The Dual Role of the Innate Immune System in the Effectiveness of mRNA Therapeutics
Abstract
:1. Introduction
2. Innate Immune Response Pathways Recognizing mRNA
2.1. RIG-I and MDA5 Signaling
2.2. TLR Signaling
2.3. NOD-like Receptors
2.4. NLRP3 Inflammasome Activation
2.5. C-Type Lectin Receptors
2.6. The cGAS/STING Pathway
2.7. Endonuclease Activity
3. Applied Aspects: mRNA “Adjuvants” and “Tolerogens”
| Name | Molecular Function | Main Signaling Pathway | Ref. |
|---|---|---|---|
| MyD88 | Adaptor protein | MyD88/TRAF6/NFkB | [285,286] |
| TRIF | Adaptor protein | TRIF/TRAF3/IRF3 | [285,287] |
| IRF1 | Transcriptional factor | IRF1 targets | [288,289] |
| IRF3 | Transcriptional factor | IRF3 targets | [288,290] |
| IRF7 | Transcriptional factor | IRF7 targets | [288,290] |
| HSP70 | Chaperone | HSP70-TLR4-NLRP3 | [291] |
| PD-1 | Membrane receptor | PD-1/SHP2/STAT1/T-bet | [292] |
| MDA5 | Intracellular receptor | MDA5-TBK1-IRF3 | [293] |
| NF-κB | Transcriptional factor | NF-κB targets | [294] |
| T-bet | Transcriptional factor | NF-κB targets | [294,295] |
| RIG-I | Intracellular receptor | RIG-I/TBK1/IRF3 | [293] |
| TBK1 | Kinase | TBK1-IRF3 | [296] |
| HMGB1 | Signaling protein | HMGB1/TLR4/NF-κB | [297,298] |
| DAI/ZBP1 | Transcriptional factor | DAI/ZBP1 targets | [299] |
| GM-CSF | Cytokine | GM-CSF/GM-CSFR | [300,301,302,303] |
| IL-4 | Cytokine | IL4/IL4R | [300] |
| IL-12 | Cytokine | IL-12/IL-12R | [304,305,306,307,308,309,310] |
| IL-2 | Cytokine | IL-2/IL-2R | [304,311,312,313] |
| IL-22 | Cytokine | IL-22/IL-22R | [314] |
| IL-21 | Cytokine | IL-21/IL-21R | [315,316] |
| IL-15 | Cytokine | IL-15/IL-15R | [315,316,317,318,319] |
| IL-6 | Cytokine | IL-6/IL-6R | [319] |
| IL-7 | Cytokine | IL-7/IL-7R | [319] |
| IL-23 | Cytokine | IL-23/IL-23R | [320] |
| CCL3 (MIP-1α) | Chemokine | MIP-1α/CCR1 | [321] |
| CCL20 (MIP-3α) | Chemokine | CCL20/CCR6 | [321] |
| CCL19 (MIP-3β) | Chemokine | CCL19/CCR7 | [321,322] |
| CCL5 (RANTES) | Chemokine | CCL5/CCR5 | [319] |
| CXCL10 (IP-10) | Chemokine | CXCL10/CXCR3 | [323] |
| CCR7 | Membrane receptor | CCL19/CCR7 | [324,325] |
| CCL21 | Chemokine | CCL19/CCR7 | [326] |
| CD80 | Membrane receptor | CD28-CD80 | [327,328] |
| CD86 | Membrane receptor | CD28-CD80 | [327,328,329] |
| CD40L | Cytokine | CD40L/CD40 | [330,331] |
| Flagellin | Bacterial protein | TLR5/MyD88/NF-κB | [281] |
4. Final Remarks
Author Contributions
Funding
Conflicts of Interest
References
- Maruggi, G.; Zhang, C.; Li, J.; Ulmer, J.B.; Yu, D. MRNA as a Transformative Technology for Vaccine Development to Control Infectious Diseases. Mol. Ther. 2019, 27, 757–772. [Google Scholar] [CrossRef] [PubMed]
- Verbeke, R.; Lentacker, I.; De Smedt, S.C.; Dewitte, H. Three Decades of Messenger RNA Vaccine Development. Nano Today 2019, 28, 100766. [Google Scholar] [CrossRef]
- Xu, S.; Yang, K.; Li, R.; Zhang, L. Mrna Vaccine Era—Mechanisms, Drug Platform and Clinical Prospection. Int. J. Mol. Sci. 2020, 21, 6582. [Google Scholar] [CrossRef] [PubMed]
- Fang, E.; Liu, X.; Li, M.; Zhang, Z.; Song, L.; Zhu, B.; Wu, X.; Liu, J.; Zhao, D.; Li, Y. Advances in COVID-19 MRNA Vaccine Development. Signal Transduct. Target. Ther. 2022, 7, 94. [Google Scholar] [CrossRef]
- Prieve, M.G.; Harvie, P.; Monahan, S.D.; Roy, D.; Li, A.G.; Blevins, T.L.; Paschal, A.E.; Waldheim, M.; Bell, E.C.; Galperin, A.; et al. Targeted MRNA Therapy for Ornithine Transcarbamylase Deficiency. Mol. Ther. 2018, 26, 801–813. [Google Scholar] [CrossRef]
- Vannucci, L.; Lai, M.; Chiuppesi, F.; Ceccherini-Nelli, L.; Pistello, M. Viral Vectors: A Look Back and Ahead on Gene Transfer Technology. New Microbiol. 2013, 36, 1–22. [Google Scholar]
- Kafasla, P.; Skliris, A.; Kontoyiannis, D.L. Post-Transcriptional Coordination of Immunological Responses by RNA-Binding Proteins. Nat. Immunol. 2014, 15, 492–502. [Google Scholar] [CrossRef]
- Platnich, J.M.; Muruve, D.A. NOD-like Receptors and Inflammasomes: A Review of Their Canonical and Non-Canonical Signaling Pathways. Arch. Biochem. Biophys. 2019, 670, 4–14. [Google Scholar] [CrossRef]
- Chen, N.; Xia, P.; Li, S.; Zhang, T.; Wang, T.T.; Zhu, J. RNA Sensors of the Innate Immune System and Their Detection of Pathogens. IUBMB Life 2017, 69, 297–304. [Google Scholar] [CrossRef]
- Pfeffer, L.M. The Role of Nuclear Factor Κb in the Interferon Response. J. Interf. Cytokine Res. 2011, 31, 553–559. [Google Scholar] [CrossRef]
- Liu, T.; Zhang, L.; Joo, D.; Sun, S.C. NF-ΚB Signaling in Inflammation. Signal Transduct. Target. Ther. 2017, 2, 17023. [Google Scholar] [CrossRef] [PubMed]
- Tak, P.P.; Firestein, G.S. NF-ΚB: A Key Role in Inflammatory Diseases. J. Clin. Investig. 2001, 107, 7–11. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Lin, L.; Zhang, Z.; Zhang, H.; Hu, H. Targeting NF-ΚB Pathway for the Therapy of Diseases: Mechanism and Clinical Study. Signal Transduct. Target. Ther. 2020, 5, 209. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Ma, C.; Zhang, Z.; Zhang, H.; Hu, H. NF-ΚB Signaling in Inflammation and Cancer. MedComm 2021, 2, 618–653. [Google Scholar] [CrossRef] [PubMed]
- Ledoux, A.C.; Perkins, N.D. NF-ΚB and the Cell Cycle. Biochem. Soc. Trans. 2014, 42, 76–81. [Google Scholar] [CrossRef] [PubMed]
- Timucin, A.C.; Basaga, H. Pro-Apoptotic Effects of Lipid Oxidation Products: HNE at the Crossroads of NF-ΚB Pathway and Anti-Apoptotic Bcl-2. Free Radic. Biol. Med. 2017, 111, 209–218. [Google Scholar] [CrossRef]
- Liang, C.; Oh, B.H.; Jung, J.U. Novel Functions of Viral Anti-Apoptotic Factors. Nat. Rev. Microbiol. 2015, 13, 7–12. [Google Scholar] [CrossRef]
- Lawrence, T. The Nuclear Factor NF-KappaB Pathway in Inflammation. Cold Spring Harb. Perspect. Biol. 2009, 1, a001651. [Google Scholar] [CrossRef]
- Zhong, L.; Simard, M.J.; Huot, J. Endothelial MicroRNAs Regulating the NF-KB Pathway and Cell Adhesion Molecules during Inflammation. FASEB J. 2018, 32, 4070–4084. [Google Scholar] [CrossRef]
- Kowalski, P.S.; Rudra, A.; Miao, L.; Anderson, D.G. Delivering the Messenger: Advances in Technologies for Therapeutic MRNA Delivery. Mol. Ther. 2019, 27, 710–728. [Google Scholar] [CrossRef]
- Feng, M.; Ding, Z.; Xu, L.; Kong, L.; Wang, W.; Jiao, S.; Shi, Z.; Greene, M.I.; Cong, Y.; Zhou, Z. Structural and Biochemical Studies of RIG-I Antiviral Signaling. Protein Cell 2013, 4, 142–154. [Google Scholar] [CrossRef] [PubMed]
- Brisse, M.; Ly, H. Comparative Structure and Function Analysis of the RIG-I-like Receptors: RIG-I and MDA5. Front. Immunol. 2019, 10, 1586. [Google Scholar] [CrossRef] [PubMed]
- Rawling, D.C.; Pyle, A.M. Parts, Assembly and Operation of the RIG-I Family of Motors. Curr. Opin. Struct. Biol. 2014, 25, 25–33. [Google Scholar] [CrossRef] [PubMed]
- Oshiumi, H.; Matsumoto, M.; Hatakeyama, S.; Seya, T. Riplet/RNF135, a RING Finger Protein, Ubiquitinates RIG-I to Promote Interferon-β Induction during the Early Phase of Viral Infection. J. Biol. Chem. 2009, 284, 807–817. [Google Scholar] [CrossRef] [PubMed]
- Zeng, W.; Sun, L.; Jiang, X.; Chen, X.; Hou, F.; Adhikari, A.; Xu, M.; Chen, Z.J. Reconstitution of the RIG-I Pathway Reveals a Signaling Role of Unanchored Polyubiquitin Chains in Innate Immunity. Cell 2010, 141, 315–330. [Google Scholar] [CrossRef]
- Zheng, Y.; Gao, C. E3 Ubiquitin Ligases, the Powerful Modulator of Innate Antiviral Immunity. Cell. Immunol. 2019, 340, 103915. [Google Scholar] [CrossRef]
- Kawai, T.; Takahashi, K.; Sato, S.; Coban, C.; Kumar, H.; Kato, H.; Ishii, K.J.; Takeuchi, O.; Akira, S. IPS-1, an Adaptor Triggering RIG-I- and Mda5-Mediated Type I Interferon Induction. Nat. Immunol. 2005, 6, 981–988. [Google Scholar] [CrossRef]
- Yoboua, F.; Martel, A.; Duval, A.; Mukawera, E.; Grandvaux, N. Respiratory Syncytial Virus-Mediated NF-KB P65 Phosphorylation at Serine 536 Is Dependent on RIG-I, TRAF6, and IKKβ. J. Virol. 2010, 84, 7267–7277. [Google Scholar] [CrossRef]
- Paz, S.; Sun, Q.; Nakhaei, P.; Romieu-Mourez, R.; Goubau, D.; Julkunen, I.; Lin, R.; Hiscott, J. Induction of IRF-3 and IRF-7 Phosphorylation Following Activation of the RIG-I Pathway. Cell. Mol. Biol. 2006, 52, 17–28. [Google Scholar]
- Chow, K.T.; Gale, M.; Loo, Y.M. RIG-I and other RNA Sensors in Antiviral Immunity. Annu. Rev. Immunol. 2018, 36, 667–694. [Google Scholar] [CrossRef]
- Wu, M.Z.; Asahara, H.; Tzertzinis, G.; Roy, B. Synthesis of Low Immunogenicity RNA with High-Temperature in vitro Transcription. RNA 2020, 26, 345–360. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, K.R.; Bruns, A.M.; Horvath, C.M. MDA5 and LGP2: Accomplices and Antagonists of Antiviral Signal Transduction. J. Virol. 2014, 88, 8194–8200. [Google Scholar] [CrossRef] [PubMed]
- Stok, J.E.; Oosenbrug, T.; Haar, L.R.; Gravekamp, D.; Bromley, C.P.; Zelenay, S.; Reis e Sousa, C.; Veen, A.G. RNA Sensing via the RIG-I-like Receptor LGP2 Is Essential for the Induction of a Type I IFN Response in ADAR1 Deficiency. EMBO J. 2022, 41, e109760. [Google Scholar] [CrossRef] [PubMed]
- Rehwinkel, J.; Gack, M.U. RIG-I-like Receptors: Their Regulation and Roles in RNA Sensing. Nat. Rev. Immunol. 2020, 20, 537–551. [Google Scholar] [CrossRef]
- Kim, D.H.; Longo, M.; Han, Y.; Lundberg, P.; Cantin, E.; Rossi, J.J. Interferon Induction by SiRNAs and SsRNAs Synthesized by Phage Polymerase. Nat. Biotechnol. 2004, 22, 321–325. [Google Scholar] [CrossRef]
- Kormann, M.S.D.; Hasenpusch, G.; Aneja, M.K.; Nica, G.; Flemmer, A.W.; Herber-Jonat, S.; Huppmann, M.; Mays, L.E.; Illenyi, M.; Schams, A.; et al. Expression of Therapeutic Proteins after Delivery of Chemically Modified MRNA in Mice. Nat. Biotechnol. 2011, 29, 154–157. [Google Scholar] [CrossRef]
- Chiang, C.; Beljanski, V.; Yin, K.; Olagnier, D.; Ben Yebdri, F.; Steel, C.; Goulet, M.-L.; DeFilippis, V.R.; Streblow, D.N.; Haddad, E.K.; et al. Sequence-Specific Modifications Enhance the Broad-Spectrum Antiviral Response Activated by RIG-I Agonists. J. Virol. 2015, 89, 8011–8025. [Google Scholar] [CrossRef]
- Claudepierre, M.-C.; Hortelano, J.; Schaedler, E.; Kleinpeter, P.; Geist, M.; Remy-Ziller, C.; Brandely, R.; Tosch, C.; Laruelle, L.; Jawhari, A.; et al. Yeast Virus-Derived Stimulator of the Innate Immune System Augments the Efficacy of Virus Vector-Based Immunotherapy. J. Virol. 2014, 88, 5242–5255. [Google Scholar] [CrossRef]
- Kato, H.; Takeuchi, O.; Mikamo-Satoh, E.; Hirai, R.; Kawai, T.; Matsushita, K.; Hiiragi, A.; Dermody, T.S.; Fujita, T.; Akira, S. Length-Dependent Recognition of Double-Stranded Ribonucleic Acids by Retinoic Acid-Inducible Gene-I and Melanoma Differentiation-Associated Gene 5. J. Exp. Med. 2008, 205, 1601–1610. [Google Scholar] [CrossRef]
- Anchisi, S.; Guerra, J.; Garcin, D. RIG-I Atpase Activity and Discrimination of Self-RNA versus Non-Self-RNA. MBio 2015, 6, e02349. [Google Scholar] [CrossRef]
- Feng, Q.; Hato, S.V.; Langereis, M.A.; Zoll, J.; Virgen-Slane, R.; Peisley, A.; Hur, S.; Semler, B.L.; van Rij, R.P.; van Kuppeveld, F.J.M. MDA5 Detects the Double-Stranded RNA Replicative form in Picornavirus-Infected Cells. Cell Rep. 2012, 2, 1187–1196. [Google Scholar] [CrossRef] [PubMed]
- Grandvaux, N.; Guan, X.; Yoboua, F.; Zucchini, N.; Fink, K.; Doyon, P.; Martin, L.; Servant, M.J.; Chartier, S. Sustained Activation of Interferon Regulatory Factor 3 during Infection by Paramyxoviruses Requires MDA5. J. Innate Immun. 2014, 6, 650–662. [Google Scholar] [CrossRef] [PubMed]
- Gitlin, L.; Benoit, L.; Song, C.; Cella, M.; Gilfillan, S.; Holtzman, M.J.; Colonna, M. Melanoma Differentiation-Associated Gene 5 (MDA5) Is Involved in the Innate Immune Response to Paramyxoviridae Infection in vivo. PLoS Pathog. 2010, 6, e1000734. [Google Scholar] [CrossRef]
- Xia, A.; Li, X.; Quan, J.; Chen, X.; Xu, Z.; Jiao, X. Mycobacterium Tuberculosis Rv0927c Inhibits NF-ΚB Pathway by Downregulating the Phosphorylation Level of IκBα and Enhances Mycobacterial Survival. Front. Immunol. 2021, 12, 721370. [Google Scholar] [CrossRef]
- Loo, Y.M.; Gale, M. Immune Signaling by RIG-I-like Receptors. Immunity 2011, 34, 680–692. [Google Scholar] [CrossRef]
- Zhai, W.; Wu, F.; Zhang, Y.; Fu, Y.; Liu, Z. The Immune Escape Mechanisms of Mycobacterium Tuberculosis. Int. J. Mol. Sci. 2019, 20, 340. [Google Scholar] [CrossRef] [PubMed]
- Minnaert, A.K.; Vanluchene, H.; Verbeke, R.; Lentacker, I.; De Smedt, S.C.; Raemdonck, K.; Sanders, N.N.; Remaut, K. Strategies for Controlling the Innate Immune Activity of Conventional and Self-Amplifying MRNA Therapeutics: Getting the Message Across. Adv. Drug Deliv. Rev. 2021, 176, 113900. [Google Scholar] [CrossRef]
- Devarkar, S.C.; Wang, C.; Miller, M.T.; Ramanathan, A.; Jiang, F.; Khan, A.G.; Patel, S.S.; Marcotrigiano, J. Structural Basis for M7G Recognition and 2′-O-Methyl Discrimination in Capped RNAs by the Innate Immune Receptor RIG-I. Proc. Natl. Acad. Sci. USA 2016, 113, 596–601. [Google Scholar] [CrossRef]
- Züst, R.; Cervantes-Barragan, L.; Habjan, M.; Maier, R.; Neuman, B.W.; Ziebuhr, J.; Szretter, K.J.; Baker, S.C.; Barchet, W.; Diamond, M.S.; et al. Ribose 2′-O-Methylation Provides a Molecular Signature for the Distinction of Self and Non-Self MRNA Dependent on the RNA Sensor Mda5. Nat. Immunol. 2011, 12, 137–143. [Google Scholar] [CrossRef]
- Rückle, A.; Haasbach, E.; Julkunen, I.; Planz, O.; Ehrhardt, C.; Ludwig, S. The NS1 Protein of Influenza A Virus Blocks RIG-I-Mediated Activation of the Noncanonical NF-ΚB Pathway and P52/RelB-Dependent Gene Expression in Lung Epithelial Cells. J. Virol. 2012, 86, 10211–10217. [Google Scholar] [CrossRef]
- Motz, C.; Schuhmann, K.M.; Kirchhofer, A.; Moldt, M.; Witte, G.; Conzelmann, K.K.; Hopfner, K.P. Paramyxovirus V Proteins Disrupt the Fold of the RNA Sensor MDA5 to Inhibit Antiviral Signaling. Science 2013, 339, 690–693. [Google Scholar] [CrossRef]
- Kasumba, D.M.; Grandvaux, N. Therapeutic Targeting of RIG-I and MDA5 Might Not Lead to the Same Rome. Trends Pharmacol. Sci. 2019, 40, 116–127. [Google Scholar] [CrossRef] [PubMed]
- Broquet, A.H.; Hirata, Y.; McAllister, C.S.; Kagnoff, M.F. RIG-I/MDA5/MAVS Are Required to Signal a Protective IFN Response in Rotavirus-Infected Intestinal Epithelium. J. Immunol. 2011, 186, 1618–1626. [Google Scholar] [CrossRef] [PubMed]
- Fredericksen, B.L.; Keller, B.C.; Fornek, J.; Katze, M.G.; Gale, M. Establishment and Maintenance of the Innate Antiviral Response to West Nile Virus Involves Both RIG-I and MDA5 Signaling through IPS-1. J. Virol. 2008, 82, 609–616. [Google Scholar] [CrossRef] [PubMed]
- Linehan, M.M.; Dickey, T.H.; Molinari, E.S.; Fitzgerald, M.E.; Potapova, O.; Iwasaki, A.; Pyle, A.M. A Minimal RNA Ligand for Potent RIG-I Activation in Living Mice. Sci. Adv. 2018, 4, e1701854. [Google Scholar] [CrossRef]
- Sleijfer, S.; Bannink, M.; Van Gool, A.R.; Kruit, W.H.J.; Stoter, G. Side Effects of Interferon-α Therapy. Pharm. World Sci. 2005, 27, 423–431. [Google Scholar] [CrossRef]
- Lemaitre, B.; Nicolas, E.; Michaut, L.; Reichhart, J.M.; Hoffmann, J.A. The Dorsoventral Regulatory Gene Cassette Spatzle/Toll/Cactus Controls the Potent Antifungal Response in Drosophila Adults. Cell 1996, 86, 973–983. [Google Scholar] [CrossRef]
- González-Crespo, S.; Levine, M. Related Target Enhancers for Dorsal and NF-ΚB Signaling Pathways. Science 1994, 264, 255–258. [Google Scholar] [CrossRef]
- Medzhitov, R.; Preston-Hurlburt, P.; Janeway, C.A. A Human Homologue of the Drosophila Toll Protein Signals Activation of Adaptive Immunity. Nature 1997, 388, 394–397. [Google Scholar] [CrossRef]
- Gay, N.J.; Keith, F.J. Drosophila Toll and IL-1 Receptor. Nature 1991, 351, 355–356. [Google Scholar] [CrossRef]
- Akira, S.; Uematsu, S.; Takeuchi, O. Pathogen Recognition and Innate Immunity. Cell 2006, 124, 783–801. [Google Scholar] [CrossRef] [PubMed]
- Fitzgerald, K.A.; Kagan, J.C. Toll-like Receptors and the Control of Immunity. Cell 2020, 180, 1044–1066. [Google Scholar] [CrossRef] [PubMed]
- Medzhitov, R. Toll-like Receptors and Innate Immunity. Nat. Rev. Immunol. 2001, 1, 135–145. [Google Scholar] [CrossRef] [PubMed]
- Iwasaki, A.; Medzhitov, R. Regulation of Adaptive Immunity by the Innate Immune System. Science 2010, 327, 291–295. [Google Scholar] [CrossRef] [PubMed]
- Bell, J.K.; Mullen, G.E.D.; Leifer, C.A.; Mazzoni, A.; Davies, D.R.; Segal, D.M. Leucine-Rich Repeats and Pathogen Recognition in Toll-like Receptors. Trends Immunol. 2003, 24, 528–533. [Google Scholar] [CrossRef]
- Chang, Z.L. Important Aspects of Toll-like Receptors, Ligands and Their Signaling Pathways. Inflamm. Res. 2010, 59, 791–808. [Google Scholar] [CrossRef]
- Matsushima, N.; Tachi, N.; Kuroki, Y.; Enkhbayar, P.; Osaki, M.; Kamiya, M.; Kretsinger, R.H. Structural Analysis of Leucine-Rich-Repeat Variants in Proteins Associated with Human Diseases. Cell. Mol. Life Sci. 2005, 62, 2771–2791. [Google Scholar] [CrossRef]
- Boehm, T.; McCurley, N.; Sutoh, Y.; Schorpp, M.; Kasahara, M.; Cooper, M.D. VLR-Based Adaptive Immunity. Annu. Rev. Immunol. 2012, 30, 203–220. [Google Scholar] [CrossRef]
- Szalkowski, A.M.; Anisimova, M. Graph-Based Modeling of Tandem Repeats Improves Global Multiple Sequence Alignment. Nucleic Acids Res. 2013, 41, e162. [Google Scholar] [CrossRef]
- Leister, D. Tandem and Segmental Gene Duplication and Recombination in the Evolution of Plant Disease Resistance Genes. Trends Genet. 2004, 20, 116–122. [Google Scholar] [CrossRef]
- Schaper, E.; Anisimova, M. The Evolution and Function of Protein Tandem Repeats in Plants. New Phytol. 2015, 206, 397–410. [Google Scholar] [CrossRef] [PubMed]
- Sato, R.; Shibata, T.; Tanaka, Y.; Kato, C.; Yamaguchi, K.; Furukawa, Y.; Shimizu, E.; Yamaguchi, R.; Imoto, S.; Miyano, S.; et al. Requirement of Glycosylation Machinery in TLR Responses Revealed by CRISPR/Cas9 Screening. Int. Immunol. 2017, 29, 347–355. [Google Scholar] [CrossRef] [PubMed]
- Gao, D.; Li, W. Structures and Recognition Modes of Toll-like Receptors. Proteins Struct. Funct. Bioinform. 2017, 85, 3–9. [Google Scholar] [CrossRef] [PubMed]
- Song, D.H.; Lee, J.O. Sensing of Microbial Molecular Patterns by Toll-like Receptors. Immunol. Rev. 2012, 250, 216–229. [Google Scholar] [CrossRef]
- El-Zayat, S.R.; Sibaii, H.; Mannaa, F.A. Toll-like Receptors Activation, Signaling, and Targeting: An Overview. Bull. Natl. Res. Cent. 2019, 43, 187. [Google Scholar] [CrossRef]
- Lee, S.M.Y.; Yip, T.F.; Yan, S.; Jin, D.Y.; Wei, H.L.; Guo, R.T.; Peiris, J.S.M. Recognition of Double-Stranded RNA and Regulation of Interferon Pathway by Toll-like Receptor 10. Front. Immunol. 2018, 9, 516. [Google Scholar] [CrossRef] [PubMed]
- Kawasaki, T.; Kawai, T. Toll-like Receptor Signaling Pathways. Front. Immunol. 2014, 5, 461. [Google Scholar] [CrossRef]
- Uronen-Hansson, H.; Allen, J.; Osman, M.; Squires, G.; Klein, N.; Callard, R.E. Toll-like Receptor 2 (TLR2) and TLR4 Are Present inside Human Dendritic Cells, Associated with Microtubules and the Golgi Apparatus but Are Not Detectable on the Cell Surface: Integrity of Microtubules Is Required for Interleukin-12 Production in Response to Internalized Bacteria. Immunology 2004, 111, 173–178. [Google Scholar] [CrossRef]
- De Zoete, M.R.; Bouwman, L.I.; Keestra, A.M.; Van Putten, J.P.M. Cleavage and Activation of a Toll-like Receptor by Microbial Proteases. Proc. Natl. Acad. Sci. USA 2011, 108, 4968–4973. [Google Scholar] [CrossRef]
- O’Neill, L.A.J.; Bowie, A.G. The Family of Five: TIR-Domain-Containing Adaptors in Toll-like Receptor Signalling. Nat. Rev. Immunol. 2007, 7, 353–364. [Google Scholar] [CrossRef]
- Yamamoto, M.; Takeda, K.; Akira, S. TIR Domain-Containing Adaptors Define the Specificity of TLR Signaling. Mol. Immunol. 2004, 40, 861–868. [Google Scholar] [CrossRef]
- Kaiko, G.E.; Horvat, J.C.; Beagley, K.W.; Hansbro, P.M. Immunological Decision-Making: How Does the Immune System Decide to Mount a Helper T-Cell Response? Immunology 2008, 123, 326–338. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Zhao, Y. Toll-like Receptors and Immune Regulation: Their Direct and Indirect Modulation on Regulatory CD4+ CD25+ T Cells. Immunology 2007, 122, 149–156. [Google Scholar] [CrossRef] [PubMed]
- Honda, K.; Takeda, K. Regulatory Mechanisms of Immune Responses to Intestinal Bacteria. Mucosal Immunol. 2009, 2, 187–196. [Google Scholar] [CrossRef] [PubMed]
- Honda, K.; Taniguchi, T. Toll-like Receptor Signaling and IRF Transcription Factors. IUBMB Life 2006, 58, 290–295. [Google Scholar] [CrossRef] [PubMed]
- Iwanaszko, M.; Kimmel, M. NF-ΚB and IRF Pathways: Cross-Regulation on Target Genes Promoter Level. BMC Genomics 2015, 16, 307. [Google Scholar] [CrossRef]
- De Haro, C.; Méndez, R.; Santoyo, J. The EIF-2α Kinases and the Control of Protein Synthesis 1. FASEB J. 1996, 10, 1378–1387. [Google Scholar] [CrossRef]
- Liang, S.L.; Quirk, D.; Zhou, A. RNase L: Its Biological Roles and Regulation. IUBMB Life 2006, 58, 508–514. [Google Scholar] [CrossRef]
- Isaacs, A.; Cox, R.A.; Rotem, Z. Foreign nucleic acids as the stimulus to make interferon. Lancet 1963, 282, 113–116. [Google Scholar] [CrossRef]
- Mosaheb, M.M.; Reiser, M.L.; Wetzler, L.M. Toll-like Receptor Ligand-Based Vaccine Adjuvants Require Intact MyD88 Signaling in Antigen-Presenting Cells for Germinal Center Formation and Antibody Production. Front. Immunol. 2017, 8, 225. [Google Scholar] [CrossRef]
- Shen, C.F.; Yen, C.L.; Fu, Y.C.; Cheng, C.M.; Shen, T.C.; De Chang, P.; Cheng, K.H.; Liu, C.C.; Chang, Y.T.; Chen, P.L.; et al. Innate Immune Responses of Vaccinees Determine early Neutralizing Antibody Production after ChAdOx1nCoV-19 Vaccination. Front. Immunol. 2022, 13, 807454. [Google Scholar] [CrossRef] [PubMed]
- Carty, M.; Guy, C.; Bowie, A.G. Detection of Viral Infections by Innate Immunity. Biochem. Pharmacol. 2021, 183, 114316. [Google Scholar] [CrossRef] [PubMed]
- Song, W.S.; Jeon, Y.J.; Namgung, B.; Hong, M.; Yoon, S. Il A Conserved TLR5 Binding and Activation Hot Spot on Flagellin. Sci. Rep. 2017, 7, srep40878. [Google Scholar] [CrossRef] [PubMed]
- Karikó, K.; Muramatsu, H.; Ludwig, J.; Weissman, D. Generating the Optimal MRNA for Therapy: HPLC Purification Eliminates Immune Activation and Improves Translation of Nucleoside-Modified, Protein-Encoding MRNA. Nucleic Acids Res. 2011, 39, e142. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Ohto, U.; Shibata, T.; Krayukhina, E.; Taoka, M.; Yamauchi, Y.; Tanji, H.; Isobe, T.; Uchiyama, S.; Miyake, K.; et al. Structural Analysis Reveals that Toll-like Receptor 7 Is a Dual Receptor for Guanosine and Single-Stranded RNA. Immunity 2016, 45, 737–748. [Google Scholar] [CrossRef]
- Tanji, H.; Ohto, U.; Shibata, T.; Taoka, M.; Yamauchi, Y.; Isobe, T.; Miyake, K.; Shimizu, T. Toll-like Receptor 8 Senses Degradation Products of Single-Stranded RNA. Nat. Struct. Mol. Biol. 2015, 22, 109–115. [Google Scholar] [CrossRef]
- Bowie, A.G.; Unterholzner, L. Viral Evasion and Subversion of Pattern-Recognition Receptor Signalling. Nat. Rev. Immunol. 2008, 8, 911–922. [Google Scholar] [CrossRef]
- Datta, A.; Sinha-Datta, U.; Dhillon, N.K.; Buch, S.; Nicot, C. The HTLV-I P30 Interferes with TLR4 Signaling and Modulates the Release of pro- and anti-Inflammatory Cytokines from Human Macrophages. J. Biol. Chem. 2006, 281, 23414–23424. [Google Scholar] [CrossRef]
- Chase, A.J.; Sedaghat, A.R.; German, J.R.; Gama, L.; Zink, M.C.; Clements, J.E.; Siliciano, R.F. Severe Depletion of CD4 + CD25 + Regulatory T Cells from the Intestinal Lamina Propria but Not Peripheral Blood or Lymph Nodes during Acute Simian Immunodeficiency Virus Infection. J. Virol. 2007, 81, 12748–12757. [Google Scholar] [CrossRef]
- Gale, M.; Sen, G.C. Overview: Viral Evasion of the Interferon System. J. Interf. Cytokine Res. 2009, 29, 475–476. [Google Scholar] [CrossRef]
- Fekonja, O.; Avbelj, M.; Jerala, R. Suppression of TLR Signaling by Targeting TIR Domain-Containing Proteins. Curr. Protein Pept. Sci. 2013, 13, 776–788. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.; Chen, J.; Wu, M.; Chen, H.; Kato, N.; Yuan, Z. Hepatitis B Virus Polymerase Inhibits RIG-I- and Toll-like Receptor 3-Mediated Beta Interferon Induction in Human Hepatocytes through Interference with Interferon Regulatory Factor 3 Activation and Dampening of the Interaction between TBK1/IKKε and DDX3. J. Gen. Virol. 2010, 91, 2080–2090. [Google Scholar] [CrossRef]
- Xu, Y.; Hu, Y.; Shi, B.; Zhang, X.; Wang, J.; Zhang, Z.; Shen, F.; Zhang, Q.; Sun, S.; Yuan, Z. HBsAg Inhibits TLR9-Mediated Activation and IFN-α Production in Plasmacytoid Dendritic Cells. Mol. Immunol. 2009, 46, 2640–2646. [Google Scholar] [CrossRef] [PubMed]
- Vacchelli, E.; Galluzzi, L.; Eggermont, A.; Fridman, W.H.; Galon, J.; Sautès-Fridman, C.; Tartour, E.; Zitvogel, L.; Kroemer, G. Trial Watch: FDA-Approved Toll-like Receptor Agonists for Cancer Therapy. Oncoimmunology 2012, 1, 894–907. [Google Scholar] [CrossRef] [PubMed]
- Andón, F.T.; Leon, S.; Ummarino, A.; Redin, E.; Allavena, P.; Serrano, D.; Anfray, C.; Calvo, A. Innate and Adaptive Responses of Intratumoral Immunotherapy with Endosomal Toll-Like Receptor Agonists. Biomedicines 2022, 10, 1590. [Google Scholar] [CrossRef]
- Lobo, N.; Brooks, N.A.; Zlotta, A.R.; Cirillo, J.D.; Boorjian, S.; Black, P.C.; Meeks, J.J.; Bivalacqua, T.J.; Gontero, P.; Steinberg, G.D.; et al. 100 Years of Bacillus Calmette–Guérin Immunotherapy: From Cattle to COVID-19. Nat. Rev. Urol. 2021, 18, 611–622. [Google Scholar] [CrossRef]
- Polack, F.P.; Thomas, S.J.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Perez, J.L.; Pérez Marc, G.; Moreira, E.D.; Zerbini, C.; et al. Safety and Efficacy of the BNT162b2 MRNA COVID-19 Vaccine. N. Engl. J. Med. 2020, 383, 2603–2615. [Google Scholar] [CrossRef]
- Decker, C.J.; Parker, R. Mechanisms of MRNA Degradation in Eukaryotes. Trends Biochem. Sci. 1994, 19, 336–340. [Google Scholar] [CrossRef]
- Hou, X.; Zaks, T.; Langer, R.; Dong, Y. Lipid Nanoparticles for MRNA Delivery. Nat. Rev. Mater. 2021, 6, 1078–1094. [Google Scholar] [CrossRef]
- Wolff, J.A.; Malone, R.W.; Williams, P.; Chong, W.; Acsadi, G.; Jani, A.; Felgner, P.L. Direct Gene Transfer into Mouse Muscle in vivo. Science 1990, 247, 1465–1468. [Google Scholar] [CrossRef]
- Babendure, J.R.; Babendure, J.L.; Ding, J.H.; Tsien, R.Y. Control of Mammalian Translation by MRNA Structure near Caps. RNA 2006, 12, 851–861. [Google Scholar] [CrossRef]
- Furuichi, Y.; LaFiandra, A.; Shatkin, A.J. 5′-Terminal Structure and MRNA Stability. Nature 1977, 266, 235–239. [Google Scholar] [CrossRef] [PubMed]
- Cooke, C.; Alwine, J.C. The Cap and the 3′ Splice Site Similarly Affect Polyadenylation Efficiency. Mol. Cell. Biol. 1996, 16, 2579–2584. [Google Scholar] [CrossRef]
- Lockard, R.E.; Lingrel, J.B. The Synthesis of Mouse Hemoglobin Chains in a Rabbit Reticulocyte Cell-Free System Programmed with Mouse Reticulocyte 9S RNA. Biochem. Biophys. Res. Commun. 1969, 37, 204–212. [Google Scholar] [CrossRef] [PubMed]
- Sartorius, R.; Trovato, M.; Manco, R.; D’Apice, L.; De Berardinis, P. Exploiting Viral Sensing Mediated by Toll-like Receptors to Design Innovative Vaccines. Npj Vaccines 2021, 6, 127. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, S.; Kandimalla, E.R. Synthetic Agonists of Toll-like Receptors 7, 8 and 9. Biochem. Soc. Trans. 2007, 35, 1461–1467. [Google Scholar] [CrossRef]
- Owen, A.M.; Fults, J.B.; Patil, N.K.; Hernandez, A.; Bohannon, J.K. TLR Agonists as Mediators of Trained Immunity: Mechanistic Insight and Immunotherapeutic Potential to Combat Infection. Front. Immunol. 2021, 11, 622614. [Google Scholar] [CrossRef]
- Schlake, T.; Thess, A.; Fotin-Mleczek, M.; Kallen, K.J. Developing MRNA-Vaccine Technologies. RNA Biol. 2012, 9, 1319–1330. [Google Scholar] [CrossRef]
- Mauger, D.M.; Joseph Cabral, B.; Presnyak, V.; Su, S.V.; Reid, D.W.; Goodman, B.; Link, K.; Khatwani, N.; Reynders, J.; Moore, M.J.; et al. MRNA Structure Regulates Protein Expression through Changes in Functional Half-Life. Proc. Natl. Acad. Sci. USA 2019, 116, 24075–24083. [Google Scholar] [CrossRef]
- Karikó, K.; Buckstein, M.; Ni, H.; Weissman, D. Suppression of RNA Recognition by Toll-like Receptors: The Impact of Nucleoside Modification and the Evolutionary Origin of RNA. Immunity 2005, 23, 165–175. [Google Scholar] [CrossRef]
- Nance, K.D.; Meier, J.L. Modifications in an Emergency: The Role of N1-Methylpseudouridine in COVID-19 Vaccines. ACS Cent. Sci. 2021, 7, 748–756. [Google Scholar] [CrossRef] [PubMed]
- Mu, X.; Greenwald, E.; Ahmad, S.; Hur, S. An Origin of the Immunogenicity of in vitro Transcribed RNA. Nucleic Acids Res. 2018, 46, 5239–5249. [Google Scholar] [CrossRef] [PubMed]
- Nelson, J.; Sorensen, E.W.; Mintri, S.; Rabideau, A.E.; Zheng, W.; Besin, G.; Khatwani, N.; Su, S.V.; Miracco, E.J.; Issa, W.J.; et al. Impact of MRNA Chemistry and Manufacturing Process on Innate Immune Activation. Sci. Adv. 2020, 6, eaaz6893. [Google Scholar] [CrossRef]
- Vaidyanathan, S.; Azizian, K.T.; Haque, A.K.M.A.; Henderson, J.M.; Hendel, A.; Shore, S.; Antony, J.S.; Hogrefe, R.I.; Kormann, M.S.D.; Porteus, M.H.; et al. Uridine Depletion and Chemical Modification Increase Cas9 MRNA Activity and Reduce Immunogenicity without HPLC Purification. Mol. Ther. Nucleic Acids 2018, 12, 530–542. [Google Scholar] [CrossRef] [PubMed]
- Svitkin, Y.V.; Cheng, Y.M.; Chakraborty, T.; Presnyak, V.; John, M.; Sonenberg, N. N1-Methyl-Pseudouridine in MRNA Enhances Translation through EIF2α-Dependent and Independent Mechanisms by Increasing Ribosome Density. Nucleic Acids Res. 2017, 45, 6023–6036. [Google Scholar] [CrossRef] [PubMed]
- Andries, O.; Mc Cafferty, S.; De Smedt, S.C.; Weiss, R.; Sanders, N.N.; Kitada, T. N1-Methylpseudouridine-Incorporated MRNA Outperforms Pseudouridine-Incorporated MRNA by Providing Enhanced Protein Expression and Reduced Immunogenicity in Mammalian Cell Lines and Mice. J. Control. Release 2015, 217, 337–344. [Google Scholar] [CrossRef] [PubMed]
- Kozak, M. Regulation of Translation via MRNA Structure in Prokaryotes and Eukaryotes. Gene 2005, 361, 13–37. [Google Scholar] [CrossRef]
- Lind, C.; Esguerra, M.; Jespers, W.; Satpati, P.; Gutierrez-de-Terán, H.; Åqvist, J. Free Energy Calculations of RNA Interactions. Methods 2019, 162–163, 85–95. [Google Scholar] [CrossRef]
- Kierzek, E.; Malgowska, M.; Lisowiec, J.; Turner, D.H.; Gdaniec, Z.; Kierzek, R. The Contribution of Pseudouridine to Stabilities and Structure of RNAs. Nucleic Acids Res. 2014, 42, 3492–3501. [Google Scholar] [CrossRef]
- Thess, A.; Grund, S.; Mui, B.L.; Hope, M.J.; Baumhof, P.; Fotin-Mleczek, M.; Schlake, T. Sequence-Engineered MRNA Without Chemical Nucleoside Modifications Enables an Effective Protein Therapy in Large Animals. Mol. Ther. 2015, 23, 1456–1464. [Google Scholar] [CrossRef]
- Heit, A.; Schmitz, F.; Haas, T.; Busch, D.H.; Wagner, H. Antigen Co-Encapsulated with Adjuvants Efficiently Drive Protective T Cell Immunity. Eur. J. Immunol. 2007, 37, 2063–2074. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, D.N.; Mahon, K.P.; Chikh, G.; Kim, P.; Chung, H.; Vicari, A.P.; Love, K.T.; Goldberg, M.; Chen, S.; Krieg, A.M.; et al. Lipid-Derived Nanoparticles for Immunostimulatory RNA Adjuvant Delivery. Proc. Natl. Acad. Sci. USA 2012, 109, E797–E803. [Google Scholar] [CrossRef] [PubMed]
- Duthie, M.S.; Windish, H.P.; Fox, C.B.; Reed, S.G. Use of Defined TLR Ligands as Adjuvants within Human Vaccines. Immunol. Rev. 2011, 239, 178–196. [Google Scholar] [CrossRef] [PubMed]
- Ebrahimian, M.; Hashemi, M.; Maleki, M.; Hashemitabar, G.; Abnous, K.; Ramezani, M.; Haghparast, A. Co-Delivery of Dual Toll-like Receptor Agonists and Antigen in Poly(Lactic-Co-Glycolic) Acid/Polyethylenimine Cationic Hybrid Nanoparticles Promote Efficient in vivo Immune Responses. Front. Immunol. 2017, 8, 1077. [Google Scholar] [CrossRef]
- Proell, M.; Riedl, S.J.; Fritz, J.H.; Rojas, A.M.; Schwarzenbacher, R. The Nod-Like Receptor (NLR) Family: A Tale of Similarities and Differences. PLoS ONE 2008, 3, e2119. [Google Scholar] [CrossRef]
- Jin, J.; Zhou, T.-J.; Ren, G.-L.; Cai, L.; Meng, X.-M. Novel Insights into NOD-like Receptors in Renal Diseases. Acta Pharmacol. Sin. 2022, 43, 2789–2806. [Google Scholar] [CrossRef]
- Kim, Y.K.; Shin, J.S.; Nahm, M.H. NOD-like Receptors in Infection, Immunity, and Diseases. Yonsei Med. J. 2016, 57, 5–14. [Google Scholar] [CrossRef]
- Askari, N.; Correa, R.G.; Zhai, D.; Reed, J.C. Expression, Purification, and Characterization of Recombinant NOD1 (NLRC1): A NLR Family Member. J. Biotechnol. 2012, 157, 75–81. [Google Scholar] [CrossRef]
- Faustin, B.; Reed, J.C. Sunburned Skin Activates Inflammasomes. Trends Cell Biol. 2008, 18, 4–8. [Google Scholar] [CrossRef]
- Schroder, K.; Tschopp, J. The Inflammasomes. Cell 2010, 140, 821–832. [Google Scholar] [CrossRef]
- Chu, J.Q.; Gao, F.F.; Wu, W.; Li, C.; Pan, Z.; Sun, J.; Wang, H.; Huang, C.; Lee, S.H.; Quan, J.-H.; et al. Expression profiles of NOD-like receptors and regulation of NLRP3 inflammasome activation in Toxoplasma gondii-infected human small intestinal epithelial cells. Parasites Vectors 2021, 14, 153. [Google Scholar] [CrossRef] [PubMed]
- Lupfer, C.; Kanneganti, T.D. Unsolved Mysteries in NLR Biology. Front. Immunol. 2013, 4, 285. [Google Scholar] [CrossRef] [PubMed]
- León Machado, J.A.; Steimle, V. The Mhc Class Ii Transactivator Ciita: Not (Quite) the Odd-One-out Anymore among Nlr Proteins. Int. J. Mol. Sci. 2021, 22, 1074. [Google Scholar] [CrossRef] [PubMed]
- Okamoto, H.; Asamitsu, K.; Nishimura, H.; Kamatani, N.; Okamoto, T. Reciprocal Modulation of Transcriptional Activities between HIV-1 Tat and MHC Class II Transactivator CIITA. Biochem. Biophys. Res. Commun. 2000, 279, 494–499. [Google Scholar] [CrossRef] [PubMed]
- Abendroth, A.; Slobedman, B.; Lee, E.; Mellins, E.; Wallace, M.; Arvin, A.M. Modulation of Major Histocompatibility Class II Protein Expression by Varicella-Zoster Virus. J. Virol. 2000, 74, 1900–1907. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.H.; Lin, J.Y.; Chou, Y.C.; Chen, M.R.; Yeh, T.H.; Lin, C.W.; Lin, S.J.; Tsai, C.H. Epstein-Barr Virus LMP2A Suppresses MHC Class II Expression by Regulating the B-Cell Transcription Factors E47 and PU.1. Blood 2015, 125, 2228–2238. [Google Scholar] [CrossRef]
- Sandhu, P.K.; Buchkovich, N.J. Human Cytomegalovirus Decreases Major Histocompatibility Complex Class II by Regulating Class II Transactivator Transcript Levels in a Myeloid Cell Line. J. Virol. 2020, 94, 10–1128. [Google Scholar] [CrossRef]
- Pai, R.K.; Convery, M.; Hamilton, T.A.; Boom, W.H.; Harding, C.V. Inhibition of IFN-γ-Induced Class II Transactivator Expression by a 19-KDa Lipoprotein from Mycobacterium Tuberculosis: A Potential Mechanism for Immune Evasion. J. Immunol. 2003, 171, 175–184. [Google Scholar] [CrossRef]
- Benko, S.; Magalhaes, J.G.; Philpott, D.J.; Girardin, S.E. NLRC5 Limits the Activation of Inflammatory Pathways. J. Immunol. 2010, 185, 1681–1691. [Google Scholar] [CrossRef]
- Vijayan, S.; Sidiq, T.; Yousuf, S.; van den Elsen, P.J.; Kobayashi, K.S. Class I Transactivator, NLRC5: A Central Player in the MHC Class I Pathway and Cancer Immune Surveillance. Immunogenetics 2019, 71, 273–282. [Google Scholar] [CrossRef]
- Meissner, T.B.; Li, A.; Biswas, A.; Lee, K.H.; Liu, Y.J.; Bayir, E.; Iliopoulos, D.; Van Den Elsen, P.J.; Kobayashi, K.S. NLR Family Member NLRC5 Is a Transcriptional Regulator of MHC Class I Genes. Proc. Natl. Acad. Sci. USA 2010, 107, 13794–13799. [Google Scholar] [CrossRef] [PubMed]
- Kuenzel, S.; Till, A.; Winkler, M.; Häsler, R.; Lipinski, S.; Jung, S.; Grötzinger, J.; Fickenscher, H.; Schreiber, S.; Rosenstiel, P. The Nucleotide-Binding Oligomerization Domain-like Receptor NLRC5 Is Involved in IFN-Dependent Antiviral Immune Responses. J. Immunol. 2010, 184, 1990–2000. [Google Scholar] [CrossRef] [PubMed]
- Ting, J.P.Y.; Duncan, J.A.; Lei, Y. How the Noninflammasome NLRs Function in the Innate Immune System. Science 2010, 327, 286–290. [Google Scholar] [CrossRef] [PubMed]
- Sabbah, A.; Chang, T.H.; Harnack, R.; Frohlich, V.; Tominaga, K.; Dube, P.H.; Xiang, Y.; Bose, S. Activation of Innate Immune Antiviral Responses by Nod2. Nat. Immunol. 2009, 10, 1073–1080. [Google Scholar] [CrossRef] [PubMed]
- Philpott, D.J.; Sorbara, M.T.; Robertson, S.J.; Croitoru, K.; Girardin, S.E. NOD Proteins: Regulators of Inflammation in Health and Disease. Nat. Rev. Immunol. 2013, 14, 9–23, Erratum in 2014, 14, 131. [Google Scholar] [CrossRef]
- Moore, C.B.; Bergstralh, D.T.; Duncan, J.A.; Lei, Y.; Morrison, T.E.; Zimmermann, A.G.; Accavitti-Loper, M.A.; Madden, V.J.; Sun, L.; Ye, Z.; et al. NLRX1 Is a Regulator of Mitochondrial Antiviral Immunity. Nature 2008, 451, 573–577. [Google Scholar] [CrossRef]
- Maisonneuve, C.; Bertholet, S.; Philpott, D.J.; De Gregorio, E. Unleashing the Potential of NOD- and Toll-like Agonists as Vaccine Adjuvants. Proc. Natl. Acad. Sci. USA 2014, 111, 12294–12299. [Google Scholar] [CrossRef]
- Martinon, F.; Agostini, L.; Meylan, E.; Tschopp, J. Identification of Bacterial Muramyl Dipeptide as Activator of the NALP3/Cryopyrin Inflammasome. Curr. Biol. 2004, 14, 1929–1934. [Google Scholar] [CrossRef]
- Magalhaes, J.G.; Fritz, J.H.; Le Bourhis, L.; Sellge, G.; Travassos, L.H.; Selvanantham, T.; Girardin, S.E.; Gommerman, J.L.; Philpott, D.J. Nod2-Dependent Th2 Polarization of Antigen-Specific Immunity. J. Immunol. 2008, 181, 7925–7935. [Google Scholar] [CrossRef]
- Barazzone, G.C.; Teixeira, A.F.; Azevedo, B.O.P.; Damiano, D.K.; Oliveira, M.P.; Nascimento, A.L.T.O.; Lopes, A.P.Y. Revisiting the Development of Vaccines Against Pathogenic Leptospira: Innovative Approaches, Present Challenges, and Future Perspectives. Front. Immunol. 2022, 12, 760291. [Google Scholar] [CrossRef]
- Rice, T.A.; Brenner, T.A.; Percopo, C.M.; Ma, M.; Keicher, J.D.; Domachowske, J.B.; Rosenberg, H.F. Signaling via Pattern Recognition Receptors NOD2 and TLR2 Contributes to Immunomodulatory Control of Lethal Pneumovirus Infection. Antivir. Res. 2016, 132, 131–140. [Google Scholar] [CrossRef] [PubMed]
- Lemesre, J.L.; Holzmuller, P.; Gonçalves, R.B.; Bourdoiseau, G.; Hugnet, C.; Cavaleyra, M.; Papierok, G. Long-Lasting Protection against Canine Visceral Leishmaniasis Using the LiESAp-MDP Vaccine in Endemic Areas of France: Double-Blind Randomised Efficacy Field Trial. Vaccine 2007, 25, 4223–4234. [Google Scholar] [CrossRef] [PubMed]
- Netea, M.G.; Domínguez-Andrés, J.; Barreiro, L.B.; Chavakis, T.; Divangahi, M.; Fuchs, E.; Joosten, L.A.B.; van der Meer, J.W.M.; Mhlanga, M.M.; Mulder, W.J.M.; et al. Defining Trained Immunity and Its Role in Health and Disease. Nat. Rev. Immunol. 2020, 20, 375–388. [Google Scholar] [CrossRef] [PubMed]
- Reinke, S.; Thakur, A.; Gartlan, C.; Bezbradica, J.S.; Milicic, A. Inflammasome-Mediated Immunogenicity of Clinical and Experimental Vaccine Adjuvants. Vaccines 2020, 8, 554. [Google Scholar] [CrossRef] [PubMed]
- Brueggeman, J.M.; Zhao, J.; Schank, M.; Yao, Z.Q.; Moorman, J.P. Trained Immunity: An Overview and the Impact on COVID-19. Front. Immunol. 2022, 13, 837524. [Google Scholar] [CrossRef]
- Swanson, K.V.; Deng, M.; Ting, J.P.-Y. The NLRP3 Inflammasome: Molecular Activation and Regulation to Therapeutics. Nat. Rev. Immunol. 2019, 19, 477–489. [Google Scholar] [CrossRef]
- Dick, M.S.; Sborgi, L.; Rühl, S.; Hiller, S.; Broz, P. ASC Filament Formation Serves as a Signal Amplification Mechanism for Inflammasomes. Nat. Commun. 2016, 7, 11929. [Google Scholar] [CrossRef]
- Bauernfeind, F.; Hornung, V. Of Inflammasomes and Pathogens—Sensing of Microbes by the Inflammasome. EMBO Mol. Med. 2013, 5, 814–826. [Google Scholar] [CrossRef]
- Lu, A.; Magupalli, V.G.; Ruan, J.; Yin, Q.; Atianand, M.K.; Vos, M.R.; Schröder, G.F.; Fitzgerald, K.A.; Wu, H.; Egelman, E.H. Unified Polymerization Mechanism for the Assembly of Asc-Dependent Inflammasomes. Cell 2014, 156, 1193–1206. [Google Scholar] [CrossRef]
- Sharma, B.R.; Kanneganti, T.D. NLRP3 Inflammasome in Cancer and Metabolic Diseases. Nat. Immunol. 2021, 22, 550–559. [Google Scholar] [CrossRef]
- Bauernfeind, F.G.; Horvath, G.; Stutz, A.; Alnemri, E.S.; MacDonald, K.; Speert, D.; Fernandes-Alnemri, T.; Wu, J.; Monks, B.G.; Fitzgerald, K.A.; et al. Cutting Edge: NF-ΚB Activating Pattern Recognition and Cytokine Receptors License NLRP3 Inflammasome Activation by Regulating NLRP3 Expression. J. Immunol. 2009, 183, 787–791. [Google Scholar] [CrossRef] [PubMed]
- Franchi, L.; Muñoz-Planillo, R.; Núñez, G. Sensing and Reacting to Microbes through the Inflammasomes. Nat. Immunol. 2012, 13, 325–332. [Google Scholar] [CrossRef] [PubMed]
- Zahid, A.; Li, B.; Kombe, A.J.K.; Jin, T.; Tao, J. Pharmacological Inhibitors of the Nlrp3 Inflammasome. Front. Immunol. 2019, 10, 2538. [Google Scholar] [CrossRef] [PubMed]
- Magupalli, V.G.; Negro, R.; Tian, Y.; Hauenstein, A.V.; Di Caprio, G.; Skillern, W.; Deng, Q.; Orning, P.; Alam, H.B.; Maliga, Z.; et al. HDAC6 Mediates an Aggresome-like Mechanism for NLRP3 and Pyrin Inflammasome Activation. Science 2020, 369, eaas8995. [Google Scholar] [CrossRef] [PubMed]
- Beckwith, K.S.; Beckwith, M.S.; Ullmann, S.; Sætra, R.S.; Kim, H.; Marstad, A.; Åsberg, S.E.; Strand, T.A.; Haug, M.; Niederweis, M.; et al. Plasma Membrane Damage Causes NLRP3 Activation and Pyroptosis during Mycobacterium Tuberculosis Infection. Nat. Commun. 2020, 11, 2270. [Google Scholar] [CrossRef]
- Di, A.; Xiong, S.; Ye, Z.; Malireddi, R.K.S.; Kometani, S.; Zhong, M.; Mittal, M.; Hong, Z.; Kanneganti, T.D.; Rehman, J.; et al. The TWIK2 Potassium Efflux Channel in Macrophages Mediates NLRP3 Inflammasome-Induced Inflammation. Immunity 2018, 49, 56–65.e4. [Google Scholar] [CrossRef]
- Lee, G.S.; Subramanian, N.; Kim, A.I.; Aksentijevich, I.; Goldbach-Mansky, R.; Sacks, D.B.; Germain, R.N.; Kastner, D.L.; Chae, J.J. The Calcium-Sensing Receptor Regulates the NLRP3 Inflammasome through Ca2+ and CAMP. Nature 2012, 492, 123–127. [Google Scholar] [CrossRef]
- Yu, X.; Lan, P.; Hou, X.; Han, Q.; Lu, N.; Li, T.; Jiao, C.; Zhang, J.; Zhang, C.; Tian, Z. HBV Inhibits LPS-Induced NLRP3 Inflammasome Activation and IL-1β Production via Suppressing the NF-ΚB Pathway and ROS Production. J. Hepatol. 2017, 66, 693–702. [Google Scholar] [CrossRef]
- Zhong, Z.; Liang, S.; Sanchez-Lopez, E.; He, F.; Shalapour, S.; Lin, X.J.; Wong, J.; Ding, S.; Seki, E.; Schnabl, B.; et al. New Mitochondrial DNA Synthesis Enables NLRP3 Inflammasome Activation. Nature 2018, 560, 198–203. [Google Scholar] [CrossRef]
- Sha, W.; Mitoma, H.; Hanabuchi, S.; Bao, M.; Weng, L.; Sugimoto, N.; Liu, Y.; Zhang, Z.; Zhong, J.; Sun, B.; et al. Human NLRP3 Inflammasome Senses Multiple Types of Bacterial RNAs. Proc. Natl. Acad. Sci. USA 2014, 111, 16059–16064. [Google Scholar] [CrossRef]
- Mitoma, H.; Hanabuchi, S.; Kim, T.; Bao, M.; Zhang, Z.; Sugimoto, N.; Liu, Y.J. The DHX33 RNA Helicase Senses Cytosolic RNA and Activates the NLRP3 Inflammasome. Immunity 2013, 39, 123–135. [Google Scholar] [CrossRef] [PubMed]
- Tarallo, V.; Hirano, Y.; Gelfand, B.D.; Dridi, S.; Kerur, N.; Kim, Y.; Cho, W.G.; Kaneko, H.; Fowler, B.J.; Bogdanovich, S.; et al. DICER1 Loss and Alu RNA Induce Age-Related Macular Degeneration via the NLRP3 Inflammasome and MyD88. Cell 2012, 149, 847–859. [Google Scholar] [CrossRef] [PubMed]
- Milner, M.T.; Maddugoda, M.; Götz, J.; Burgener, S.S.; Schroder, K. The NLRP3 Inflammasome Triggers Sterile Neuroinflammation and Alzheimer’s Disease. Curr. Opin. Immunol. 2021, 68, 116–124. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, W.T.; Pham, L.; Symons, G.F.; Monif, M.; Shultz, S.R.; McDonald, S.J. The NLRP3 Inflammasome in Traumatic Brain Injury: Potential as a Biomarker and Therapeutic Target. J. Neuroinflamm. 2020, 17, 104. [Google Scholar] [CrossRef]
- Forster, J.; Nandi, D.; Kulkarni, A. MRNA-Carrying Lipid Nanoparticles That Induce Lysosomal Rupture Activate NLRP3 Inflammasome and Reduce MRNA Transfection Efficiency. Biomater. Sci. 2022, 10, 5566–5582. [Google Scholar] [CrossRef]
- Won, T.; Gilotra, N.A.; Wood, M.K.; Hughes, D.M.; Talor, M.V.; Lovell, J.; Milstone, A.M.; Steenbergen, C.; Čiháková, D. Increased Interleukin 18-Dependent Immune Responses Are Associated with Myopericarditis after COVID-19 MRNA Vaccination. Front. Immunol. 2022, 13, 851620. [Google Scholar] [CrossRef]
- Zhang, R.; Hong, F.; Zhao, M.; Cai, X.; Jiang, X.; Ye, N.; Su, K.; Li, N.; Tang, M.; Ma, X.; et al. New Highly Potent NLRP3 Inhibitors: Furanochalcone Velutone F Analogues. ACS Med. Chem. Lett. 2022, 13, 560–569. [Google Scholar] [CrossRef]
- Awe, J.P.; Crespo, A.V.; Li, Y.; Kiledjian, M.; Byrne, J.A. BAY11 Enhances OCT4 Synthetic MRNA Expression in Adult Human Skin Cells. Stem Cell Res. Ther. 2013, 4, 15. [Google Scholar] [CrossRef]
- Martinon, F.; Burns, K.; Tschopp, J. The Inflammasome: A Molecular Platform Triggering Activation of Inflammatory Caspases and Processing of ProIL-β. Mol. Cell 2002, 10, 417–426. [Google Scholar] [CrossRef]
- Den Brok, M.H.; Büll, C.; Wassink, M.; De Graaf, A.M.; Wagenaars, J.A.; Minderman, M.; Thakur, M.; Amigorena, S.; Rijke, E.O.; Schrier, C.C.; et al. Saponin-Based Adjuvants Induce Cross-Presentation in Dendritic Cells by Intracellular Lipid Body Formation. Nat. Commun. 2016, 7, 13324. [Google Scholar] [CrossRef]
- Kayagaki, N.; Warming, S.; Lamkanfi, M.; Walle, L.V.; Louie, S.; Dong, J.; Newton, K.; Qu, Y.; Liu, J.; Heldens, S.; et al. Non-Canonical Inflammasome Activation Targets Caspase-11. Nature 2011, 479, 117–121. [Google Scholar] [CrossRef]
- Muñoz-Wolf, N.; McCluskey, S.; Lavelle, E.C. The Role of Inflammasomes in Adjuvant-Driven Humoral and Cellular Immune Responses. In Immunopotentiators in Modern Vaccines, 2nd ed.; Academic Press: New York, NY, USA, 2017. [Google Scholar]
- Davidsen, J.; Rosenkrands, I.; Christensen, D.; Vangala, A.; Kirby, D.; Perrie, Y.; Agger, E.M.; Andersen, P. Characterization of Cationic Liposomes Based on Dimethyldioctadecylammonium and Synthetic Cord Factor from M. tuberculosis (Trehalose 6,6′-Dibehenate)—A Novel Adjuvant Inducing Both Strong CMI and Antibody Responses. Biochim. Biophys. Acta Biomembr. 2005, 1718, 22–31. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Wang, X.; Huang, X.; Zhang, J.; Xia, N.; Zhao, Q. Calcium Phosphate Nanoparticles as a New Generation Vaccine Adjuvant. Expert Rev. Vaccines 2017, 16, 895–906. [Google Scholar] [CrossRef] [PubMed]
- Sharp, F.A.; Ruane, D.; Claass, B.; Creagh, E.; Harris, J.; Malyala, P.; Singh, M.; O’Hagan, D.T.; Pétrilli, V.; Tschopp, J.; et al. Uptake of Particulate Vaccine Adjuvants by Dendritic Cells Activates the NALP3 Inflammasome. Proc. Natl. Acad. Sci. USA 2009, 106, 870–875. [Google Scholar] [CrossRef] [PubMed]
- Zelensky, A.N.; Gready, J.E. The C-Type Lectin-like Domain Superfamily. FEBS J. 2005, 272, 6179–6217. [Google Scholar] [CrossRef] [PubMed]
- Drouin, M.; Saenz, J.; Chiffoleau, E. C-Type Lectin-like Receptors: Head or Tail in Cell Death Immunity. Front. Immunol. 2020, 11, 251. [Google Scholar] [CrossRef] [PubMed]
- Sancho, D.; Reis e Sousa, C. Sensing of Cell Death by Myeloid C-Type Lectin Receptors. Curr. Opin. Immunol. 2013, 25, 46–52. [Google Scholar] [CrossRef]
- Bermejo-Jambrina, M.; Eder, J.; Helgers, L.C.; Hertoghs, N.; Nijmeijer, B.M.; Stunnenberg, M.; Geijtenbeek, T.B.H. C-Type Lectin Receptors in Antiviral Immunity and Viral Escape. Front. Immunol. 2018, 9, 590. [Google Scholar] [CrossRef]
- Meyer-Wentrup, F.; Cambi, A.; Joosten, B.; Looman, M.W.; de Vries, I.J.M.; Figdor, C.G.; Adema, G.J. DCIR Is Endocytosed into Human Dendritic Cells and Inhibits TLR8-Mediated Cytokine Production. J. Leukoc. Biol. 2009, 85, 518–525. [Google Scholar] [CrossRef]
- Zhao, X.; Shen, Y.; Hu, W.; Chen, J.; Wu, T.; Sun, X.; Yu, J.; Wu, T.; Chen, W. DCIR Negatively Regulates CpG-ODN-Induced IL-1β and IL-6 Production. Mol. Immunol. 2015, 68, 641–647. [Google Scholar] [CrossRef]
- Troegeler, A.; Mercier, I.; Cougoule, C.; Pietretti, D.; Colom, A.; Duval, C.; Vu Manh, T.P.; Capilla, F.; Poincloux, R.; Pingris, K.; et al. C-Type Lectin Receptor DCIR Modulates Immunity to Tuberculosis by Sustaining Type I Interferon Signaling in Dendritic Cells. Proc. Natl. Acad. Sci. USA 2017, 114, E540–E549. [Google Scholar] [CrossRef] [PubMed]
- Cao, W.; Zhang, L.; Rosen, D.B.; Bover, L.; Watanabe, G.; Bao, M.; Lanier, L.L.; Liu, Y.J. BDCA2/FcεRIγ Complex Signals through a Novel BCR-like Pathway in Human Plasmacytoid Dendritic Cells. PLoS Biol. 2007, 5, e248. [Google Scholar] [CrossRef] [PubMed]
- Geijtenbeek, T.B.H.; Gringhuis, S.I. Signalling through C-Type Lectin Receptors: Shaping Immune Responses. Nat. Rev. Immunol. 2009, 9, 465–479. [Google Scholar] [CrossRef] [PubMed]
- Dzionek, A.; Sohma, Y.; Nagafune, J.; Cella, M.; Colonna, M.; Facchetti, F.; Günther, G.; Johnston, I.; Lanzavecchia, A.; Nagasaka, T.; et al. BDCA-2, a Novel Plasmacytoid Dendritic Cell-Specific Type II C-Type Lectin, Mediates Antigen Capture and Is a Potent Inhibitor of Interferon α/β Induction. J. Exp. Med. 2001, 194, 1823–1834. [Google Scholar] [CrossRef] [PubMed]
- Ludwig, I.S.; Lekkerkerker, A.N.; Depla, E.; Bosman, F.; Musters, R.J.P.; Depraetere, S.; van Kooyk, Y.; Geijtenbeek, T.B.H. Hepatitis C Virus Targets DC-SIGN and L-SIGN To Escape Lysosomal Degradation. J. Virol. 2004, 78, 8322–8332. [Google Scholar] [CrossRef]
- Ablasser, A.; Chen, Z.J. CGAS in Action: Expanding Roles in Immunity and Inflammation. Science 2019, 363, eaat8657. [Google Scholar] [CrossRef] [PubMed]
- Ablasser, A.; Hur, S. Regulation of CGAS- and RLR-Mediated Immunity to Nucleic Acids. Nat. Immunol. 2020, 21, 17–29. [Google Scholar] [CrossRef]
- Ishikawa, H.; Ma, Z.; Barber, G.N. STING Regulates Intracellular DNA-Mediated, Type i Interferon-Dependent Innate Immunity. Nature 2009, 461, 788–792. [Google Scholar] [CrossRef]
- Burdette, D.L.; Monroe, K.M.; Sotelo-Troha, K.; Iwig, J.S.; Eckert, B.; Hyodo, M.; Hayakawa, Y.; Vance, R.E. STING Is a Direct Innate Immune Sensor of Cyclic Di-GMP. Nature 2011, 478, 515–518. [Google Scholar] [CrossRef]
- Ni, G.; Ma, Z.; Damania, B. CGAS and STING: At the Intersection of DNA and RNA Virus-Sensing Networks. PLoS Pathog. 2018, 14, e1007148. [Google Scholar] [CrossRef]
- Civril, F.; Deimling, T.; De Oliveira Mann, C.C.; Ablasser, A.; Moldt, M.; Witte, G.; Hornung, V.; Hopfner, K.P. Structural Mechanism of Cytosolic DNA Sensing by CGAS. Nature 2013, 498, 332–337. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, H.; Barber, G.N. STING Is an Endoplasmic Reticulum Adaptor that Facilitates Innate Immune Signalling. Nature 2008, 455, 674–678. [Google Scholar] [CrossRef] [PubMed]
- Zhong, B.; Yang, Y.; Li, S.; Wang, Y.Y.; Li, Y.; Diao, F.; Lei, C.; He, X.; Zhang, L.; Tien, P.; et al. The Adaptor Protein MITA Links Virus-Sensing Receptors to IRF3 Transcription Factor Activation. Immunity 2008, 29, 538–550. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Goulet, M.-L.; Sze, A.; Hadj, S.B.; Belgnaoui, S.M.; Lababidi, R.R.; Zheng, C.; Fritz, J.H.; Olagnier, D.; Lin, R. RIG-I-Mediated STING Upregulation Restricts Herpes Simplex Virus 1 Infection. J. Virol. 2016, 90, 9406–9419. [Google Scholar] [CrossRef] [PubMed]
- Franz, K.M.; Neidermyer, W.J.; Tan, Y.J.; Whelan, S.P.J.; Kagan, J.C. STING-Dependent Translation Inhibition Restricts RNA Virus Replication. Proc. Natl. Acad. Sci. USA 2018, 115, E2058–E2067. [Google Scholar] [CrossRef]
- Ding, Q.; Cao, X.; Lu, J.; Huang, B.; Liu, Y.J.; Kato, N.; Shu, H.B.; Zhong, J. Hepatitis C Virus NS4B Blocks the Interaction of STING and TBK1 to Evade Host Innate Immunity. J. Hepatol. 2013, 59, 52–58. [Google Scholar] [CrossRef]
- Ma, Z.; Damania, B. The CGAS-STING Defense Pathway and Its Counteraction by Viruses. Cell Host Microbe 2016, 19, 150–158. [Google Scholar] [CrossRef]
- Vincent, J.; Adura, C.; Gao, P.; Luz, A.; Lama, L.; Asano, Y.; Okamoto, R.; Imaeda, T.; Aida, J.; Rothamel, K.; et al. Small Molecule Inhibition of CGAS Reduces Interferon Expression in Primary Macrophages from Autoimmune Mice. Nat. Commun. 2017, 8, 750. [Google Scholar] [CrossRef]
- Hall, J.; Brault, A.; Vincent, F.; Weng, S.; Wang, H.; Dumlao, D.; Aulabaugh, A.; Aivazian, D.; Castro, D.; Chen, M.; et al. Discovery of PF-06928215 as a High Affinity Inhibitor of CGAS Enabled by a Novel Fluorescence Polarization Assay. PLoS ONE 2017, 12, e0184843. [Google Scholar] [CrossRef]
- Lama, L.; Adura, C.; Xie, W.; Tomita, D.; Kamei, T.; Kuryavyi, V.; Gogakos, T.; Steinberg, J.I.; Miller, M.; Ramos-Espiritu, L.; et al. Development of Human CGAS-Specific Small-Molecule Inhibitors for Repression of DsDNA-Triggered Interferon Expression. Nat. Commun. 2019, 10, 2261. [Google Scholar] [CrossRef]
- Padilla-Salinas, R.; Sun, L.; Anderson, R.; Yang, X.; Zhang, S.; Chen, Z.J.; Yin, H. Discovery of Small-Molecule Cyclic GMP-AMP Synthase Inhibitors. J. Org. Chem. 2020, 85, 1579–1600. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Xiong, M.; Yuan, X.; Li, M.; Sun, H.; Xu, Y. In Silico Screening-Based Discovery of Novel Inhibitors of Human Cyclic GMP-AMP Synthase: A Cross-Validation Study of Molecular Docking and Experimental Testing. J. Chem. Inf. Model. 2020, 60, 3265–3276. [Google Scholar] [CrossRef] [PubMed]
- An, J.; Woodward, J.J.; Sasaki, T.; Minie, M.; Elkon, K.B. Cutting Edge: Antimalarial Drugs Inhibit IFN-β Production through Blockade of Cyclic GMP-AMP Synthase–DNA Interaction. J. Immunol. 2015, 194, 4089–4093. [Google Scholar] [CrossRef] [PubMed]
- An, J.; Minie, M.; Sasaki, T.; Woodward, J.J.; Elkon, K.B. Antimalarial Drugs as Immune Modulators: New Mechanisms for Old Drugs. Annu. Rev. Med. 2017, 68, 317–330. [Google Scholar] [CrossRef] [PubMed]
- An, J.; Woodward, J.J.; Lai, W.; Minie, M.; Sun, X.; Tanaka, L.; Snyder, J.M.; Sasaki, T.; Elkon, K.B. Inhibition of Cyclic GMP-AMP Synthase Using a Novel Antimalarial Drug Derivative in Trex1-Deficient Mice. Arthritis Rheumatol. 2018, 70, 1807–1819. [Google Scholar] [CrossRef]
- Steinhagen, F.; Zillinger, T.; Peukert, K.; Fox, M.; Thudium, M.; Barchet, W.; Putensen, C.; Klinman, D.; Latz, E.; Bode, C. Suppressive Oligodeoxynucleotides Containing TTAGGG Motifs Inhibit CGAS Activation in Human Monocytes. Eur. J. Immunol. 2018, 48, 605–611. [Google Scholar] [CrossRef]
- Wang, M.; Sooreshjani, M.A.; Mikek, C.; Opoku-Temeng, C.; Sintim, H.O. Suramin Potently Inhibits CGAMP Synthase, CGAS, in THP1 Cells to Modulate IFN-β Levels. Future Med. Chem. 2018, 10, 1301–1317. [Google Scholar] [CrossRef]
- Decout, A.; Katz, J.D.; Venkatraman, S.; Ablasser, A. The CGAS–STING Pathway as a Therapeutic Target in Inflammatory Diseases. Nat. Rev. Immunol. 2021, 21, 548–569. [Google Scholar] [CrossRef]
- Van Herck, S.; Feng, B.; Tang, L. Delivery of STING Agonists for Adjuvanting Subunit Vaccines. Adv. Drug Deliv. Rev. 2021, 179, 114020. [Google Scholar] [CrossRef]
- Tse, S.W.; McKinney, K.; Walker, W.; Nguyen, M.; Iacovelli, J.; Small, C.; Hopson, K.; Zaks, T.; Huang, E. MRNA-Encoded, Constitutively Active STINGV155M Is a Potent Genetic Adjuvant of Antigen-Specific CD8+ T Cell Response. Mol. Ther. 2021, 29, 2227–2238. [Google Scholar] [CrossRef]
- Won, J.K.; Bakhoum, S.F. The Cytosolic DNA-Sensing CGAS–Sting Pathway in Cancer. Cancer Discov. 2020, 10, 26–39. [Google Scholar]
- Miao, L.; Li, L.; Huang, Y.; Delcassian, D.; Chahal, J.; Han, J.; Shi, Y.; Sadtler, K.; Gao, W.; Lin, J.; et al. Delivery of MRNA Vaccines with Heterocyclic Lipids Increases anti-Tumor Efficacy by STING-Mediated Immune Cell Activation. Nat. Biotechnol. 2019, 37, 1174–1185. [Google Scholar] [CrossRef] [PubMed]
- Parker, R.; Song, H. The Enzymes and Control of Eukaryotic MRNA Turnover. Nat. Struct. Mol. Biol. 2004, 11, 121–127. [Google Scholar] [CrossRef]
- Schoenberg, D.R. Mechanisms of Endonuclease-Mediated MRNA Decay. Wiley Interdiscip. Rev. RNA 2011, 2, 582–600. [Google Scholar] [CrossRef] [PubMed]
- Houseley, J.; LaCava, J.; Tollervey, D. RNA-Quality Control by the Exosome. Nat. Rev. Mol. Cell Biol. 2006, 7, 529–539. [Google Scholar] [CrossRef] [PubMed]
- Guerra, G.M.; May, D.; Kroll, T.; Koch, P.; Groth, M.; Wang, Z.Q.; Li, T.L.; Grigaravičius, P. Cell Type-Specific Role of RNA Nuclease SMG6 in Neurogenesis. Cells 2021, 10, 3365. [Google Scholar] [CrossRef]
- Li, W.M.; Barnes, T.; Lee, C.H. Endoribonucleases-Enzymes Gaining Spotlight in MRNA Metabolism. FEBS J. 2010, 277, 627–641. [Google Scholar] [CrossRef]
- Gómez-Aguado, I.; Rodríguez-Castejón, J.; Vicente-Pascual, M.; Rodríguez-Gascón, A.; Solinís, M.Á.; Del Pozo-Rodríguez, A. Nanomedicines to Deliver MRNA: State of the Art and Future Perspectives. Nanomaterials 2020, 10, 364. [Google Scholar] [CrossRef]
- Noubissi, F.K.; Elcheva, I.; Bhatia, N.; Shakoori, A.; Ougolkov, A.; Liu, J.; Minamoto, T.; Ross, J.; Fuchs, S.Y.; Spiegelman, V.S. CRD-BP Mediates Stabilization of ΒTrCP1 and c-Myc MRNA in Response to β-Catenin Signalling. Nature 2006, 441, 898–901. [Google Scholar] [CrossRef]
- Sparanese, D.; Lee, C.H. CRD-BP Shields c-Myc and MDR-1 RNA from Endonucleolytic Attack by a Mammalian Endoribonuclease. Nucleic Acids Res. 2007, 35, 1209–1221. [Google Scholar] [CrossRef]
- Linares-Fernández, S.; Lacroix, C.; Exposito, J.Y.; Verrier, B. Tailoring MRNA Vaccine to Balance Innate/Adaptive Immune Response. Trends Mol. Med. 2020, 26, 311–323. [Google Scholar] [CrossRef] [PubMed]
- Tahtinen, S.; Tong, A.J.; Himmels, P.; Oh, J.; Paler-Martinez, A.; Kim, L.; Wichner, S.; Oei, Y.; McCarron, M.J.; Freund, E.C.; et al. IL-1 and IL-1ra Are Key Regulators of the Inflammatory Response to RNA Vaccines. Nat. Immunol. 2022, 23, 532–542. [Google Scholar] [CrossRef] [PubMed]
- Karpov, T.; Postovalova, A.; Akhmetova, D.; Muslimov, A.R.; Eletskaya, E.; Zyuzin, M.V.; Timin, A.S. Universal Chelator-Free Radiolabeling of Organic and Inorganic-Based Nanocarriers with Diagnostic and Therapeutic Isotopes for Internal Radiotherapy. Chem. Mater. 2022, 34, 6593–6605. [Google Scholar] [CrossRef]
- Diebold, S.S.; Kaisho, T.; Hemmi, H.; Akira, S.; Reis, E.; Sousa, C. Innate Antiviral Responses by Means of TLR7-Mediated Recognition of Single-Stranded RNA. Science 2004, 303, 1529–1531. [Google Scholar] [CrossRef] [PubMed]
- Heil, F.; Hemmi, H.; Hochrein, H.; Ampenberger, F.; Kirschning, C.; Akira, S.; Lipford, G.; Wagner, H.; Bauer, S. Species-Specific Recognition of Single-Stranded RNA via Till-like Receptor 7 and 8. Science 2004, 303, 1526–1529. [Google Scholar] [CrossRef] [PubMed]
- Hornung, V.; Ellegast, J.; Kim, S.; Brzózka, K.; Jung, A.; Kato, H.; Poeck, H.; Akira, S.; Conzelmann, K.K.; Schlee, M.; et al. 5′-Triphosphate RNA Is the Ligand for RIG-I. Science 2006, 314, 994–997. [Google Scholar] [CrossRef] [PubMed]
- Anderson, B.R.; Muramatsu, H.; Nallagatla, S.R.; Bevilacqua, P.C.; Sansing, L.H.; Weissman, D.; Karikó, K. Incorporation of Pseudouridine into MRNA Enhances Translation by Diminishing PKR Activation. Nucleic Acids Res. 2010, 38, 5884–5892. [Google Scholar] [CrossRef]
- Devoldere, J.; Dewitte, H.; De Smedt, S.C.; Remaut, K. Evading Innate Immunity in Nonviral MRNA Delivery: Don’t Shoot the Messenger. Drug Discov. Today 2016, 21, 11–25. [Google Scholar] [CrossRef]
- Baiersdörfer, M.; Boros, G.; Muramatsu, H.; Mahiny, A.; Vlatkovic, I.; Sahin, U.; Karikó, K. A Facile Method for the Removal of DsRNA Contaminant from In Vitro-Transcribed MRNA. Mol. Ther. Nucleic Acids 2019, 15, 26–35. [Google Scholar] [CrossRef]
- Piao, X.; Yadav, V.; Wang, E.; Chang, W.; Tau, L.; Lindenmuth, B.E.; Wang, S.X. Double-Stranded RNA Reduction by Chaotropic Agents during in vitro Transcription of Messenger RNA. Mol. Ther. Nucleic Acids 2022, 29, 618–624. [Google Scholar] [CrossRef]
- Podda, A.; Del Giudice, G. MF59-Adjuvanted Vaccines: Increased Immunogenicity with an Optimal Safety Profile. Expert Rev. Vaccines 2003, 2, 197–204. [Google Scholar] [CrossRef]
- Pardi, N.; Hogan, M.J.; Porter, F.W.; Weissman, D. MRNA Vaccines—A New Era in Vaccinology. Nat. Rev. Drug Discov. 2018, 17, 261–279. [Google Scholar] [CrossRef] [PubMed]
- Saunders, K.O.; Pardi, N.; Parks, R.; Santra, S.; Mu, Z.; Sutherland, L.; Scearce, R.; Barr, M.; Eaton, A.; Hernandez, G.; et al. Lipid Nanoparticle Encapsulated Nucleoside-Modified MRNA Vaccines Elicit Polyfunctional HIV-1 Antibodies Comparable to Proteins in Nonhuman Primates. Npj Vaccines 2021, 6, 50. [Google Scholar] [CrossRef] [PubMed]
- Ndeupen, S.; Qin, Z.; Jacobsen, S.; Bouteau, A.; Estanbouli, H.; Igyártó, B.Z. The MRNA-LNP Platform’s Lipid Nanoparticle Component Used in Preclinical Vaccine Studies Is Highly Inflammatory. iScience 2021, 24, 103479. [Google Scholar] [CrossRef] [PubMed]
- Parhiz, H.; Brenner, J.S.; Patel, P.N.; Papp, T.E.; Shahnawaz, H.; Li, Q.; Shi, R.; Zamora, M.E.; Yadegari, A.; Marcos-Contreras, O.A.; et al. Added to Pre-Existing Inflammation, MRNA-Lipid Nanoparticles Induce Inflammation Exacerbation (IE). J. Control. Release 2022, 344, 50–61. [Google Scholar] [CrossRef]
- Abrams, M.T.; Koser, M.L.; Seitzer, J.; Williams, S.C.; Dipietro, M.A.; Wang, W.; Shaw, A.W.; Mao, X.; Jadhav, V.; Davide, J.P.; et al. Evaluation of Efficacy, Biodistribution, and Inflammation for a Potent SiRNA Nanoparticle: Effect of Dexamethasone Co-Treatment. Mol. Ther. 2010, 18, 171–180. [Google Scholar] [CrossRef]
- Bevers, S.; Kooijmans, S.A.A.; Van de Velde, E.; Evers, M.J.W.; Seghers, S.; Gitz-Francois, J.J.J.M.; van Kronenburg, N.C.H.; Fens, M.H.A.M.; Mastrobattista, E.; Hassler, L.; et al. MRNA-LNP Vaccines Tuned for Systemic Immunization Induce Strong Antitumor Immunity by Engaging Splenic Immune Cells. Mol. Ther. 2022, 30, 3078–3094. [Google Scholar] [CrossRef]
- Kon, E.; Elia, U.; Peer, D. Principles for Designing an Optimal MRNA Lipid Nanoparticle Vaccine. Curr. Opin. Biotechnol. 2022, 73, 329–336. [Google Scholar] [CrossRef]
- Chatzikleanthous, D.; O’Hagan, D.T.; Adamo, R. Lipid-Based Nanoparticles for Delivery of Vaccine Adjuvants and Antigens: Toward Multicomponent Vaccines. Mol. Pharm. 2021, 18, 2867–2888. [Google Scholar] [CrossRef]
- Fan, J.; Jin, S.; Gilmartin, L.; Toth, I.; Hussein, W.M.; Stephenson, R.J. Advances in Infectious Disease Vaccine Adjuvants. Vaccines 2022, 10, 1120. [Google Scholar] [CrossRef]
- Eusébio, D.; Neves, A.R.; Costa, D.; Biswas, S.; Alves, G.; Cui, Z.; Sousa, Â. Methods to Improve the Immunogenicity of Plasmid DNA Vaccines. Drug Discov. Today 2021, 26, 2575–2592. [Google Scholar] [CrossRef]
- Wu, Z.; Liu, K. Overview of Vaccine Adjuvants. Med. Drug Discov. 2021, 11, 100103. [Google Scholar] [CrossRef]
- Garçon, N.; Leroux-Roels, G.; Cheng, W.-F. Vaccine Adjuvants. Perspect. Vaccinol. 2011, 1, 89–113. [Google Scholar] [CrossRef]
- Miller, S.M.; Cybulski, V.; Whitacre, M.; Bess, L.S.; Livesay, M.T.; Walsh, L.; Burkhart, D.; Bazin, H.G.; Evans, J.T. Novel Lipidated Imidazoquinoline TLR7/8 Adjuvants Elicit Influenza-Specific Th1 Immune Responses and Protect Against Heterologous H3N2 Influenza Challenge in Mice. Front. Immunol. 2020, 11, 406. [Google Scholar] [CrossRef] [PubMed]
- Pifferi, C.; Fuentes, R.; Fernández-Tejada, A. Natural and Synthetic Carbohydrate-Based Vaccine Adjuvants and Their Mechanisms of Action. Nat. Rev. Chem. 2021, 5, 197–216. [Google Scholar] [CrossRef] [PubMed]
- Aucouturier, J.; Dupuis, L.; Ganne, V. Adjuvants Designed for Veterinary and Human Vaccines. Vaccine 2001, 19, 2666–2672. [Google Scholar] [CrossRef]
- Díaz-Dinamarca, D.A.; Salazar, M.L.; Castillo, B.N.; Manubens, A.; Vasquez, A.E.; Salazar, F.; Becker, M.I. Protein-Based Adjuvants for Vaccines as Immunomodulators of the Innate and Adaptive Immune Response: Current Knowledge, Challenges, and Future Opportunities. Pharmaceutics 2022, 14, 1671. [Google Scholar] [CrossRef]
- Draper, S.J.; Angov, E.; Horii, T.; Miller, L.H.; Srinivasan, P.; Theisen, M.; Biswas, S. Recent Advances in Recombinant Protein-Based Malaria Vaccines. Vaccine 2015, 33, 7433–7443. [Google Scholar] [CrossRef] [PubMed]
- Schijns, V.E.; Lavelle, E.C. Trends in Vaccine Adjuvants. Expert Rev. Vaccines 2011, 10, 539–550. [Google Scholar] [CrossRef]
- Nguyen-Contant, P.; Sangster, M.Y.; Topham, D.J. Squalene-Based Influenza Vaccine Adjuvants and Their Impact on the Hemagglutinin-Specific B Cell Response. Pathogens 2021, 10, 355. [Google Scholar] [CrossRef]
- Brito, L.A.; Chan, M.; Shaw, C.A.; Hekele, A.; Carsillo, T.; Schaefer, M.; Archer, J.; Seubert, A.; Otten, G.R.; Beard, C.W.; et al. A Cationic Nanoemulsion for the Delivery of Next-Generation RNA Vaccines. Mol. Ther. 2014, 22, 2118–2129. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Petrovsky, N. Molecular Adjuvants for DNA Vaccines. Curr. Issues Mol. Biol. 2017, 22, 17–40. [Google Scholar] [CrossRef] [PubMed]
- Kobiyama, K.; Jounai, N.; Aoshi, T.; Tozuka, M.; Takeshita, F.; Coban, C.; Ishii, K. Innate Immune Signaling by, and Genetic Adjuvants for DNA Vaccination. Vaccines 2013, 1, 278–292. [Google Scholar] [CrossRef]
- Floros, T.; Tarhini, A.A. Anticancer Cytokines: Biology and Clinical Effects of Interferon-A2, Interleukin (IL)-2, IL-15, IL-21, and IL-12. In Seminars in Oncology; WB Saunders: Philadelphia, PA, USA, 2015; Volume 42. [Google Scholar]
- Konjević, G.M.; Vuletić, A.M.; Mirjačić Martinović, K.M.; Larsen, A.K.; Jurišić, V.B. The Role of Cytokines in the Regulation of NK Cells in the Tumor Environment. Cytokine 2019, 117, 30–40. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Liu, C.H.; Roberts, A.I.; Das, J.; Xu, G.; Ren, G.; Zhang, Y.; Zhang, L.; Zeng, R.Y.; Tan, H.S.W.; et al. Granulocyte-Macrophage Colony-Stimulating Factor (GM-CSF) and T-Cell Responses: What We Do and Don’t Know. Cell Res. 2006, 16, 126–133. [Google Scholar] [CrossRef]
- Luster, A.D. The Role of Chemokines in Linking Innate and Adaptive Immunity. Curr. Opin. Immunol. 2002, 14, 129–135. [Google Scholar] [CrossRef]
- Van Kooten, C.; Banchereau, J. Functions of CD40 on B Cells, Dendritic Cells and Other Cells. Curr. Opin. Immunol. 1997, 9, 330–337. [Google Scholar] [CrossRef]
- Gool, S.W.; Vandenberghe, P.; de Boer, M.; Ceuppens, J.L. CD80, CD86 and CD40 Provide Accessory Signals in a Multiple-Step T-Cell Activation Model. Immunol. Rev. 1996, 153, 47–83. [Google Scholar] [CrossRef]
- Applequist, S.E.; Rollman, E.; Wareing, M.D.; Lidén, M.; Rozell, B.; Hinkula, J.; Ljunggren, H.-G. Activation of Innate Immunity, Inflammation, and Potentiation of DNA Vaccination through Mammalian Expression of the TLR5 Agonist Flagellin. J. Immunol. 2005, 175, 3882–3891. [Google Scholar] [CrossRef]
- Leal, L.; Guardo, A.C.; Moron-Lopez, S.; Salgado, M.; Mothe, B.; Heirman, C.; Pannus, P.; Vanham, G.; Van Den Ham, H.J.; Gruters, R.; et al. Phase i Clinical Trial of an Intranodally Administered MRNA-Based Therapeutic Vaccine against HIV-1 Infection. AIDS 2018, 32, 2533–2545. [Google Scholar] [CrossRef]
- Bonehill, A.; Tuyaerts, S.; Van Nuffel, A.M.T.; Heirman, C.; Bos, T.J.; Fostier, K.; Neyns, B.; Thielemans, K. Enhancing the T-Cell Stimulatory Capacity of Human Dendritic Cells by Co-Electroporation with CD40L, CD70 and Constitutively Active TLR4 Encoding MRNA. Mol. Ther. 2008, 16, 1170–1180. [Google Scholar] [CrossRef]
- Van Lint, S.; Goyvaerts, C.; Maenhout, S.; Goethals, L.; Disy, A.; Benteyn, D.; Pen, J.; Bonehill, A.; Heirman, C.; Breckpot, K.; et al. Preclinical Evaluation of TriMix and Antigen MRNA-Based Antitumor Therapy. Cancer Res. 2012, 72, 1661–1671. [Google Scholar] [CrossRef] [PubMed]
- Takeshita, F.; Tanaka, T.; Matsuda, T.; Tozuka, M.; Kobiyama, K.; Saha, S.; Matsui, K.; Ishii, K.J.; Coban, C.; Akira, S.; et al. Toll-Like Receptor Adaptor Molecules Enhance DNA-Raised Adaptive Immune Responses against Influenza and Tumors through Activation of Innate Immunity. J. Virol. 2006, 80, 6218–6224. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Cai, S.; Pang, H.; Jian, J.; Wu, Z. Immunogenicity and Efficacy of DNA Vaccine Encoding Antigenic AcfA via Addition of the Molecular Adjuvant Myd88 against Vibrio Alginolyticus in Epinephelus Coioides. Fish Shellfish. Immunol. 2017, 66, 71–77. [Google Scholar] [CrossRef] [PubMed]
- Wan, C.; Yi, L.; Yang, Z.; Yang, J.; Shao, H.; Zhang, C.; Pan, Z. The Toll-like Receptor Adaptor Molecule TRIF Enhances DNA Vaccination against Classical Swine Fever. Vet. Immunol. Immunopathol. 2010, 137, 47–53. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, S.; Amara, R.R.; Yeow, W.-S.; Pitha, P.M.; Robinson, H.L. Regulation of DNA-Raised Immune Responses by Cotransfected Interferon Regulatory Factors. J. Virol. 2002, 76, 6652–6659. [Google Scholar] [CrossRef]
- Castaldello, A.; Sgarbanti, M.; Marsili, G.; Brocca-Cofano, E.; Remoli, A.L.; Caputo, A.; Battistini, A. Interferon Regulatory Factor-1 Acts as a Powerful Adjuvant in Tat DNA Based Vaccination. J. Cell. Physiol. 2010, 224, 702–709. [Google Scholar] [CrossRef]
- Bramson, J.L.; Dayball, K.; Hall, J.R.; Millar, J.B.; Miller, M.; Wan, Y.H.; Lin, R.; Hiscott, J. Super-Activated Interferon-Regulatory Factors Can Enhance Plasmid Immunization. Vaccine 2003, 21, 1363–1370. [Google Scholar] [CrossRef]
- Chu, D.; Moroda, M.; Piao, L.X.; Aosai, F. CTL Induction by DNA Vaccine with Toxoplasma Gondii-HSP70 Gene. Parasitol. Int. 2014, 63, 408–416. [Google Scholar] [CrossRef]
- Zhou, J.; Cheung, A.K.L.; Tan, Z.; Wang, H.; Yu, W.; Du, Y.; Kang, Y.; Lu, X.; Liu, L.; Yuen, K.Y.; et al. PD1-Based DNA Vaccine Amplifies HIV-1 GAG-Specific CD8+ T Cells in Mice. J. Clin. Investig. 2013, 123, 2629–2642. [Google Scholar] [CrossRef]
- Liniger, M.; Summerfield, A.; Ruggli, N. MDA5 Can Be Exploited as Efficacious Genetic Adjuvant for DNA Vaccination against Lethal H5N1 Influenza Virus Infection in Chickens. PLoS ONE 2012, 7, e49952. [Google Scholar] [CrossRef]
- Shedlock, D.J.; Tingey, C.; Mahadevan, L.; Hutnick, N.; Reuschel, E.L.; Kudchodkar, S.; Flingai, S.; Yan, J.; Kim, J.J.; Ugen, K.E.; et al. Co-Administration of Molecular Adjuvants Expressing NF-Kappa B Subunit P65/RelA or Type-1 Transactivator T-Bet Enhance Antigen Specific DNA Vaccine-Induced Immunity. Vaccines 2014, 2, 196–215. [Google Scholar] [CrossRef] [PubMed]
- Hu, D.; Wu, J.; Zhang, R.; Chen, L. T-Bet Acts as a Powerful Adjuvant in Ag85B DNA-Based Vaccination against Tuberculosis. Mol. Med. Rep. 2012, 6, 139–144. [Google Scholar] [CrossRef] [PubMed]
- Coban, C.; Kobiyama, K.; Aoshi, T.; Takeshita, F.; Horii, T.; Akira, S.; Ishii, K.J. Novel Strategies to Improve DNA Vaccine Immunogenicity. Curr. Gene Ther. 2011, 11, 479–484. [Google Scholar] [CrossRef] [PubMed]
- Muthumani, G.; Laddy, D.J.; Sundaram, S.G.; Fagone, P.; Shedlock, D.J.; Kannan, S.; Wu, L.; Chung, C.W.; Lankaraman, K.M.; Burns, J.; et al. Co-Immunization with an Optimized Plasmid-Encoded Immune Stimulatory Interleukin, High-Mobility Group Box 1 Protein, Results in Enhanced Interferon-β Secretion by Antigen-Specific CD8 T Cells. Immunology 2009, 128, e612–e620. [Google Scholar] [CrossRef] [PubMed]
- Fagone, P.; Shedlock, D.J.; Bao, H.; Kawalekar, O.U.; Yan, J.; Gupta, D.; Morrow, M.P.; Patel, A.; Kobinger, G.P.; Muthumani, K.; et al. Molecular Adjuvant HMGB1 Enhances Anti-Influenza Immunity during DNA Vaccination. Gene Ther. 2011, 18, 1070–1077. [Google Scholar] [CrossRef] [PubMed]
- Lladser, A.; Mougiakakos, D.; Tufvesson, H.; Ligtenberg, M.A.; Quest, A.F.G.; Kiessling, R.; Ljungberg, K. DAI (DLM-1/ZBP1) as a Genetic Adjuvant for DNA Vaccines That Promotes Effective Antitumor CTL Immunity. Mol. Ther. 2011, 19, 594–601. [Google Scholar] [CrossRef]
- Siddiqui, A.A.; Phillips, T.; Charest, H.; Podesta, R.B.; Quinlin, M.L.; Pinkston, J.R.; Lloyd, J.D.; Paz, M.; Villalovos, R.M.; Pompa, J. Induction of Protective Immunity against Schistosoma Mansoni via DNA Priming and Boosting with the Large Subunit of Calpain (Sm-P80): Adjuvant Effects of Granulocyte-Macrophage Colony-Stimulating Factor and Interleukin-4. Infect. Immun. 2003, 71, 3844–3851. [Google Scholar] [CrossRef]
- Sedegah, M.; Weiss, W.; Sacci, J.B.; Charoenvit, Y.; Hedstrom, R.; Gowda, K.; Majam, V.F.; Tine, J.; Kumar, S.; Hobart, P.; et al. Improving Protective Immunity Induced by DNA-Based Immunization: Priming with Antigen and GM-CSF-Encoding Plasmid DNA and Boosting with Antigen-Expressing Recombinant Poxvirus. J. Immunol. 2000, 164, 5905–5912. [Google Scholar] [CrossRef]
- Norell, H.; Poschke, I.; Charo, J.; Wei, W.Z.; Erskine, C.; Piechocki, M.P.; Knutson, K.L.; Bergh, J.; Lidbrink, E.; Kiessling, R. Vaccination with a Plasmid DNA Encoding HER-2/Neu Together with Low Doses of GM-CSF and IL-2 in Patients with Metastatic Breast Carcinoma: A Pilot Clinical Trial. J. Transl. Med. 2010, 8, 53. [Google Scholar] [CrossRef]
- Perales, M.A.; Yuan, J.; Powel, S.; Gallardo, H.F.; Rasalan, T.S.; Gonzalez, C.; Manukian, G.; Wang, J.; Zhang, Y.; Chapman, P.B.; et al. Phase I/II Study of GM-CSF DNA as an Adjuvant for a Multipeptide Cancer Vaccine in Patients with Advanced Melanoma. Mol. Ther. 2008, 16, 2022–2029. [Google Scholar] [CrossRef] [PubMed]
- Siddiqui, A.A.; Phillips, T.; Charest, H.; Podesta, R.B.; Quinlin, M.L.; Pinkston, J.R.; Lloyd, J.D.; Pompa, J.; Villalovos, R.M.; Paz, M. Enhancement of Sm-P80 (Large Subunit of Calpain) Induced Protective Immunity against Schistosoma Mansoni through Co-Delivery of Interleukin-2 and Interleukin-12 in a DNA Vaccine Formulation. Vaccine 2003, 21, 2882–2889. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Taylor, M.G.; Johansen, M.V.; Bickle, Q.D. Vaccination of Mice with a Cocktail DNA Vaccine Induces a Th1-Type Immune Response and Partial Protection against Schistosoma Japonicum Infection. Vaccine 2001, 20, 724–730. [Google Scholar] [CrossRef] [PubMed]
- Khosroshahi, K.H.; Ghaffarifar, F.; Sharifi, Z.; D’Souza, S.; Dalimi, A.; Hassan, Z.M.; Khoshzaban, F. Comparing the Effect of IL-12 Genetic Adjuvant and Alum Non-Genetic Adjuvant on the Efficiency of the Cocktail DNA Vaccine Containing Plasmids Encoding SAG-1 and ROP-2 of Toxoplasma Gondii. Parasitol. Res. 2012, 111, 403–411. [Google Scholar] [CrossRef]
- Maspi, N.; Ghaffarifar, F.; Sharifi, Z.; Dalimi, A. Codelivery of DNA Vaccination Encoding LeIF Gene and IL-12 Increases Protection against Leishmania Major Infection in BALB/c Mice. Parasite Immunol. 2016, 38, 228–235. [Google Scholar] [CrossRef]
- Naderi, M.; Saeedi, A.; Moradi, A.; Kleshadi, M.; Zolfaghari, M.R.; Gorji, A.; Ghaemi, A. Interleukin-12 as a Genetic Adjuvant Enhances Hepatitis C Virus NS3 DNA Vaccine Immunogenicity. Virol. Sin. 2013, 28, 167–173. [Google Scholar] [CrossRef]
- Yang, S.H.; Lee, C.G.; Park, S.H.; Im, S.J.; Kim, Y.M.; Son, J.M.; Wang, J.S.; Yoon, S.K.; Song, M.K.; Ambrozaitis, A.; et al. Correlation of Antiviral T-Cell Responses with Suppression of Viral Rebound in Chronic Hepatitis B Carriers: A Proof-of-Concept Study. Gene Ther. 2006, 13, 1110–1117. [Google Scholar] [CrossRef]
- Yang, Y.; Shao, Z.; Gao, J. Antitumor Effect of a DNA Vaccine Harboring Prostate Cancer-Specific Antigen with IL-12 as an Intramolecular Adjuvant. J. Mol. Microbiol. Biotechnol. 2017, 27, 168–174. [Google Scholar] [CrossRef]
- Tian, D.Y.; Sun, Y.; Wai, S.F.; Lee, F.K.; Meng, Q.L.; Suen, K.M.; Wang, N.; Han, W.; Li, S.; Li, Y.F.; et al. Enhancement of the Immunogenicity of an Alphavirus Replicon-Based DNA Vaccine against Classical Swine Fever by Electroporation and Coinjection with a Plasmid Expressing Porcine Interleukin 2. Vaccine 2012, 30, 3587–3594. [Google Scholar] [CrossRef]
- Zhao, G.W.; Yan, R.F.; Muleke, C.I.; Sun, Y.M.; Xu, L.X.; Li, X.R. Vaccination of Goats with DNA Vaccines Encoding H11 and IL-2 Induces Partial Protection against Haemonchus Contortus Infection. Vet. J. 2012, 191, 94–100. [Google Scholar] [CrossRef]
- Passardi, A.; Cecconetto, L.; Dall’Agata, M.; Dazzi, C.; Pasquini, E.; Oliverio, G.; Zumaglini, F.; Zoli, W.; Nanni, O.; Milandri, C.; et al. Randomized Phase II Study with Two Gemcitabine- and Docetaxel-Based Combinations as First-Line Chemotherapy for Metastatic Non-Small Cell Lung Cancer. J. Transl. Med. 2008, 6, 65. [Google Scholar] [CrossRef] [PubMed]
- Hezarjaribi, H.Z.; Ghaffarifar, F.; Dalimi, A.; Sharifi, Z.; Jorjani, O. Effect of IL-22 on DNA Vaccine Encoding LACK Gene of Leishmania Major in BALB/c Mice. Exp. Parasitol. 2013, 134, 341–348. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.Y.; Chen, J.; Petersen, E.; Zhou, D.H.; Huang, S.Y.; Song, H.Q.; Zhu, X.Q. Synergy of MIL-21 and MIL-15 in Enhancing DNA Vaccine Efficacy against Acute and Chronic Toxoplasma Gondii Infection in Mice. Vaccine 2014, 32, 3058–3065. [Google Scholar] [CrossRef]
- Chen, J.; Li, Z.Y.; Huang, S.Y.; Petersen, E.; Song, H.Q.; Zhou, D.H.; Zhu, X.Q. Protective Efficacy of Toxoplasma Gondii Calcium-Dependent Protein Kinase 1 (TgCDPK1) Adjuvated with Recombinant IL-15 and IL-21 against Experimental Toxoplasmosis in Mice. BMC Infect. Dis. 2014, 14, 487. [Google Scholar] [CrossRef] [PubMed]
- Eickhoff, C.S.; Vasconcelos, J.R.; Sullivan, N.L.; Blazevic, A.; Bruna-Romero, O.; Rodrigues, M.M.; Hoft, D.F. Co-Administration of a Plasmid DNA Encoding IL-15 Improves Long-Term Protection of a Genetic Vaccine against Trypanosoma Cruzi. PLoS Negl. Trop. Dis. 2011, 5, e983. [Google Scholar] [CrossRef]
- Kutzler, M.A.; Robinson, T.M.; Chattergoon, M.A.; Choo, D.K.; Choo, A.Y.; Choe, P.Y.; Ramanathan, M.P.; Parkinson, R.; Kudchodkar, S.; Tamura, Y.; et al. Coimmunization with an Optimized IL-15 Plasmid Results in Enhanced Function and Longevity of CD8 T Cells That Are Partially Independent of CD4 T Cell Help. J. Immunol. 2005, 175, 112–123. [Google Scholar] [CrossRef]
- Su, B.; Wang, J.; Zhao, G.; Wang, X.; Li, J.; Wang, B. Sequential Administration of Cytokine Genes to Enhance Cellular Immune Responses and CD4+ T Memory Cells during DNA Vaccination. Hum. Vaccines Immunother. 2012, 8, 1659–1667. [Google Scholar] [CrossRef]
- Williman, J.; Young, S.; Buchan, G.; Slobbe, L.; Wilson, M.; Pang, P.; Austyn, J.; Preston, S.; Baird, M. DNA Fusion Vaccines Incorporating IL-23 or RANTES for Use in Immunization against Influenza. Vaccine 2008, 26, 5153–5158. [Google Scholar] [CrossRef]
- Song, R.; Liu, S.; Leong, K.W. Effects of MIP-1α, MIP-3α, and MIP-3β on the Induction of HIV Gag-Specific Immune Response with DNA Vaccines. Mol. Ther. 2007, 15, 1007–1015. [Google Scholar] [CrossRef]
- Westermann, J.; Nguyen-Hoai, T.; Baldenhofer, G.; Höpken, U.E.; Lipp, M.; Dörken, B.; Pezzutto, A. CCL19 (ELC) as an Adjuvant for DNA Vaccination: Induction of a TH1-Type T-Cell Response and Enhancement of Antitumor Immunity. Cancer Gene Ther. 2007, 14, 523–532. [Google Scholar] [CrossRef]
- Kang, T.H.; Kim, K.W.; Bae, H.C.; Seong, S.Y.; Kim, T.W. Enhancement of DNA Vaccine Potency by Antigen Linkage to IFN-Iγ-Inducible Protein-10. Int. J. Cancer 2011, 128, 702–714. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.W.; Aleyas, A.G.; George, J.A.; Kim, S.J.; Kim, H.K.; Yoo, D.J.; Kang, S.H.; Eo, S.K. Genetic Co-Transfer of CCR7 Ligands Enhances Immunity and Prolongs Survival against Virulent Challenge of Pseudorabies Virus. Immunol. Cell Biol. 2009, 87, 91–99. [Google Scholar] [CrossRef]
- Toka, F.N.; Gierynska, M.; Rouse, B.T. Codelivery of CCR7 Ligands as Molecular Adjuvants Enhances the Protective Immune Response against Herpes Simplex Virus Type 1. J. Virol. 2003, 77, 12742–12752. [Google Scholar] [CrossRef] [PubMed]
- Yamano, T.; Kaneda, Y.; Huang, S.; Hiramatsu, S.H.; Hoon, D.S.B. Enhancement of Immunity by a DNA Melanoma Vaccine against TRP2 with CCL21 as an Adjuvant. Mol. Ther. 2006, 13, 194–202. [Google Scholar] [CrossRef] [PubMed]
- Fló, J.; Tisminetzky, S.; Baralle, F. Modulation of the Immune Response to DNA Vaccine by Co-Delivery of Costimulatory Molecules. Immunology 2000, 100, 259–267. [Google Scholar] [CrossRef] [PubMed]
- de Andrés, X.; Reina, R.; Ciriza, J.; Crespo, H.; Glaria, I.; Ramírez, H.; Grilló, M.J.; Pérez, M.M.; Andrésdóttir, V.; Rosati, S.; et al. Use of B7 Costimulatory Molecules as Adjuvants in a Prime-Boost Vaccination against Visna/Maedi Ovine Lentivirus. Vaccine 2009, 27, 4591–4600. [Google Scholar] [CrossRef] [PubMed]
- Xue, H.; Liang, F.; Liu, N.; Song, X.; Yuan, F.; Luo, Y.; Zhao, X.; Long, J.; Sun, Y.; Xi, Y. Potent Antirheumatic Activity of a New DNA Vaccine Targeted to B7-2/CD28 Costimulatory Signaling Pathway in Autoimmune Arthritis. Hum. Gene Ther. 2011, 22, 65–76. [Google Scholar] [CrossRef]
- Stone, G.W.; Barzee, S.; Snarsky, V.; Kee, K.; Spina, C.A.; Yu, X.-F.; Kornbluth, R.S. Multimeric Soluble CD40 Ligand and GITR Ligand as Adjuvants for Human Immunodeficiency Virus DNA Vaccines. J. Virol. 2006, 80, 1762–1772. [Google Scholar] [CrossRef]
- Dybul, M.; Mercier, G.; Belson, M.; Hallahan, C.W.; Liu, S.; Perry, C.; Herpin, B.; Ehler, L.; Davey, R.T.; Metcalf, J.A.; et al. CD40 Ligand Trimer and IL-12 Enhance Peripheral Blood Mononuclear Cells and CD4 + T Cell Proliferation and Production of IFN-γ in Response to P24 Antigen in HIV-Infected Individuals: Potential Contribution of Anergy to HIV-Specific Unresponsiveness. J. Immunol. 2000, 165, 1685–1691. [Google Scholar] [CrossRef]
- Elia, U.; Ramishetti, S.; Rosenfeld, R.; Dammes, N.; Bar-Haim, E.; Naidu, G.S.; Makdasi, E.; Yahalom-Ronen, Y.; Tamir, H.; Paran, N.; et al. Design of SARS-CoV-2 HFc-Conjugated Receptor-Binding Domain MRNA Vaccine Delivered via Lipid Nanoparticles. ACS Nano 2021, 15, 9627–9637. [Google Scholar] [CrossRef]
- Elia, U.; Rotem, S.; Bar-Haim, E.; Ramishetti, S.; Naidu, G.S.; Gur, D.; Aftalion, M.; Israeli, M.; Bercovich-Kinori, A.; Alcalay, R.; et al. Lipid Nanoparticle RBD-HFc MRNA Vaccine Protects HACE2 Transgenic Mice against a Lethal SARS-CoV-2 Infection. Nano Lett. 2021, 21, 4774–4779. [Google Scholar] [CrossRef] [PubMed]
- Duivelshof, B.L.; Murisier, A.; Camperi, J.; Fekete, S.; Beck, A.; Guillarme, D.; D’Atri, V. Therapeutic Fc-Fusion Proteins: Current Analytical Strategies. J. Sep. Sci. 2021, 44, 35–62. [Google Scholar] [CrossRef] [PubMed]
- Goswami, R.; Chatzikleanthous, D.; Lou, G.; Giusti, F.; Bonci, A.; Taccone, M.; Brazzoli, M.; Gallorini, S.; Ferlenghi, I.; Berti, F.; et al. Mannosylation of LNP Results in Improved Potency for Self-Amplifying RNA (SAM) Vaccines. ACS Infect. Dis. 2019, 5, 1546–1558. [Google Scholar] [CrossRef] [PubMed]
- Satija, R.; Shalek, A.K. Heterogeneity in Immune Responses: From Populations to Single Cells. Trends Immunol. 2014, 35, 219–229. [Google Scholar] [CrossRef]
- Hervé, C.; Laupèze, B.; Del Giudice, G.; Didierlaurent, A.M.; Da Silva, F.T. The How’s and What’s of Vaccine Reactogenicity. Npj Vaccines 2019, 4, 39. [Google Scholar] [CrossRef]
- Dabo, S.; Meurs, E.F. DsRNA-Dependent Protein Kinase PKR and Its Role in Stress, Signaling and HCV Infection. Viruses 2012, 4, 2598–2635. [Google Scholar] [CrossRef]
- Kim, Y.; Park, J.; Kim, S.; Kim, M.A.; Kang, M.G.; Kwak, C.; Kang, M.; Kim, B.; Rhee, H.W.; Kim, V.N. PKR Senses Nuclear and Mitochondrial Signals by Interacting with Endogenous Double-Stranded RNAs. Mol. Cell 2018, 71, 1051–1063.e6. [Google Scholar] [CrossRef]
- Drappier, M.; Michiels, T. Inhibition of the OAS/RNase L Pathway by Viruses. Curr. Opin. Virol. 2015, 15, 19–26. [Google Scholar] [CrossRef]
- Shevyrev, D.; Blinova, E.A.; Kozlov, V.A. The influence of humoral factors of homeostatistic proliferation on t-regulatory cells in vitro. Bull. Sib. Med. 2019, 18, 286–293. [Google Scholar] [CrossRef]
- Bolze, A.; Neveux, I.; Schiabor Barrett, K.M.; White, S.; Isaksson, M.; Dabe, S.; Lee, W.; Grzymski, J.J.; Washington, N.L.; Cirulli, E.T. HLA-A*03:01 Is Associated with Increased Risk of Fever, Chills, and Stronger Side Effects from Pfizer-BioNTech COVID-19 Vaccination. Hum. Genet. Genom. Adv. 2022, 3, 100084. [Google Scholar] [CrossRef]
- Mouliou, D.S.; Dardiotis, E. Current Evidence in SARS-CoV-2 MRNA Vaccines and Post-Vaccination Adverse Reports: Knowns and Unknowns. Diagnostics 2022, 12, 1555. [Google Scholar] [CrossRef] [PubMed]
- Seneff, S.; Nigh, G.; Kyriakopoulos, A.M.; McCullough, P.A. Innate Immune Suppression by SARS-CoV-2 MRNA Vaccinations: The Role of G-Quadruplexes, Exosomes, and MicroRNAs. Food Chem. Toxicol. 2022, 164, 113008. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhao, M.; Zhou, W.; Chen, L.; Li, M.; Jiang, B.; Zhao, X.; Wu, H.; Shen, L.; Zhang, N.; et al. The Link between Genetic Variation and Variability in Vaccine Responses: A Narrative Review. J. Bio-X Res. 2022, 5, 49–54. [Google Scholar] [CrossRef]
- Newport, M.J. The Genetic Regulation of Infant Immune Responses to Vaccination. Front. Immunol. 2015, 6, 18. [Google Scholar] [CrossRef]
- Nishibayashi, H.; Kishimoto, S.; Sekiguchi, K.; Okada, S.; Ihara, K. Myocarditis in 13-Year-Old Monochorionic Diamniotic Twins After COVID-19 Vaccination. J. Clin. Immunol. 2022, 42, 1351–1353. [Google Scholar] [CrossRef]
- Heymans, S.; Cooper, L.T. Myocarditis after COVID-19 MRNA Vaccination: Clinical Observations and Potential Mechanisms. Nat. Rev. Cardiol. 2022, 19, 75–77. [Google Scholar] [CrossRef]
- Monge, S.; Pastor-Barriuso, R.; Hernán, M.A. The imprinting effect of COVID-19 vaccines: An expected selection bias in observational studies. BMJ 2023, 381, e074404. [Google Scholar] [CrossRef]
- Jankowsky, E.; Fairman, M.E. RNA helicases—One fold for many functions. Curr. Opin. Struct. Biol. 2007, 17, 316–324. [Google Scholar] [CrossRef]
- Chavda, V.P.; Hossain, M.K.; Beladiya, J.; Apostolopoulos, V. Nucleic Acid Vaccines for COVID-19: A Paradigm Shift in the Vaccine Development Arena. Biologics 2021, 1, 337–356. [Google Scholar] [CrossRef]
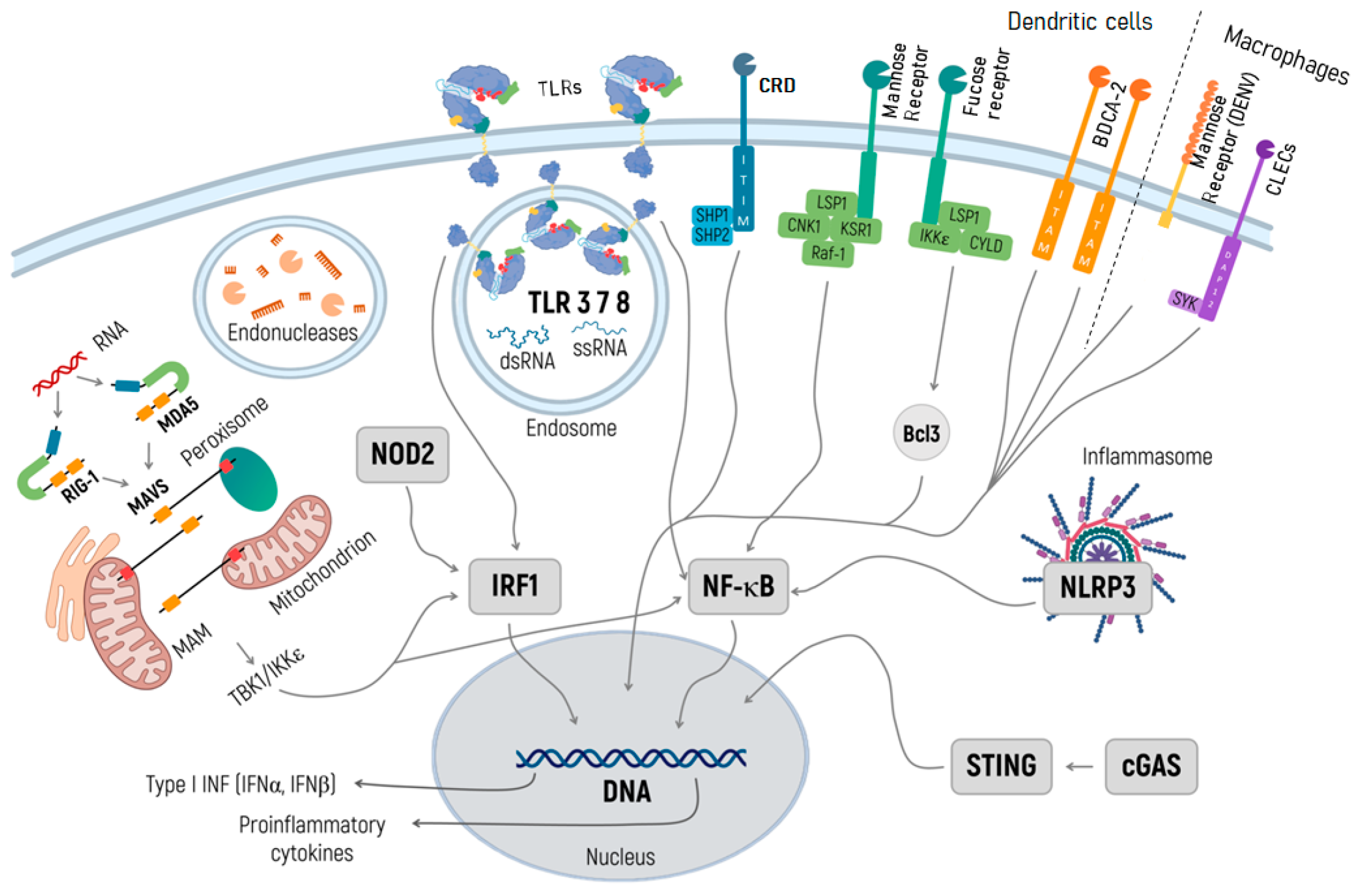
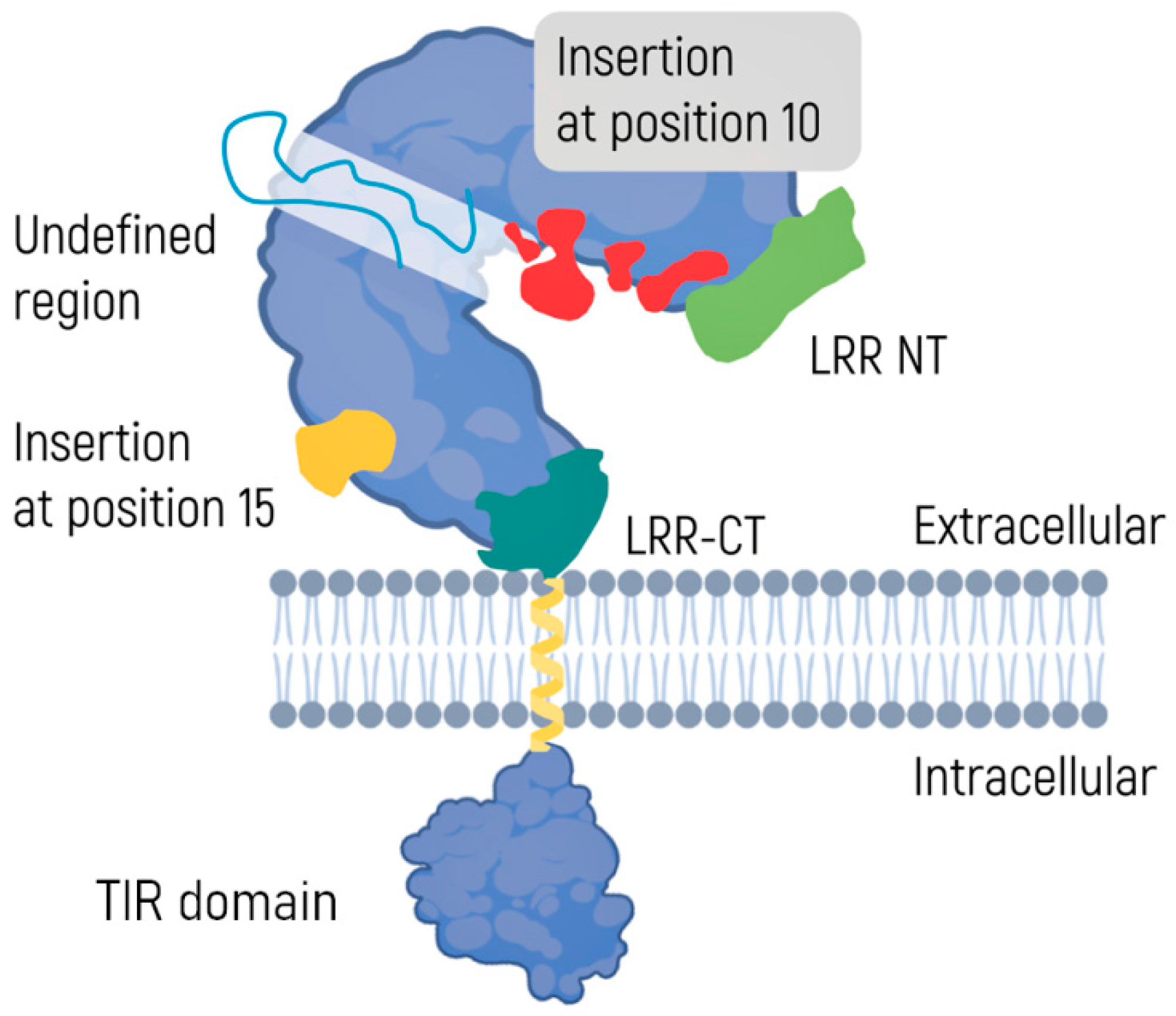
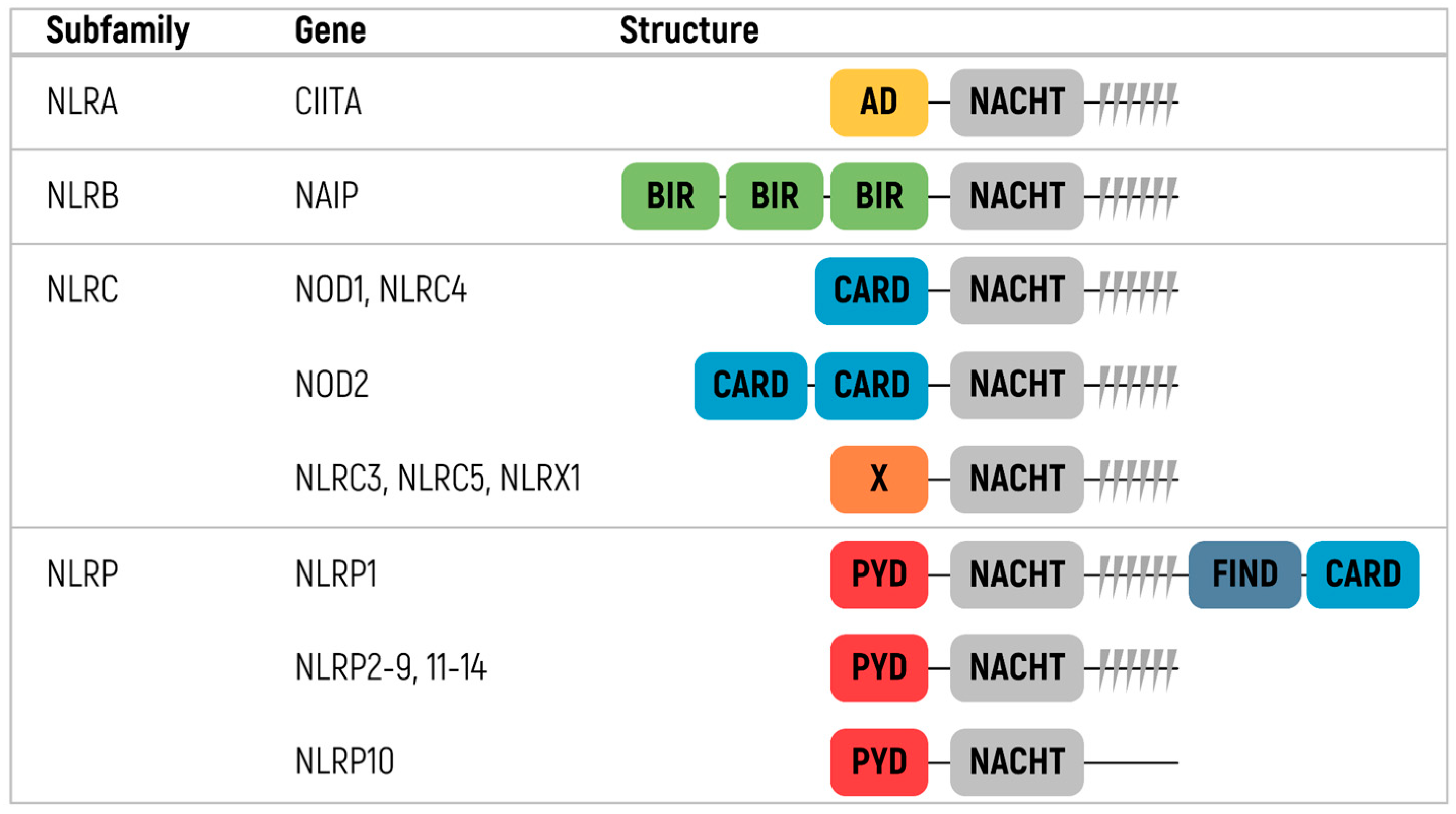
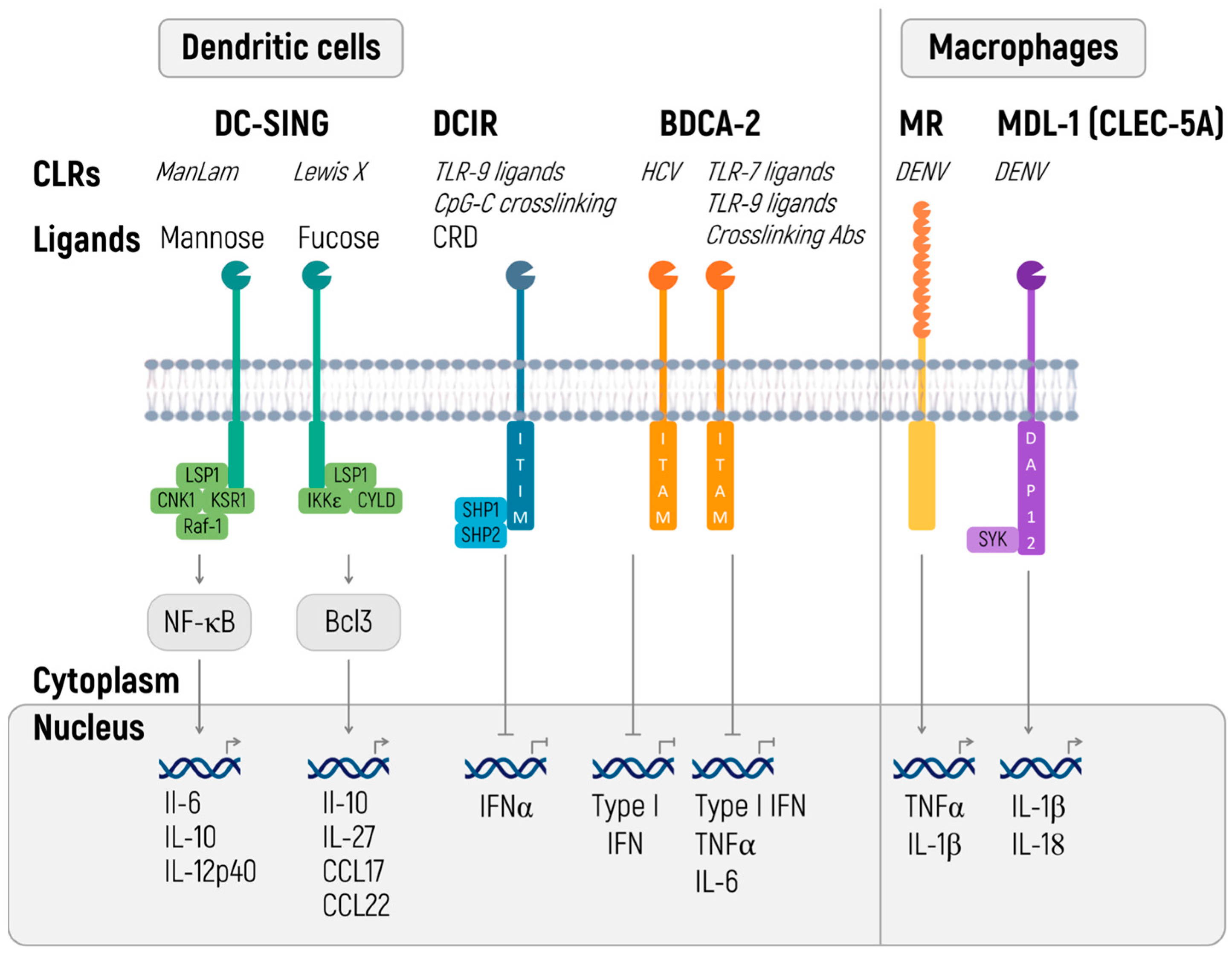
| Biomolecules | Specific Proteins | Roles | Ref. |
|---|---|---|---|
| Cytokines | IL-1, IL-2, IL-5, IL-6, IL-8, Il-12, TNF, G-CSF, GM-CSF, IFN-β, etc. | Mediate the innate immune response | [13] |
| Chemokines | MCP-1, IL-18, RANTES, MIP-2, CXCL 10, CXCL1 | Promote angiogenesis | [14] |
| Cell cycle regulators | Bcl-2L1, PAI2, cyclins | Stimulate cell proliferation | [15] |
| Anti-apoptotic factors | Fas, BCL-2, c-FLIP, caspases, IAPs, BFL-1, survivin | Apoptosis regulation | [16,17] |
| Adhesion molecules | E-selectin, ICAM-1, VCAM-1, ECAM-1, MMPs | Adhesion and cell migration | [18,19] |
| Adjuvant Category | Component | Ref. |
|---|---|---|
| Mineral salts | Alum or calcium salts | [264] |
| Chemical compounds | Imidazoquinolines, saponin and its derivatives | [265] |
| Synthetic polynucleotides adjuvants | Poly I:C, CpG oligonucleotides | [261,266] |
| Adjuvants of bacterial and viral origin | Flagellin, cholera toxin, Bacillus Calmette–Guérin (BCG), virus-like particles, LPS | [267] |
| Protein-based adjuvants | Proteases, cytokines, chemokines | [268,269] |
| Emulsions adjuvants | Freund’s adjuvant, incomplete Freund’s adjuvant, and montanides (MF59 and others) | [270,271] |
| Name | Main Signaling Pathway | Ref. |
|---|---|---|
| Chromatography purification | NLRP3 | [94,188] |
| mRNA capping | RIG-I and MDA5, RLR signaling | [36,47,48,49] |
| Reduction of mRNA uridine content | RIG-I and MDA5, RLR | [36,47] |
| Incorporation of modified nucleosides | RIG-I and MDA5, RLR | [36,47,94,119,120,121,122,123,124] |
| mRNA polyadenylation | RIG-I and MDA5, RLR | [36,47] |
| Reduction of double-stranded IVT products | RIG-I and MDA5, RLR | [36,47] |
| Inhibition of PRR | RIG-I and MDA5, RLR | [36,47] |
| Enhanced DCIR stimulation | TLR | [199] |
| Increased BDCA-2 stimulation | TLR | [199] |
| cGAS antagonists | cGAS | [221,222,223,224,225] |
| Glyburide | NLRP3 | [188] |
| 16673-34-0 | NLRP3 | [188] |
| FC11A-2 | NLRP3 | [188] |
| VX-765 | NLRP3 | [188] |
| VX-740 | NLRP3 | [188] |
| BHB | NLRP3 | [188] |
| MCC950 | NLRP3 | [188] |
| CY-09 | NLRP3 | [188] |
| Tranilast | NLRP3 | [188] |
| OLT1177 | NLRP3 | [188] |
| Oridonin | NLRP3 | [188] |
| Parthenolide | NLRP3 | [188] |
| Furanochalcone Veluton | NLRP3 | [188] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Muslimov, A.; Tereshchenko, V.; Shevyrev, D.; Rogova, A.; Lepik, K.; Reshetnikov, V.; Ivanov, R. The Dual Role of the Innate Immune System in the Effectiveness of mRNA Therapeutics. Int. J. Mol. Sci. 2023, 24, 14820. https://doi.org/10.3390/ijms241914820
Muslimov A, Tereshchenko V, Shevyrev D, Rogova A, Lepik K, Reshetnikov V, Ivanov R. The Dual Role of the Innate Immune System in the Effectiveness of mRNA Therapeutics. International Journal of Molecular Sciences. 2023; 24(19):14820. https://doi.org/10.3390/ijms241914820
Chicago/Turabian StyleMuslimov, Albert, Valeriy Tereshchenko, Daniil Shevyrev, Anna Rogova, Kirill Lepik, Vasiliy Reshetnikov, and Roman Ivanov. 2023. "The Dual Role of the Innate Immune System in the Effectiveness of mRNA Therapeutics" International Journal of Molecular Sciences 24, no. 19: 14820. https://doi.org/10.3390/ijms241914820
APA StyleMuslimov, A., Tereshchenko, V., Shevyrev, D., Rogova, A., Lepik, K., Reshetnikov, V., & Ivanov, R. (2023). The Dual Role of the Innate Immune System in the Effectiveness of mRNA Therapeutics. International Journal of Molecular Sciences, 24(19), 14820. https://doi.org/10.3390/ijms241914820








