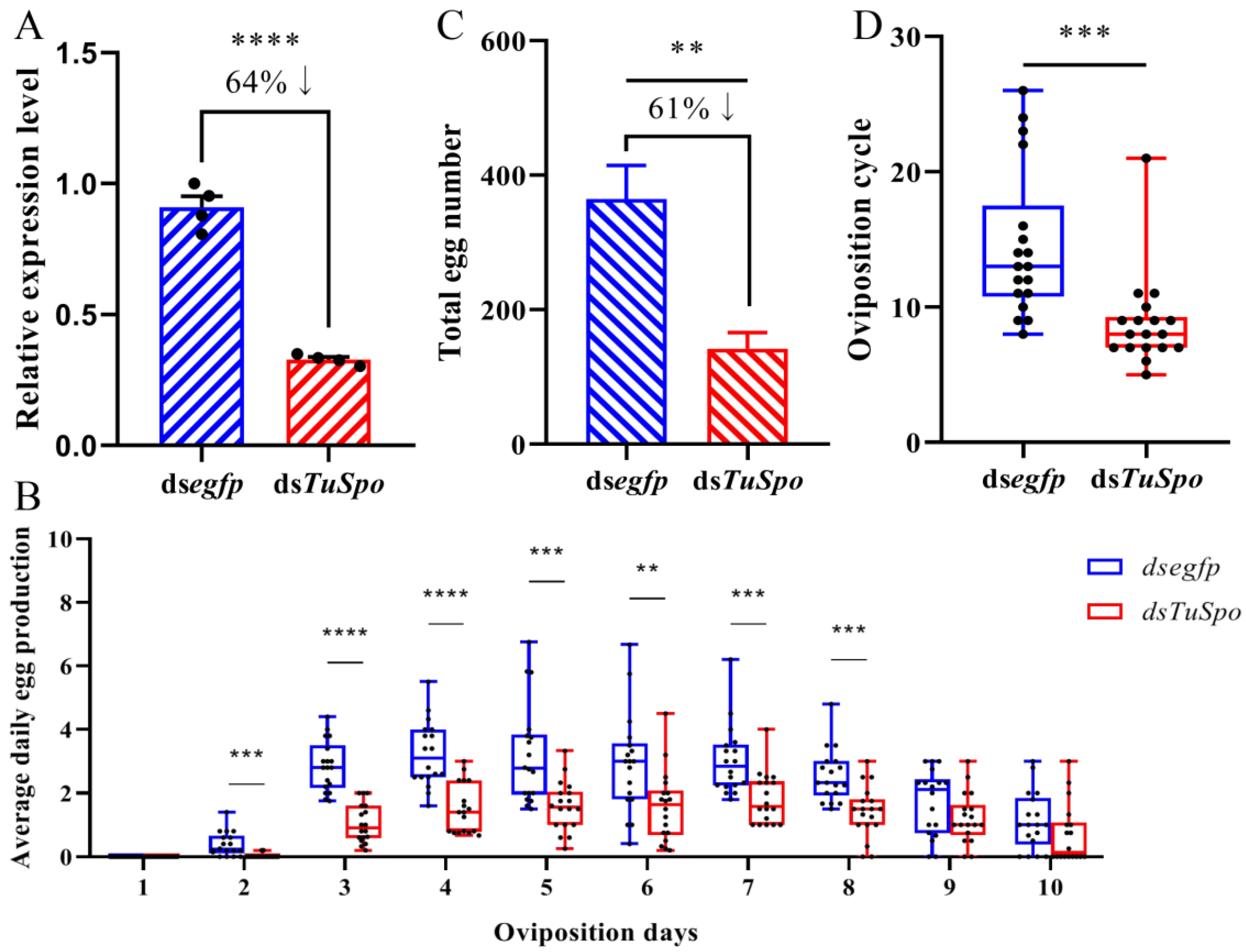
2.2. RNAi of Halloween Gene Spook Reduced the Female Oviposition of T. urticae
2.3. Effect of Halloween Gene Spook Silencing on the Transcript Levels of Other Genes Downstream
3. Discussion
4. Materials and Methods
4.1. Culturing of Mites and Oviposition Statistics
4.2. Expression Dynamics of the Halloween Genes in the Oviposition Period
4.3. RNAi of TuSpo
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Niwa, R.; Niwa, Y.S. Enzymes for ecdysteroid biosynthesis: Their biological functions in insects and beyond. Biosci. Biotechnol. Biochem. 2014, 78, 1283–1292. [Google Scholar] [CrossRef] [PubMed]
- Schwedes, C.C.; Carney, G.E. Ecdysone signalling in adult Drosophila melanogaster. J. Insect Physiol. 2012, 58, 293–302. [Google Scholar] [CrossRef] [PubMed]
- Belles, X.; Piulachs, M.D. Ecdysone signalling and ovarian development in insects: From stem cells to ovarian follicle formation. Biochim. Biophys. Acta 2015, 1849, 181–186. [Google Scholar] [CrossRef] [PubMed]
- Morris, L.X.; Spradling, A.C. Steroid signaling within Drosophila ovarian epithelial cells sex-specifically modulates early germ cell development and meiotic entry. PLoS ONE 2012, 7, e46109. [Google Scholar] [CrossRef] [PubMed]
- Swevers, L. An update on ecdysone signaling during insect oogenesis. Curr. Opin. Insect Sci. 2019, 31, 8–13. [Google Scholar] [CrossRef] [PubMed]
- Parthasarathy, R.; Sheng, Z.; Sun, Z.; Palli, S.R. Ecdysteroid regulation of ovarian growth and oocyte maturation in the red flour beetle, Tribolium castaneum. Insect Biochem. Mol. Biol. 2010, 40, 429–439. [Google Scholar] [CrossRef] [PubMed]
- Swevers, L.; Iatrou, K. The ecdysone regulatory cascade and ovarian development in lepidopteran insects: Insights from the silkmoth paradigm. Insect Biochem. Mol. Biol. 2003, 33, 1285–1297. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Ye, Y.Z.; Ogihara, M.H.; Takeshima, M.; Fujinaga, D.; Liu, C.W.; Zhu, Z.; Kataoka, H.; Bao, Y.Y. Functional analysis of ecdysteroid biosynthetic enzymes of the rice planthopper, Nilaparvata lugens. Insect Biochem. Mol. Biol. 2020, 123, 103428. [Google Scholar] [CrossRef]
- Lenaerts, C.; Van Wielendaele, P.; Peeters, P.; Vanden Broeck, J.; Marchal, E. Ecdysteroid signalling components in metamorphosis and development of the desert locust, Schistocerca gregaria. Insect Biochem. Mol. Biol. 2016, 75, 10–23. [Google Scholar] [CrossRef]
- Jia, S.; Wan, P.J.; Li, G.Q. Molecular cloning and characterization of the putative Halloween gene Phantom from the small brown planthopper Laodelphax striatellus. Insect Sci. 2015, 22, 707–718. [Google Scholar] [CrossRef]
- Jia, S.; Wan, P.J.; Zhou, L.T.; Mu, L.L.; Li, G.Q. Molecular cloning and RNA interference-mediated functional characterization of a Halloween gene spook in the white-backed planthopper Sogatella furcifera. BMC Mol. Biol. 2013, 14, 19. [Google Scholar] [CrossRef] [PubMed]
- Niwa, R.; Matsuda, T.; Yoshiyama, T.; Namiki, T.; Mita, K.; Fujimoto, Y.; Kataoka, H. CYP306A1, a cytochrome P450 enzyme, is essential for ecdysteroid biosynthesis in the prothoracic glands of Bombyx and Drosophila. J. Biol. Chem. 2004, 279, 35942–35949. [Google Scholar] [CrossRef] [PubMed]
- Rewitz, K.F.; O’Connor, M.B.; Gilbert, L.I. Molecular evolution of the insect Halloween family of cytochrome P450s: Phylogeny, gene organization and functional conservation. Insect Biochem. Mol. Biol. 2007, 37, 741–753. [Google Scholar] [CrossRef] [PubMed]
- Rewitz, K.F.; Rybczynski, R.; Warren, J.T.; Gilbert, L.I. The Halloween genes code for cytochrome P450 enzymes mediating synthesis of the insect moulting hormone. Biochem. Soc. Trans. 2006, 34, 1256–1260. [Google Scholar] [CrossRef] [PubMed]
- Schumann, I.; Kenny, N.; Hui, J.; Hering, L.; Mayer, G. Halloween genes in panarthropods and the evolution of the early moulting pathway in Ecdysozoa. Royal Soc. Open Sci. 2018, 5, 180888. [Google Scholar] [CrossRef] [PubMed]
- Marchal, E.; Vandersmissen, H.P.; Badisco, L.; Van de Velde, S.; Van Wielendaele, P.; Verlinden, H.; Iga, M.; Huybrechts, R.; Simonet, G.; Smagghe, G.; et al. Control of ecdysteroidogenesis in prothoracic glands of insects: A review. Peptides 2010, 31, 506–519. [Google Scholar] [CrossRef] [PubMed]
- Christiaens, O.; Iga, M.; Velarde, R.; Rougé, P.; Smagghe, G. Halloween genes and nuclear receptors in ecdysteroid biosynthesis and signaling in the pea aphid. Insect Mol. Biol. 2010, 19, 187–200. [Google Scholar] [CrossRef] [PubMed]
- Grbic, M.; Van Leeuwen, T.; Clark, R.M.; Rombauts, S.; Rouze, P.; Grbic, V.; Osborne, E.J.; Dermauw, W.; Ngoc, P.C.; Ortego, F.; et al. The International spider mite genome consortium, The genome of Tetranychus urticae reveals herbivorous pest adaptations. Nature 2011, 479, 487–492. [Google Scholar] [CrossRef]
- Elgendy, A.M.; Elmogy, M.; Takeda, M. Molecular cloning, characterization, and expression pattern of the Ultraspiracle gene homolog (RXR/USP) from the hemimetabolous insect Periplaneta americana (Dictyoptera, Blattidae) during vitellogenesis. Mol. Biotechnol. 2014, 56, 126–135. [Google Scholar] [CrossRef]
- Hult, E.F.; Huang, J.; Marchal, E.; Lam, J.; Tobe, S.S. RXR/USP and EcR are critical for the regulation of reproduction and the control of JH biosynthesis in Diploptera punctata. J. Insect Physiol. 2015, 80, 48–60. [Google Scholar] [CrossRef]
- Tarrant, A.M.; Behrendt, L.; Stegeman, J.J.; Verslycke, T. Ecdysteroid receptor from the American lobster Homarus americanus: EcR/RXR isoform cloning and ligand-binding properties. Gen. Comp. Endocrinol. 2011, 173, 346–355. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Liu, X.Y.; Smagghe, G.; Niu, J.Z.; Wang, J.J. Molting process revealed by the detailed expression profiles of RXR1/RXR2 and mining the associated genes in a spider mite, Panonychus citri. Insect Sci. 2022, 29, 430–442. [Google Scholar] [CrossRef]
- Li, G.; Liu, X.Y.; Han, X.; Niu, J.Z.; Wang, J.J. RNAi of the nuclear receptor HR3 suggests a role in the molting process of the spider mite Panonychus citri. Exp. Appl. Acarol. 2020, 81, 75–83. [Google Scholar] [CrossRef] [PubMed]
- Schwedes, C.; Tulsiani, S.; Carney, G.E. Ecdysone receptor expression and activity in adult Drosophila melanogaster. J. Insect Physiol. 2011, 57, 899–907. [Google Scholar] [CrossRef] [PubMed]
- Iga, M.; Smagghe, G. Identification and expression profile of Halloween genes involved in ecdysteroid biosynthesis in Spodoptera littoralis. Peptides 2010, 31, 456–467. [Google Scholar] [CrossRef] [PubMed]
- Sugahara, R.; Tanaka, S.; Shiotsuki, T. RNAi-mediated knockdown of Spook reduces ecdysteroid titers and causes precocious metamorphosis in the desert locust Schistocerca gregaria. Dev. Biol. 2017, 429, 71–80. [Google Scholar] [CrossRef] [PubMed]
- Schellens, S.; Lenaerts, C.; Perez Baca, M.D.R.; Cools, D.; Peeters, P.; Marchal, E.; Vanden Broeck, J. Knockdown of the Halloween genes Spook, Shadow and Shade influences oocyte development, egg shape, oviposition and hatching in the desert locust. Int. J. Mol. Sci. 2022, 23, 9232. [Google Scholar] [CrossRef] [PubMed]
- Knapp, E.; Sun, J. Steroid signalling in mature follicles is important for Drosophila ovulation. Proc. Natl. Acad. Sci. USA 2017, 114, 699–704. [Google Scholar] [CrossRef]
- Zhang, C.; Wan, B.; Jin, M.R.; Wang, J.; Xin, T.R.; Zou, Z.W.; Xia, B. The loss of Halloween gene function seriously affects the development and reproduction of Diaphorina citri (Hemiptera: Liviidae) and increases its susceptibility to pesticides. Pestic. Biochem. Physiol. 2023, 191, 105361. [Google Scholar] [CrossRef]
- Ogihara, M.H.; Hikiba, J.; Suzuki, Y.; Taylor, D.; Kataoka, H. Ovarian ecdysteroidogenesis in both immature and mature stages of an Acari, Ornithodoros moubata. PLoS ONE 2015, 10, e0124953. [Google Scholar] [CrossRef]
- Cabrera, A.R.; Shirk, P.D.; Evans, J.D.; Hung, K.; Sims, J.; Alborn, H.; Teal, P.E. Three Halloween genes from the Varroa mite, Varroa destructor (Anderson & Trueman) and their expression during reproduction. Insect Mol. Biol. 2015, 24, 277–292. [Google Scholar]
- Gijbels, M.; Schellens, S.; Schellekens, T.; Bruyninckx, E.; Marchal, E.; Vanden Broeck, J. Precocious downregulation of Kruppel-Homolog 1 in the migratory locust, Locusta migratoria, gives rise to an adultoid phenotype with accelerated ovarian development but disturbed mating and oviposition. Int. J. Mol. Sci. 2020, 21, 6058. [Google Scholar] [CrossRef] [PubMed]
- Friesen, K.J.; Reuben Kaufman, W. Quantification of vitellogenesis and its control by 20-hydroxyecdysone in the ixodid tick, Amblyomma hebraeum. J. Insect Physiol. 2002, 48, 773–782. [Google Scholar] [CrossRef] [PubMed]
- Thompson, D.M.; Khalil, S.M.; Jeffers, L.A.; Ananthapadmanaban, U.; Sonenshine, D.E.; Mitchell, R.D.; Osgood, C.J.; Apperson, C.S.; Michael Roe, R. In vivo role of 20-hydroxyecdysone in the regulation of the vitellogenin mRNA and egg development in the American dog tick, Dermacentor variabilis (Say). J. Insect Physiol. 2005, 51, 1105–1116. [Google Scholar] [CrossRef] [PubMed]
- Van Leeuwen, T.; Vontas, J.; Tsagkarakou, A.; Dermauw, W.; Tirry, L. Acaricide resistance mechanisms in the two-spotted spider mite Tetranychus urticae and other important Acari: A review. Insect Biochem. Mol. Biol. 2010, 40, 563–572. [Google Scholar] [CrossRef] [PubMed]
- Wei, P.; Chen, M.; Nan, C.; Feng, K.; Shen, G.; Cheng, J.; He, L. Downregulation of carboxylesterase contributes to cyflumetofen resistance in Tetranychus cinnabarinus (Boisduval). Pest Manag. Sci. 2019, 75, 2166–2173. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Yang, L.; He, Q.; Zhou, S. Regulatory mechanisms of vitellogenesis in insects. Front. Cell Dev. Biol. 2020, 8, 593613. [Google Scholar] [CrossRef] [PubMed]
- Roy, S.; Saha, T.T.; Zou, Z.; Raikhel, A.S. Regulatory pathways controlling female insect reproduction. Annu. Rev. Entomol. 2018, 63, 489–511. [Google Scholar] [CrossRef]
- Ameku, T.; Niwa, R. Mating-induced increase in germline stem cells via the neuroendocrine system in female Drosophila. PLoS Genet. 2016, 12, e1006123. [Google Scholar] [CrossRef]
- Jang, A.C.; Chang, Y.C.; Bai, J.; Montell, D. Border-cell migration requires integration of spatial and temporal signals by the BTB protein Abrupt. Nat. Cell Biol. 2009, 11, 569–579. [Google Scholar] [CrossRef]
- Mello, T.R.P.; Aleixo, A.C.; Pinheiro, D.G.; Nunes, F.M.F.; Cristino, A.S.; Bitondi, M.M.G.; Barchuk, A.R.; Simoes, Z.L.P. Hormonal control and target genes of Ftz-f1 expression in the honeybee Apis mellifera: A positive loop linking juvenile hormone, Ftz-f1, and vitellogenin. Insect Mol. Biol. 2019, 28, 145–159. [Google Scholar] [CrossRef] [PubMed]
- Tan, Y.A.; Zhao, X.D.; Sun, H.J.; Zhao, J.; Xiao, L.B.; Hao, D.J.; Jiang, Y.P. Phospholipase C gamma (PLCgamma) regulates soluble trehalase in the 20E-induced fecundity of Apolygus lucorum. Insect Sci. 2021, 28, 430–444. [Google Scholar] [CrossRef] [PubMed]
- Tan, Y.A.; Zhao, X.D.; Sun, Y.; Hao, D.J.; Zhao, J.; Jiang, Y.P.; Bai, L.X.; Xiao, L.B. The nuclear hormone receptor E75A regulates vitellogenin gene (Al-Vg) expression in the mirid bug Apolygus lucorum. Insect Mol. Biol. 2018, 27, 188–197. [Google Scholar] [CrossRef]
- Parthasarathy, R.; Palli, S.R. Molecular analysis of nutritional and hormonal regulation of female reproduction in the red flour beetle, Tribolium castaneum. Insect Biochem. Mol. Biol. 2011, 41, 294–305. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Liu, T.Y.; Jiang, H.B.; Liu, X.Q.; Dou, W.; Park, Y.; Smagghe, G.; Wang, J.J. The ecdysis triggering hormone system, via ETH/ETHR-B, is essential for successful reproduction of a major pest insect, Bactrocera dorsalis (Hendel). Front. Physiol. 2019, 10, 151. [Google Scholar] [CrossRef] [PubMed]
- Friesen, K.J.; Kaufman, W.R. Effects of 20-hydroxyecdysone and other hormones on egg development, and identification of a vitellin-binding protein in the ovary of the tick, Amblyomma hebraeum. J. Insect Physiol. 2004, 50, 519–529. [Google Scholar] [CrossRef] [PubMed]
- Seixas, A.; Friesen, K.J.; Kaufman, W.R. Effect of 20-hydroxyecdysone and haemolymph on oogenesis in the ixodid tick Amblyomma hebraeum. J. Insect Physiol. 2008, 54, 1175–1183. [Google Scholar] [CrossRef]
- Marchal, E.; Badisco, L.; Verlinden, H.; Vandersmissen, T.; Van Soest, S.; Van Wielendaele, P.; Vanden Broeck, J. Role of the Halloween genes, Spook and Phantom in ecdysteroidogenesis in the desert locust, Schistocerca gregaria. J. Insect Physiol. 2011, 57, 1240–1248. [Google Scholar] [CrossRef]
- Peng, L.; Wang, L.; Zou, M.M.; Vasseur, L.; Chu, L.N.; Qin, Y.D.; Zhai, Y.L.; You, M.S. Identification of Halloween genes and RNA interference-mediated functional characterization of a Halloween gene Shadow in Plutella xylostella. Front. Physiol. 2019, 10, 1120. [Google Scholar] [CrossRef]
- Hu, K.; Fu, B.; Wang, C.; Liu, J.; Tang, Y.; Zhang, W.; Zhu, J.; Li, Y.; Pan, Q.; Liu, F. The role of 20E biosynthesis relative gene Shadow in the reproduction of the predatory mirid bug, Cyrtorhinus lividipennis (Hemiptera: Miridae). Arch. Insect Biochem. Physiol. 2022, 109, e21854. [Google Scholar] [CrossRef]
- Liu, Z.; Nanda, S.; Yang, C.; Chen, S.; Guo, M.; Khan, M.M.; Qiu, B.; Zhang, Y.; Zhou, X.; Pan, H. RNAi suppression of the nuclear receptor Ftz-f1 impaired ecdysis, pupation, and reproduction in the 28-spotted potato ladybeetle, Henosepilachna vigintioctopunctata. Pestic. Biochem. Physiol. 2022, 182, 105029. [Google Scholar] [CrossRef] [PubMed]
- Smagghe, G.; Zotti, M.; Retnakaran, A. Targeting female reproduction in insects with biorational insecticides for pest management: A critical review with suggestions for future research. Curr. Opin. Insect Sci. 2019, 31, 65–69. [Google Scholar] [CrossRef] [PubMed]
- Smagghe, G.; Degheele, D. Action of a novel nonsteroidal ecdysteroid mimic, tebufenozide (RH-5992), on insects of different orders. Pestic. Sci. 1994, 42, 85–92. [Google Scholar] [CrossRef]
- Dhadialla, T.S.; Retnakaran, A.; Smagghe, G. Insect growth and development disrupting insecticides. In Insect Control; Gilbert, L.I., Gill, S.S., Eds.; Elsevier/Academic Press: London, UK, 2010; pp. 679–740. [Google Scholar]
- Zotti, M.J.; De Geyter, E.; Swevers, L.; Braz, A.S.K.; Scott, L.P.B.; Rougé, P.; Toledano, J.C.; Guedes, J.V.C.; Grutzmacher, A.D.; Lenardão, E.J.; et al. A cell-based reporter assay for screening for EcR agonist/antagonist activity of natural ecdysteroids in Lepidoptera (Bm5) and Diptera (S2) cell cultures, followed by modeling of EcR interactions and normal modes analysis. Pestic. Biochem. Physiol. 2013, 107, 309–320. [Google Scholar] [CrossRef] [PubMed]
- Fahrbach, S.E.; Smagghe, G.; Velarde, R.A. Insect nuclear receptors. Annu. Rev. Entomol. 2012, 57, 83–106. [Google Scholar] [CrossRef] [PubMed]
- Amdam, G.V.; Simoes, Z.L.; Hagen, A.; Norberg, K.; Schroder, K.; Mikkelsen, O.; Kirkwood, T.B.; Omholt, S.W. Hormonal control of the yolk precursor vitellogenin regulates immune function and longevity in honeybees. Exp. Gerontol. 2004, 39, 767–773. [Google Scholar] [CrossRef] [PubMed]
- Corona, M.; Velarde, R.A.; Remolina, S.; Moran-Lauter, A.; Wang, Y.; Hughes, K.A.; Robinson, G.E. Vitellogenin, juvenile hormone, insulin signaling, and queen honey bee longevity. Pro. Natl. Acad. Sci. USA 2007, 104, 7128–7133. [Google Scholar] [CrossRef] [PubMed]
- Hossain, M.S.; Liu, Y.; Zhou, S.; Li, K.; Tian, L.; Li, S. 20-Hydroxyecdysone-induced transcriptional activity of FoxO upregulates brummer and acid lipase-1 and promotes lipolysis in Bombyx fat body. Insect Biochem. Mol. Biol. 2013, 43, 829–838. [Google Scholar] [CrossRef]
- Antebi, A. Regulation of longevity by the reproductive system. Exp. Gerontol. 2013, 48, 596–602. [Google Scholar] [CrossRef]
- Cagliari, D.; Taning, C.N.T.; Christiaens, O.; De Schutter, K.; Lewille, B.; Dewettinck, K.; Zotti, M.; Smagghe, G. Parental RNA interference as a tool to study genes involved in rostrum development in the Neotropical brown stink bug, Euschistus heros. J. Insect Physiol. 2020, 128, 104161. [Google Scholar] [CrossRef]
- Wei, P.; Wang, C.; Li, C.; Chen, M.; Sun, J.; Van Leeuwen, T.; He, L. Comparing the efficiency of RNAi after feeding and injection of dsRNA in spider mites. Pestic. Biochem. Physiol. 2021, 179, 104966. [Google Scholar] [CrossRef] [PubMed]
- Taning, C.N.T.; Yu, N.; Van Eynde, B.; Ma, S.; Smagghe, G. CRISPR/Cas9 in insects: Applications, best practices and biosafety concerns. J. Insect Physiol. 2017, 98, 245–257. [Google Scholar] [CrossRef]
- Li, J.J.; Shi, Y.; Wu, J.N.; Li, H.; Smagghe, G.; Liu, T.X. CRISPR/Cas9 in Lepidopteran insects: Progress, application and prospects. J. Insect Physiol. 2021, 135, 104325. [Google Scholar] [CrossRef] [PubMed]
- Li, J.J.; Xu, H.M.; Zhao, H.Z.; Pan, M.Z.; Smagghe, G.; Li, Z.Y.; Liu, T.J.; Shi, Y. Regulating role of neuropeptide PTTH releaved in Spodoptera frugiperda using RNAi- and CRISPR/Cas9-based functional genomic tools. Entomol. Gen. 2023, 43, 451–459. [Google Scholar] [CrossRef]
- Li, G.; Sun, Q.Z.; Liu, X.Y.; Zhang, J.; Dou, W.; Niu, J.Z.; Wang, J.J. Expression dynamics of key ecdysteroid and juvenile hormone biosynthesis genes imply a coordinated regulation pattern in the molting process of a spider mite, Tetranychus urticae. Exp. Appl. Acarol. 2019, 78, 361–372. [Google Scholar] [CrossRef]
- Li, G.; Niu, J.; Zotti, M.; Sun, Q.; Zhu, L.; Zhang, J.; Liao, C.; Dou, W.; Wei, D.; Wang, J.J.; et al. Characterization and expression patterns of key ecdysteroid biosynthesis and signaling genes in a spider mite (Panonychus citri). Insect Biochem. Mol. Biol. 2017, 87, 136–146. [Google Scholar] [CrossRef] [PubMed]
- Dermauw, W.; Jonckheere, W.; Riga, M.; Livadaras, I.; Vontas, J.; Van Leeuwen, T. Targeted mutagenesis using CRISPR-Cas9 in the chelicerate herbivore Tetranychus urticae. Insect Biochem. Mol. Biol. 2020, 120, 103347. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, L.; Li, Z.; Yi, T.; Li, G.; Smagghe, G.; Jin, D. Ecdysteroid Biosynthesis Halloween Gene Spook Plays an Important Role in the Oviposition Process of Spider Mite, Tetranychus urticae. Int. J. Mol. Sci. 2023, 24, 14797. https://doi.org/10.3390/ijms241914797
Wang L, Li Z, Yi T, Li G, Smagghe G, Jin D. Ecdysteroid Biosynthesis Halloween Gene Spook Plays an Important Role in the Oviposition Process of Spider Mite, Tetranychus urticae. International Journal of Molecular Sciences. 2023; 24(19):14797. https://doi.org/10.3390/ijms241914797
Chicago/Turabian StyleWang, Liang, Zhuo Li, Tianci Yi, Gang Li, Guy Smagghe, and Daochao Jin. 2023. "Ecdysteroid Biosynthesis Halloween Gene Spook Plays an Important Role in the Oviposition Process of Spider Mite, Tetranychus urticae" International Journal of Molecular Sciences 24, no. 19: 14797. https://doi.org/10.3390/ijms241914797




