Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Spike Protein S1 Induces Methylglyoxal-Derived Hydroimidazolone/Receptor for Advanced Glycation End Products (MG-H1/RAGE) Activation to Promote Inflammation in Human Bronchial BEAS-2B Cells
Abstract
:1. Introduction
2. Results
2.1. The Effect of the SARS-CoV-2 S1 Spike Protein on Human Bronchial BEAS-2B and Alveolar A549 Cell Viability
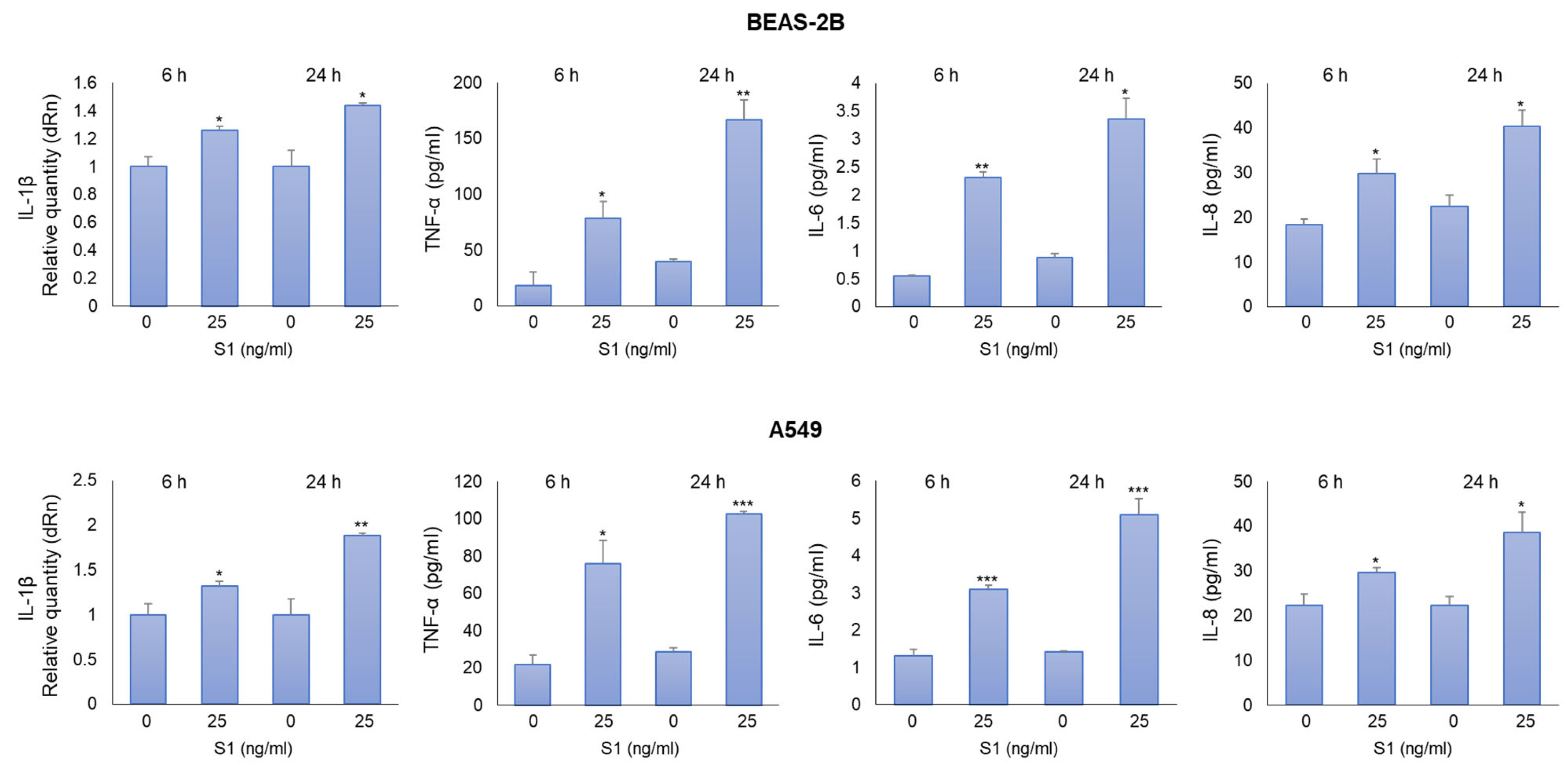
2.2. SARS-CoV-2 S1 Spike Protein Induces Inflammatory Cytokines in Human Bronchial BEAS-2B and Alveolar A549 Cells
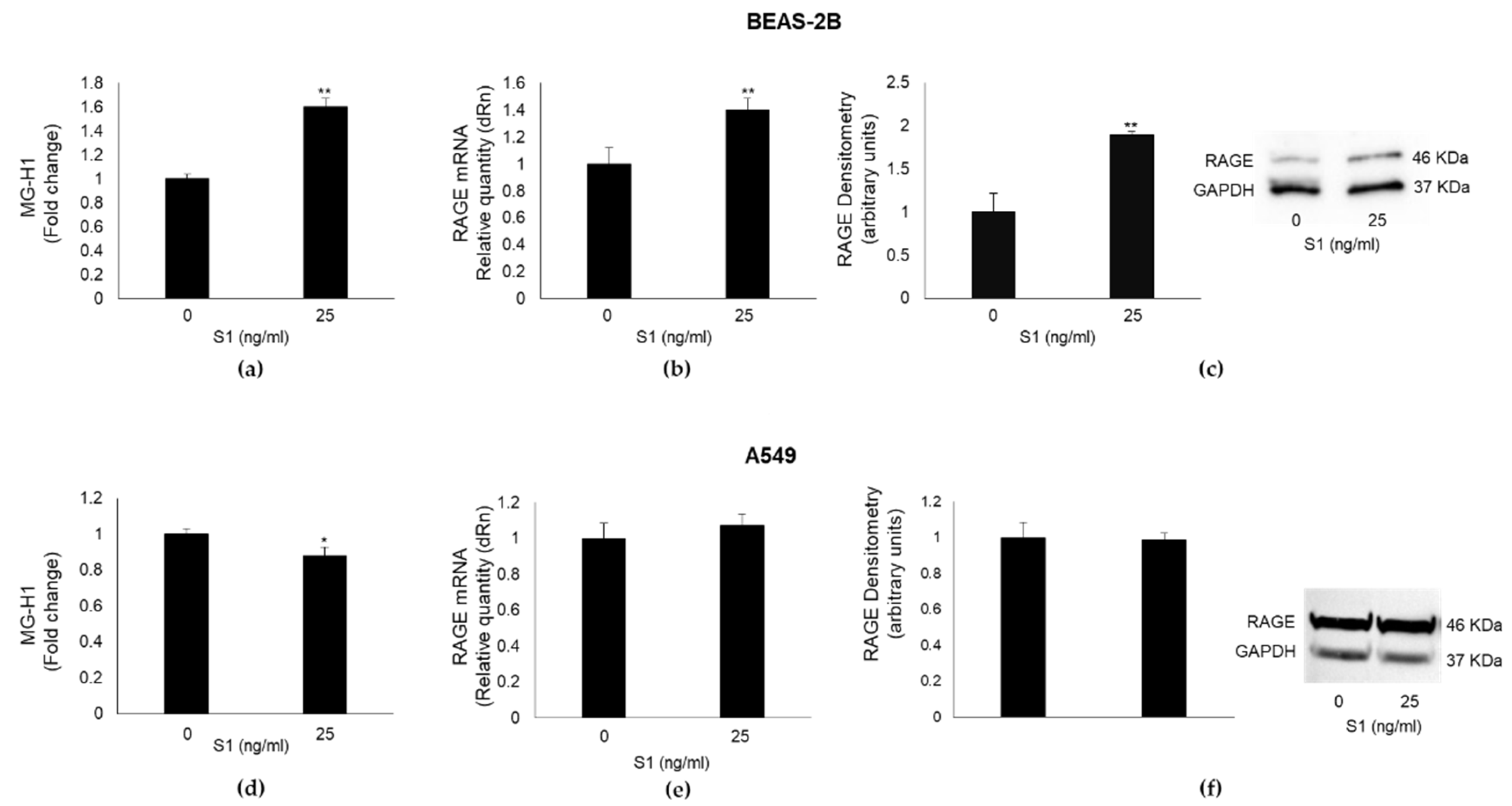
2.3. SARS-CoV-2 S1 Spike Protein Affects MG-H1 Levels and RAGE Expression in Human Bronchial BEAS-2B Cells
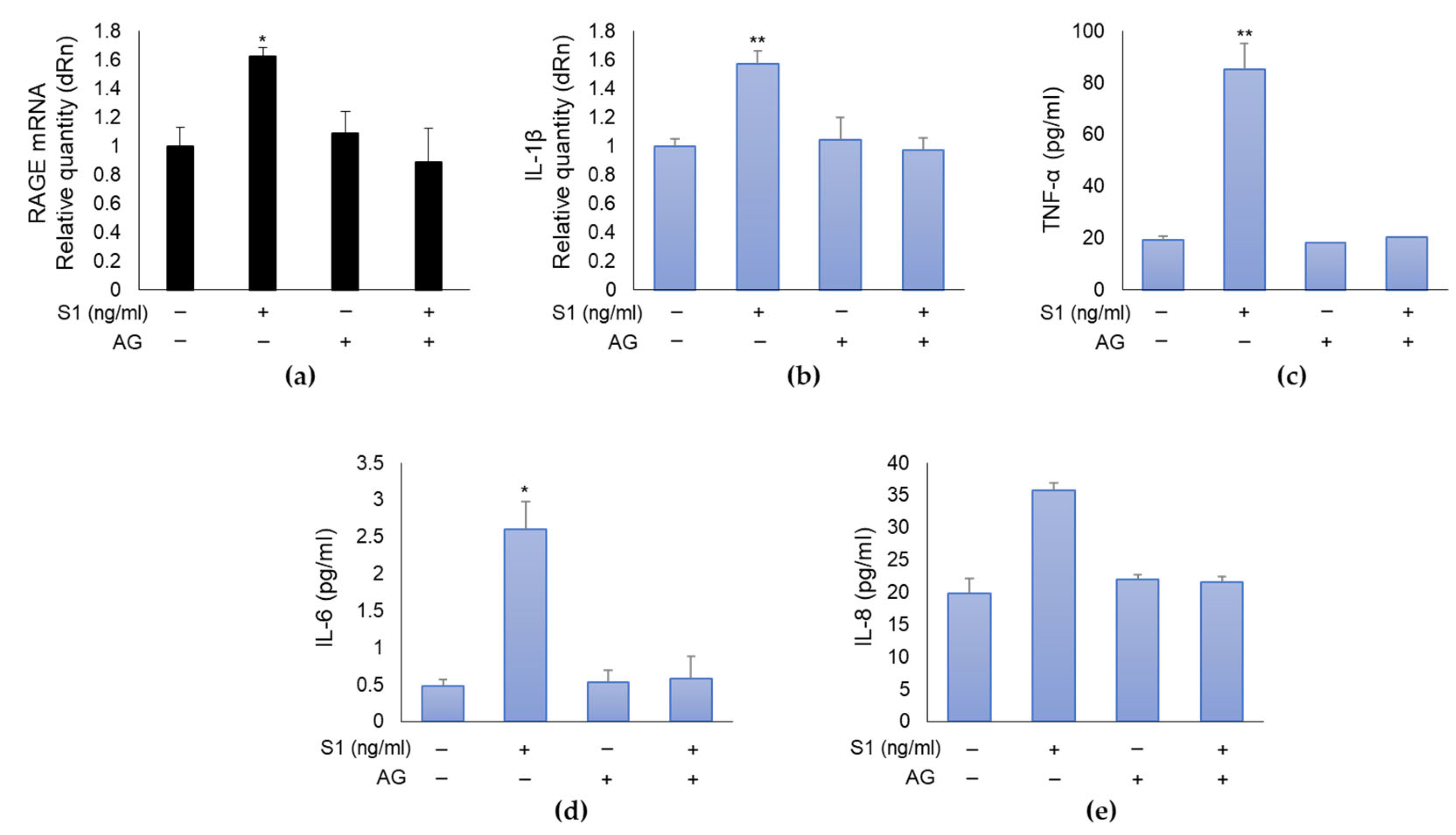
2.4. SARS-CoV-2 S1 Spike Protein Induces Inflammation in Human Bronchial BEAS-2B Cells through MG-H1/RAGE Axis
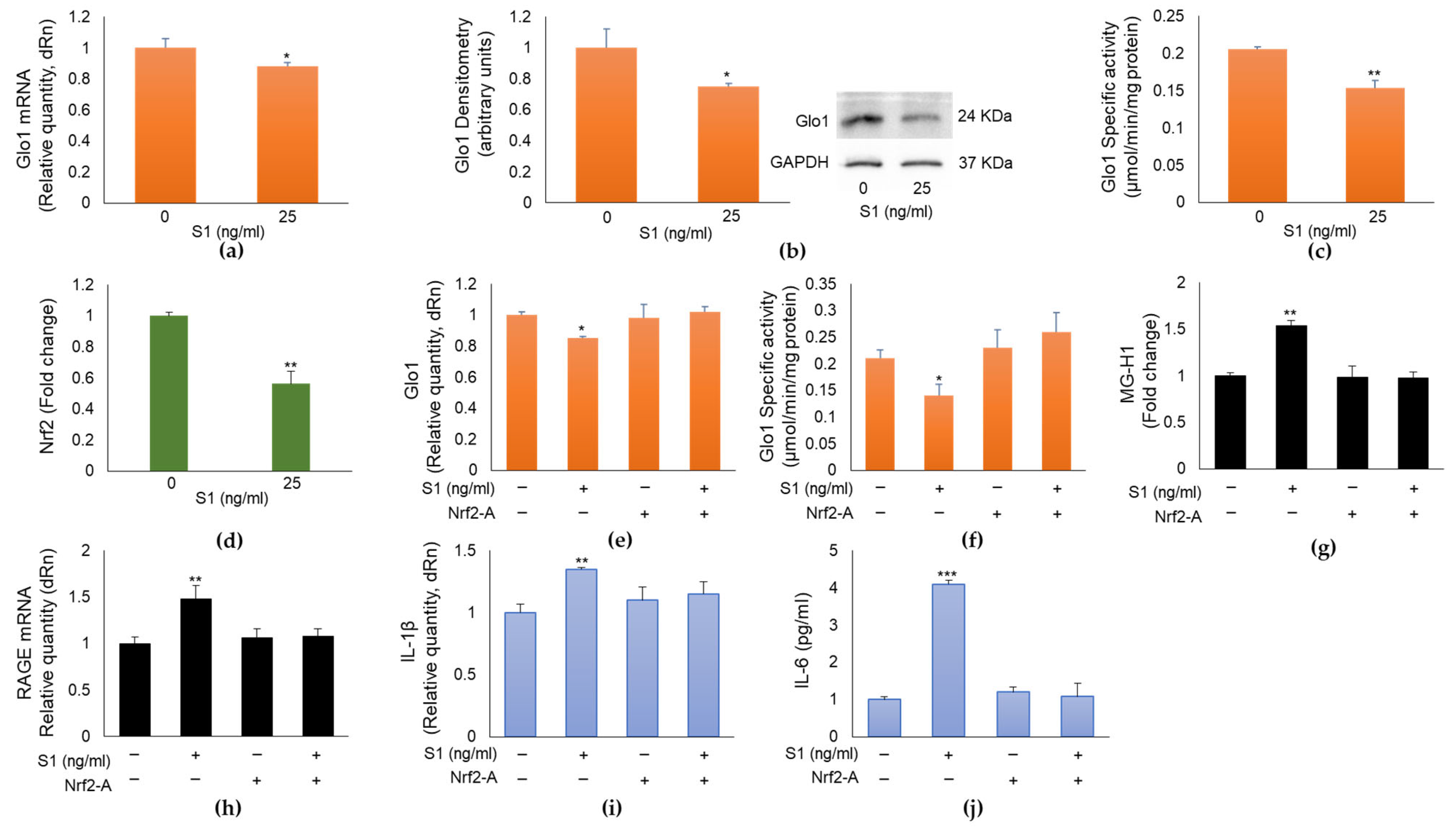
2.5. SARS-CoV-2 S1 Spike Protein Controls MG-H1/RAGE Proinflammatory Pathway through the Nuclear Factor Erythroid 2-Related Factor 2 (Nrf2)-Dependent Glo1 Downregulation in Human Bronchial BEAS-2B Cells
2.6. MG-H1 levels, RAGE, Glo1, and IL-1β Expression in Nasopharyngeal Swabs of SARS-CoV-2-Infected Patients at Different Clinical Severity of COVID-19
3. Discussion
4. Materials and Methods
4.1. Reagents
4.2. Cell Cultures
4.3. Cell Viability and Morphology
4.4. RNA Isolation, Reverse Transcription, and Real-Time Reverse Transcriptase–Polymerase Chain Reaction (RT-PCR) Analyses
4.5. TNF-α, IL-6 and IL-8 Detection
4.6. Detection of Methylglyoxal (MG)-H1
4.7. Cell and Nuclear Lysis and Western Blot
4.8. Nrf2 Activation Detection
4.9. Total Protein and Enzyme-Specific Activity of Glyoxalase 1 (Glo1) Detection
4.10. Patients
4.11. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Forsyth, C.B.; Zhang, L.; Bhushan, A.; Swanson, B.; Zhang, L.; Mamede, J.I.; Voigt, R.M.; Shaikh, M.; Engen, P.A.; Keshavarzian, A.; et al. The SARS-CoV-2 S1 Spike Protein Promotes MAPK and NF-kB Activation in Human Lung Cells and Inflammatory Cytokine Production in Human Lung and Intestinal Epithelial Cells. Microorganisms 2022, 10, 1996. [Google Scholar] [CrossRef] [PubMed]
- Boufidou, F.; Medić, S.; Lampropoulou, V.; Siafakas, N.; Tsakris, A.; Anastassopoulou, C. SARS-CoV-2 Reinfections and Long COVID in the Post-Omicron Phase of the Pandemic. Int. J. Mol. Sci. 2023, 24, 12962. [Google Scholar] [CrossRef] [PubMed]
- Hasankhani, A.; Bahrami, A.; Tavakoli-Far, B.; Iranshahi, S.; Ghaemi, F.; Akbarizadeh, M.R.; Amin, A.H.; Kiasari, B.A.; Shabestari, A.M. The role of peroxisome proliferator-activated receptors in the modulation of hyperinflammation induced by SARS-CoV-2 infection: A perspective for COVID-19 therapy. Front. Immunol. 2023, 14, 1127358. [Google Scholar] [CrossRef] [PubMed]
- Antognelli, C.; Talesa, V.N. Glyoxalases in Urological Malignancies. Int. J. Mol. Sci. 2018, 19, 415. [Google Scholar] [CrossRef] [PubMed]
- Lai, S.W.T.; Lopez Gonzalez, E.J.; Zoukari, T.; Ki, P.; Shuck, S.C. Methylglyoxal and Its Adducts: Induction, Repair, and Association with Disease. Chem. Res. Toxicol. 2022, 35, 1720–1746. [Google Scholar] [CrossRef] [PubMed]
- Antognelli, C.; Mancuso, F.; Frosini, R.; Arato, I.; Calvitti, M.; Calafiore, R.; Talesa, V.N.; Luca, G. Testosterone and Follicle Stimulating Hormone-Dependent Glyoxalase 1 Up-Regulation Sustains the Viability of Porcine Sertoli Cells through the Control of Hydroimidazolone- and Argpyrimidine-Mediated NF-κB Pathway. Am. J. Pathol. 2018, 188, 2553–2563. [Google Scholar] [CrossRef] [PubMed]
- Ishibashi, Y.; Matsui, T.; Nakamura, N.; Sotokawauchi, A.; Higashimoto, Y.; Yamagishi, S.I. Methylglyoxal-derived hydroimidazolone-1 evokes inflammatory reactions in endothelial cells via an interaction with receptor for advanced glycation end products. Diab. Vasc. Dis. Res. 2017, 14, 450–453. [Google Scholar] [CrossRef] [PubMed]
- Lim, J.M.; Yoo, H.J.; Lee, K.W. High Molecular Weight Fucoidan Restores Intestinal Integrity by Regulating Inflammation and Tight Junction Loss Induced by Methylglyoxal-Derived Hydroimidazolone-1. Mar. Drugs 2022, 20, 580. [Google Scholar] [CrossRef]
- Antognelli, C.; Ferri, I.; Bellezza, G.; Siccu, P.; Love, H.D.; Talesa, V.N.; Sidoni, A. Glyoxalase 2 drives tumorigenesis in human prostate cells in a mechanism involving androgen receptor and p53-p21 axis. Mol. Carcinog. 2017, 56, 2112–2126. [Google Scholar] [CrossRef]
- Delle Monache, S.; Pulcini, F.; Frosini, R.; Mattei, V.; Talesa, V.N.; Antognelli, C. Methylglyoxal-Dependent Glycative Stress Is Prevented by the Natural Antioxidant Oleuropein in Human Dental Pulp Stem Cells through Nrf2/Glo1 Pathway. Antioxidants 2021, 10, 716. [Google Scholar] [CrossRef]
- Khan, H.; Patel, S.; Majumdar, A. Role of NRF2 and Sirtuin activators in COVID-19. Clin. Immunol. 2021, 233, 108879. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Kumar, S.; Hassan, M.; Wu, H.; Thimmulappa, R.K.; Kumar, A.; Sharma, S.K.; Parmar, V.S.; Biswal, S.; Malhotra, S.V. Novel Chalcone Derivatives as Potent Nrf2 Activators in Mice and Human Lung Epithelial Cells. J. Med. Chem. 2011, 54, 4147–4159. [Google Scholar] [CrossRef] [PubMed]
- Cuadrado, A.; Pajares, M.; Benito, C.; Jiménez-Villegas, J.; Escoll, M.; Fernández-Ginés, R.; Garcia Yagüe, A.J.; Lastra, D.; Manda, G.; Rojo, A.I.; et al. Can Activation of NRF2 Be a Strategy against COVID-19? Trends Pharmacol. Sci. 2020, 41, 598–610. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.; Shafiei, M.S.; Longoria, C.; Schoggins, J.W.; Savani, R.C.; Zaki, H. SARS-CoV-2 spike protein induces inflammation via TLR2-dependent activation of the NF-κB pathway. Elife 2021, 10, e68563. [Google Scholar] [CrossRef] [PubMed]
- Arjsri, P.; Srisawad, K.; Mapoung, S.; Semmarath, W.; Thippraphan, P.; Umsumarng, S.; Yodkeeree, S.; Dejkriengkraikul, P. Hesperetin from Root Extract of Clerodendrum petasites S. Moore Inhibits SARS-CoV-2 Spike Protein S1 Subunit-Induced NLRP3 Inflammasome in A549 Lung Cells via Modulation of the Akt/MAPK/AP-1 Pathway. Int. J. Mol. Sci. 2022, 23, 10346. [Google Scholar] [CrossRef] [PubMed]
- Chittasupho, C.; Srisawad, K.; Arjsri, P.; Phongpradist, R.; Tingya, W.; Ampasavate, C.; Dejkriengkraikul, P. Targeting Spike Glycoprotein S1 Mediated by NLRP3 Inflammasome Machinery and the Cytokine Releases in A549 Lung Epithelial Cells by Nanocurcumin. Pharmaceuticals 2023, 16, 862. [Google Scholar] [CrossRef] [PubMed]
- Semmarath, W.; Mapoung, S.; Umsumarng, S.; Arjsri, P.; Srisawad, K.; Thippraphan, P.; Yodkeeree, S.; Dejkriengkraikul, P. Cyanidin-3-O-glucoside and Peonidin-3-O-glucoside-Rich Fraction of Black Rice Germ and Bran Suppresses Inflammatory Responses from SARS-CoV-2 Spike Glycoprotein S1-Induction In Vitro in A549 Lung Cells and THP-1 Macrophages via Inhibition of the NLRP3 Inflammasome Pathway. Nutrients 2022, 14, 2738. [Google Scholar] [CrossRef]
- Suzuki, Y.J.; Gychka, S.G. SARS-CoV-2 Spike Protein Elicits Cell Signaling in Human Host Cells: Implications for Possible Consequences of COVID-19 Vaccines. Vaccines 2021, 9, 36. [Google Scholar] [CrossRef]
- Patterson, B.K.; Francisco, E.B.; Yogendra, R.; Long, E.; Pise, A.; Rodrigues, H.; Hall, E.; Herrera, M.; Parikh, P.; Guevara-Coto, J.; et al. Persistence of SARS CoV-2 S1 Protein in CD16+ Monocytes in Post-Acute Sequelae of COVID-19 (PASC) up to 15 Months Post-Infection. Front. Immunol. 2022, 12, 746021. [Google Scholar] [CrossRef]
- Files, J.K.; Sarkar, S.; Fram, T.R.; Boppana, S.; Sterrett, S.; Qin, K.; Bansal, A.; Long, D.M.; Sabbaj, S.; Kobie, J.J.; et al. Duration of post-COVID-19 symptoms is associated with sustained SARS-CoV-2-specific immune responses. JCI Insight 2021, 6, e151544. [Google Scholar] [CrossRef]
- Swank, Z.; Senussi, Y.; Manickas-Hill, Z.; Yu, X.G.; Li, J.Z.; Alter, G.; Walt, D.R. Persistent Circulating Severe Acute Respiratory Syndrome Coronavirus 2 Spike Is Associated With Post-acute Coronavirus Disease 2019 Sequelae. Clin. Infect Dis. 2023, 76, e487–e490. [Google Scholar] [CrossRef] [PubMed]
- Martins-Gonçalves, R.; Hottz, E.D.; Bozza, P.T. Acute to post-acute COVID-19 thromboinflammation persistence: Mechanisms and potential consequences. Curr. Res. Immunol. 2023, 4, 100058. [Google Scholar] [CrossRef] [PubMed]
- Bartolomeo, C.S.; Lemes, R.M.R.; Morais, R.L.; Pereria, G.C.; Nunes, T.A.; Costa, A.J.; de Barros Maciel, R.M.; Braconi, C.T.; Maricato, J.T.; Janini, L.M.R.; et al. SARS-CoV-2 infection and replication kinetics in different human cell types: The role of autophagy, cellular metabolism and ACE2 expression. Life Sci. 2022, 308, 120930. [Google Scholar] [CrossRef] [PubMed]
- Tsou, H.H.; Wang, P.H.; Ting, T.H.; Ping, Y.H.; Liu, T.Y.; Cheng, H.W.; Wang, H.T. Effect of heated tobacco products and traditional cigarettes on pulmonary toxicity and SARS-CoV-2-induced lung injury. Toxicology 2022, 479, 153318. [Google Scholar] [CrossRef] [PubMed]
- Dai, Y.; Zhang, Y.; Zhang, L.; Song, Z. Synthesis and Biological Evaluation of Paclitaxel-aminoguanidine Conjugates for Suppressing Breast Cancer. Curr. Org. Synth. 2023, 20, 890–896. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Song, X.; Ma, T.; Li, Y.; Bai, H.; Zhang, Z.; Hu, H.; Yuan, R.; Wen, Y.; Gao, L. Aminoguanidine inhibits IL-1β-induced protein expression of iNOS and COX-2 by blocking the NF-κB signaling pathway in rat articular chondrocytes. Exp. Ther. Med. 2020, 20, 2623–2630. [Google Scholar] [CrossRef] [PubMed]
- Chiappalupi, S.; Salvadori, L.; Donato, R.; Riuzzi, F.; Sorci, G. Hyperactivated RAGE in Comorbidities as a Risk Factor for Severe COVID-19-The Role of RAGE-RAS Crosstalk. Biomolecules 2021, 11, 876. [Google Scholar] [CrossRef]
- Watanabe, Y.; Allen, J.D.; Wrapp, D.; McLellan, J.S.; Crispin, M. Site-specific glycan analysis of the SARS-CoV-2 spike. Science 2020, 369, 330–333. [Google Scholar] [CrossRef]
- Tóbon-Velasco, J.C.; Cuevas, E.; Torres-Ramos, M.A. Receptor for AGEs (RAGE) as mediator of NF-kB pathway activation in neuroinflammation and oxidative stress. CNS Neurol Disord. Drug Targets 2014, 13, 1615–1626. [Google Scholar] [CrossRef]
- Schreiber, A.; Viemann, D.; Schöning, J.; Schloer, S.; Mecate Zambrano, A.; Brunotte, L.; Faist, A.; Schöfbänker, M.; Hrincius, E.; Hoffmann, H.; et al. The MEK1/2-inhibitor ATR-002 efficiently blocks SARS-CoV-2 propagation and alleviates pro-inflammatory cytokine/chemokine responses. Cell Mol. Life Sci. 2022, 79, 65. [Google Scholar] [CrossRef]
- Paidi, R.K.; Jana, M.; Mishra, R.K.; Dutta, D.; Pahan, K. Selective Inhibition of the Interaction between SARS-CoV-2 Spike S1 and ACE2 by SPIDAR Peptide Induces Anti-Inflammatory Therapeutic Responses. J. Immunol. 2021, 207, 2521–2533. [Google Scholar] [CrossRef] [PubMed]
- Barilli, A.; Visigalli, R.; Ferrari, F.; Bianchi, M.G.; Dall’Asta, V.; Rotoli, B.M. Immune-Mediated Inflammatory Responses of Alveolar Epithelial Cells: Implications for COVID-19 Lung Pathology. Biomedicines 2022, 10, 618. [Google Scholar] [CrossRef] [PubMed]
- Alomar, F.A. Methylglyoxal in COVID-19-induced hyperglycemia and new-onset diabetes. Eur. Rev. Med. Pharmacol. Sci. 2022, 26, 8152–8171. [Google Scholar] [CrossRef]
- Xia, J.; Tang, W.; Wang, J.; Lai, D.; Xu, Q.; Huang, R.; Hu, Y.; Gong, X.; Fan, J.; Shu, Q.; et al. SARS-CoV-2 N Protein Induces Acute Lung Injury in Mice via NF-ĸB Activation. Front. Immunol. 2021, 12, 791753. [Google Scholar] [CrossRef] [PubMed]
- Moolamalla, S.T.R.; Balasubramanian, R.; Chauhan, R.; Priyakumar, U.D.; Vinod, P.K. Host metabolic reprogramming in response to SARS-CoV-2 infection: A systems biology approach. Microb Pathog. 2021, 158, 105114. [Google Scholar] [CrossRef]
- Codo, A.C.; Davanzo, G.G.; Monteiro, L.B.; de Souza, G.F.; Muraro, S.P.; Virgilio-da-Silva, J.V.; Prodonoff, J.S.; Carregari, V.C.; de Biagi Junior, C.A.O.; Crunfli, F.; et al. Elevated Glucose Levels Favor SARS-CoV-2 Infection and Monocyte Response through a HIF-1α/Glycolysis-Dependent Axis. Cell Metab. 2020, 32, 498–499. [Google Scholar] [CrossRef]
- Sun, Q.; Li, L.; Jin, F.; Liu, Y.; Yang, B.; Meng, W.; Zhang, Z.; Qi, F. SARS-CoV-2 Spike Protein S1 Exposure Increases Susceptibility to Angiotensin II-Induced Hypertension in Rats by Promoting Central Neuroinflammation and Oxidative Stress. Neurochem Res. 2023, 48, 3016–3026. [Google Scholar] [CrossRef]
- Casper, E. The crosstalk between Nrf2 and NF-κB pathways in coronary artery disease: Can it be regulated by SIRT6? Life Sci. 2023, 330, 122007. [Google Scholar] [CrossRef]
- Ahmed, S.M.U.; Luo, L.; Namani, A.; Wang, X.J.; Tang, X. Nrf2 signaling pathway: Pivotal roles in inflammation. Biochim. Biophys. Acta Mol. Basis Dis. 2017, 1863, 585–597. [Google Scholar] [CrossRef]
- Cuadrado, A.; Manda, G.; Hassan, A.; Alcaraz, M.J.; Barbas, C.; Daiber, A.; Ghezzi, P.; León, R.; López, M.G.; Oliva, B.; et al. Transcription factor NRF2 as a therapeutic target for chronic diseases: A systems medicine approach. Pharmacol. Rev. 2018, 70, 348–383. [Google Scholar] [CrossRef]
- Alomar, F.A.; Alshakhs, M.N.; Abohelaika, S.; Almarzouk, H.M.; Almualim, M.; Al-Ali, A.K.; Al-Muhanna, F.; Alomar, M.F.; Alhaddad, M.J.; Almulaify, M.S.; et al. Elevated plasma level of the glycolysis byproduct methylglyoxal on admission is an independent biomarker of mortality in ICU COVID-19 patients. Sci. Rep. 2022, 12, 9510. [Google Scholar] [CrossRef]


| NS | M/S | N | |
|---|---|---|---|
| Age (years) | |||
| Mean ± SD | 64.75 ± 19.3 | 76.05 ± 16.0 * | 59.10 ± 17.6 ** |
| Median | 65 | 80 | 59 |
| Range | 32–89 | 29–96 | 29–86 |
| Gender | |||
| Male | 45% (n = 9) | 55% (n = 22) | 42% (n = 8) |
| Female | 55% (n = 11) | 45% (n = 18) | 58% (n = 11) |
| Gene | Sense Primer (5′-3′) | Antisense Primer (5′-3′) |
|---|---|---|
| Glo1 | AGAAAGCACGGGGTGAAACTG | TACACCTTCAGTCCCGACTCC |
| IL-1β | GGACCTGGACCTCTGCCCTCTGG | GCCTGCCTGAAGCCCTTGCTGTAG |
| RAGE | TGAAGGAACAGACCAGGAGACAC | GCACAGGCTCCCAGACAC |
| GAPDH | CAAGGTCATCCATGACAACTTTG | GTCCACCACCCTGTTGCTGTAG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Manfredelli, D.; Pariano, M.; Costantini, C.; Graziani, A.; Bozza, S.; Romani, L.; Puccetti, P.; Talesa, V.N.; Antognelli, C. Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Spike Protein S1 Induces Methylglyoxal-Derived Hydroimidazolone/Receptor for Advanced Glycation End Products (MG-H1/RAGE) Activation to Promote Inflammation in Human Bronchial BEAS-2B Cells. Int. J. Mol. Sci. 2023, 24, 14868. https://doi.org/10.3390/ijms241914868
Manfredelli D, Pariano M, Costantini C, Graziani A, Bozza S, Romani L, Puccetti P, Talesa VN, Antognelli C. Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Spike Protein S1 Induces Methylglyoxal-Derived Hydroimidazolone/Receptor for Advanced Glycation End Products (MG-H1/RAGE) Activation to Promote Inflammation in Human Bronchial BEAS-2B Cells. International Journal of Molecular Sciences. 2023; 24(19):14868. https://doi.org/10.3390/ijms241914868
Chicago/Turabian StyleManfredelli, Dominga, Marilena Pariano, Claudio Costantini, Alessandro Graziani, Silvia Bozza, Luigina Romani, Paolo Puccetti, Vincenzo Nicola Talesa, and Cinzia Antognelli. 2023. "Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Spike Protein S1 Induces Methylglyoxal-Derived Hydroimidazolone/Receptor for Advanced Glycation End Products (MG-H1/RAGE) Activation to Promote Inflammation in Human Bronchial BEAS-2B Cells" International Journal of Molecular Sciences 24, no. 19: 14868. https://doi.org/10.3390/ijms241914868






