Structure of the Drosophila melanogaster Flight Muscle Myosin Filament at 4.7 Å Resolution Reveals New Details of Non-Myosin Proteins
Abstract
:1. Introduction
2. Results
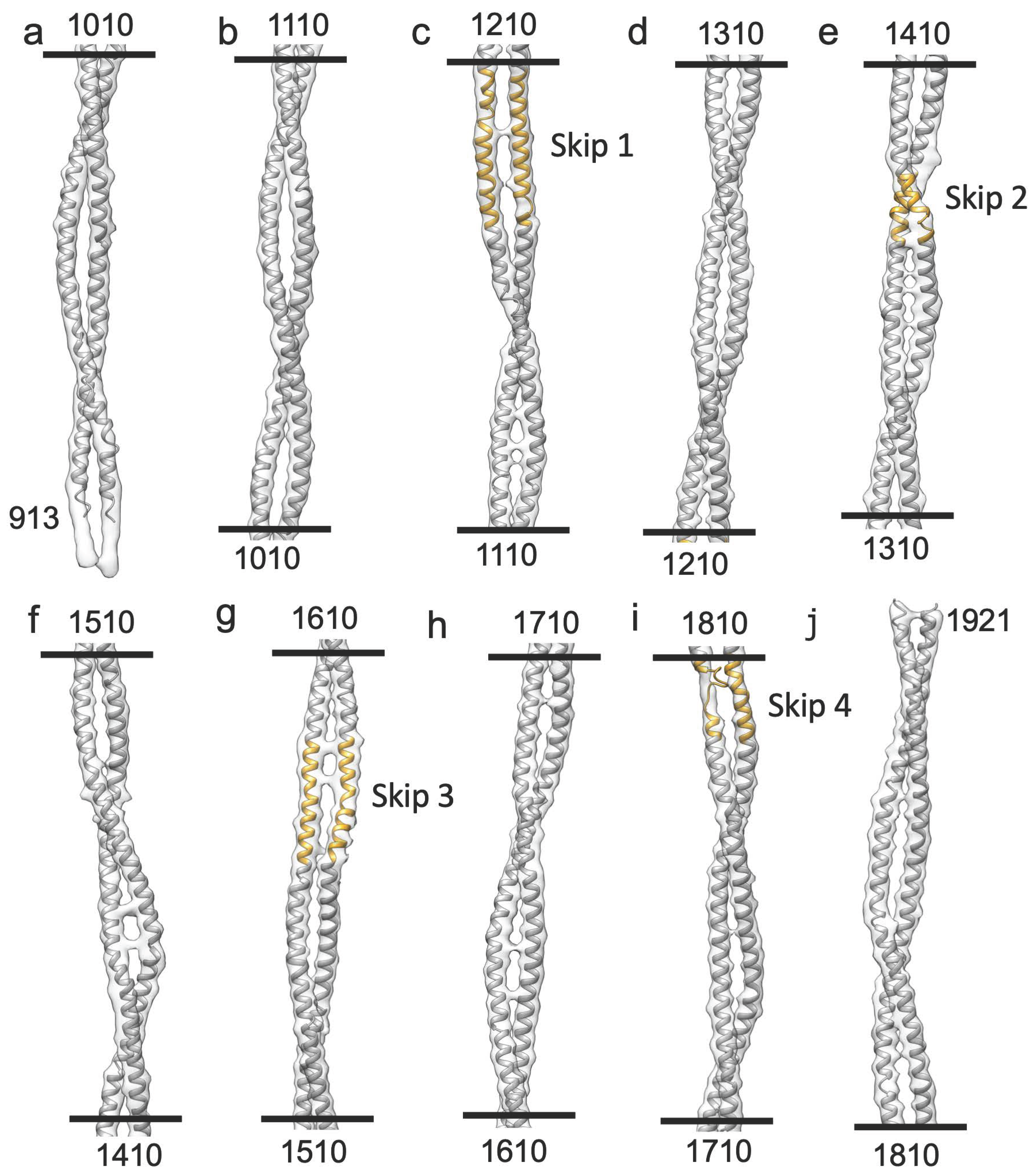
2.1. Myosin Tail Structure and Arrangement
2.2. Myosin Heads and Proximal S2 Region
2.3. Homology Model of the Myosin Coiled-Coil
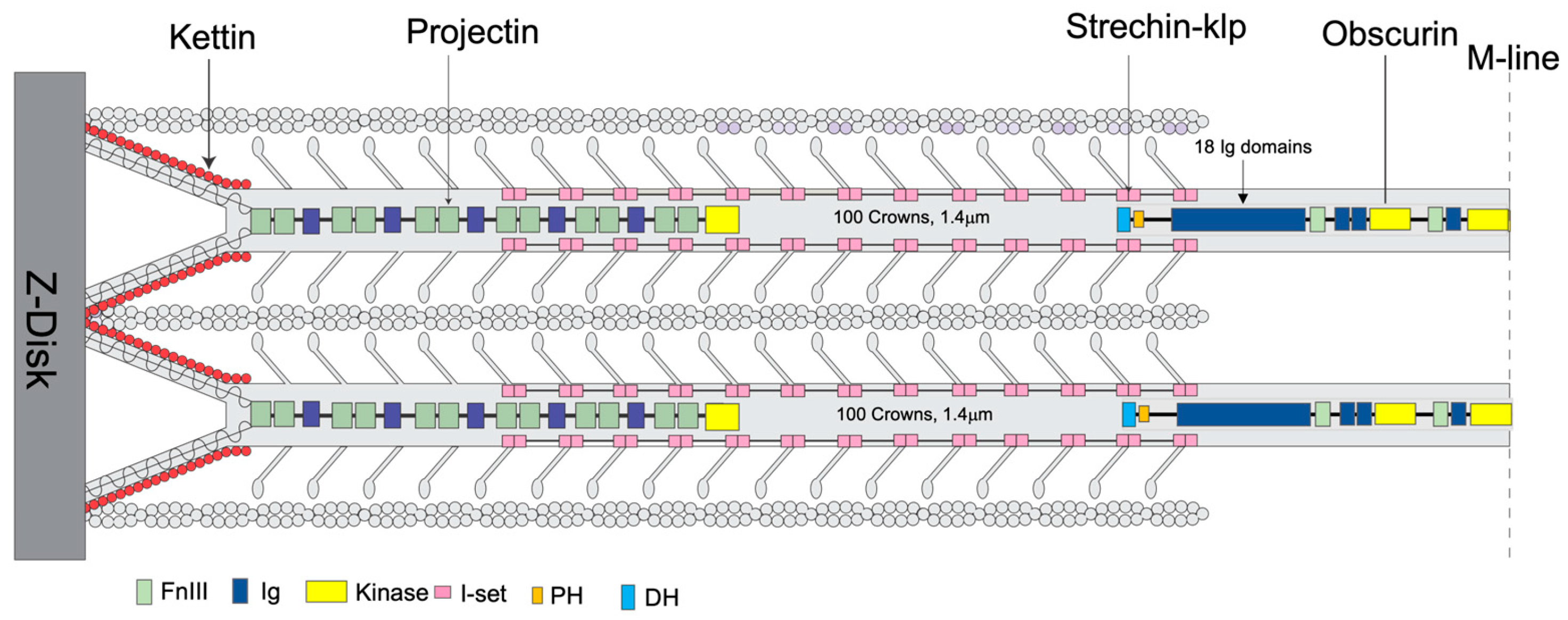
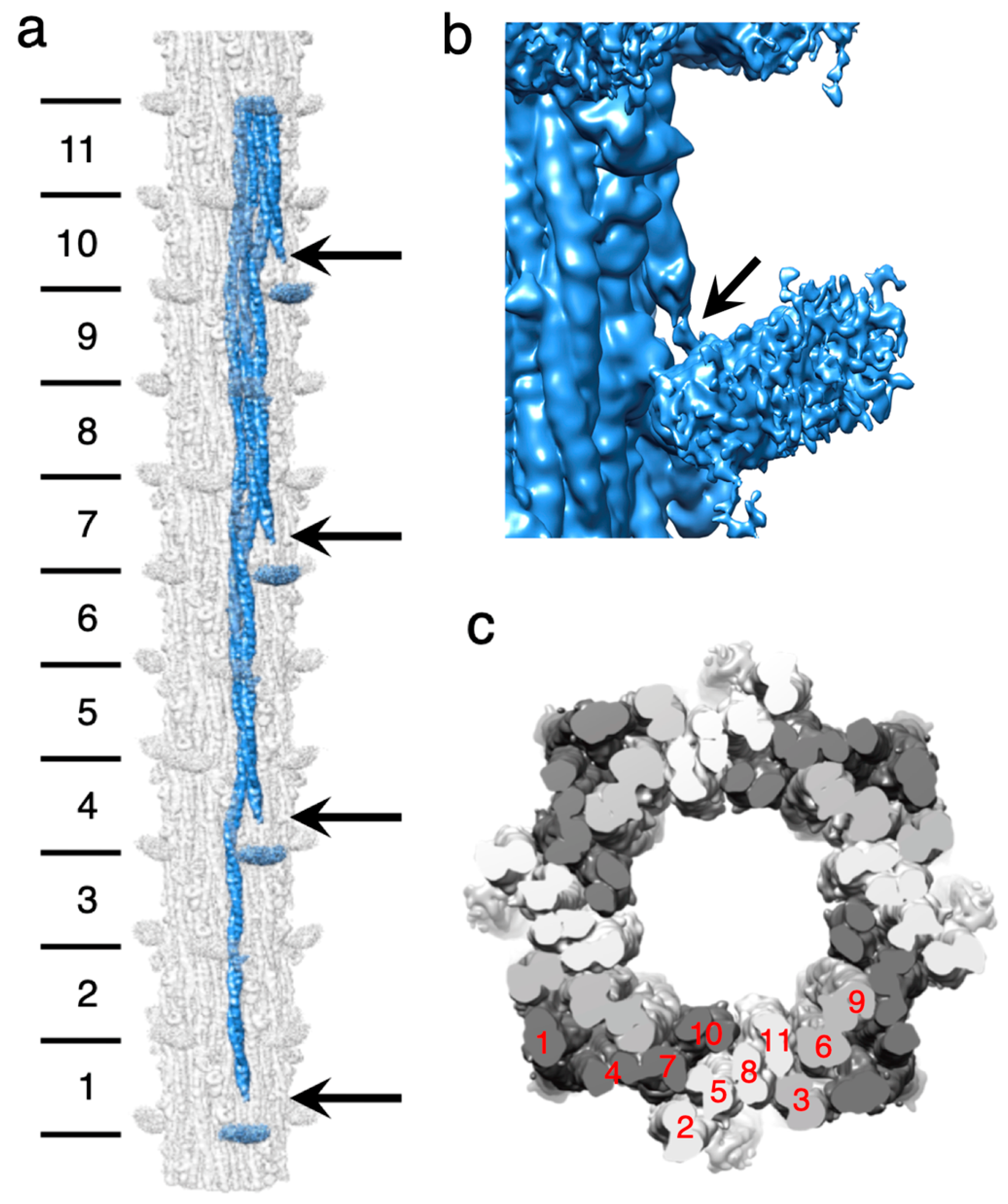
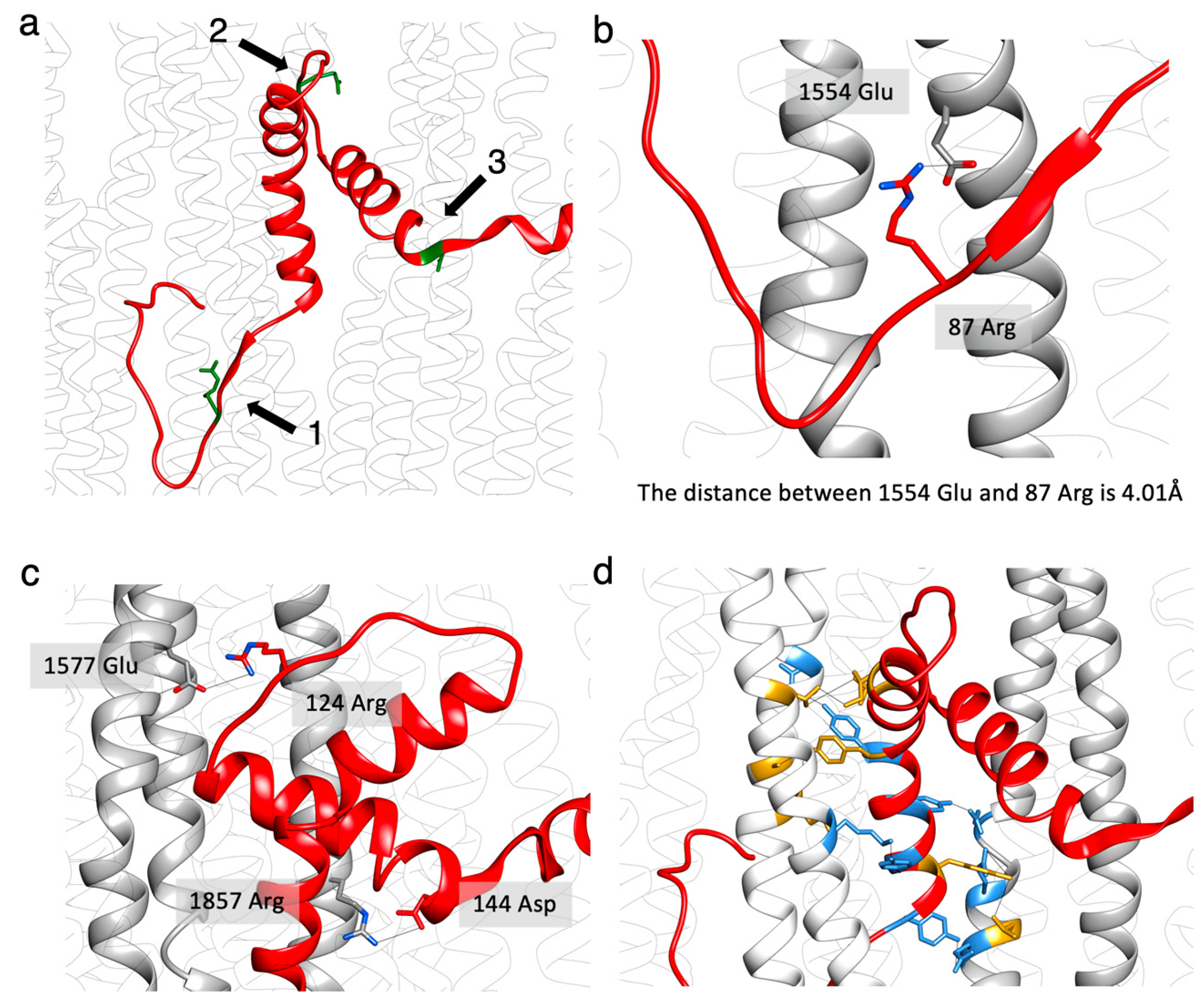
2.4. Non-Myosin Proteins
2.4.1. Myofilin
2.4.2. Stretchin-Klp
2.4.3. Flightin
3. Discussion
3.1. Disordered Myosin Heads
3.2. Role of the Proximal S2 in Muscle Contraction
4. Conclusions
5. Materials and Methods
5.1. Thick Filament Preparation
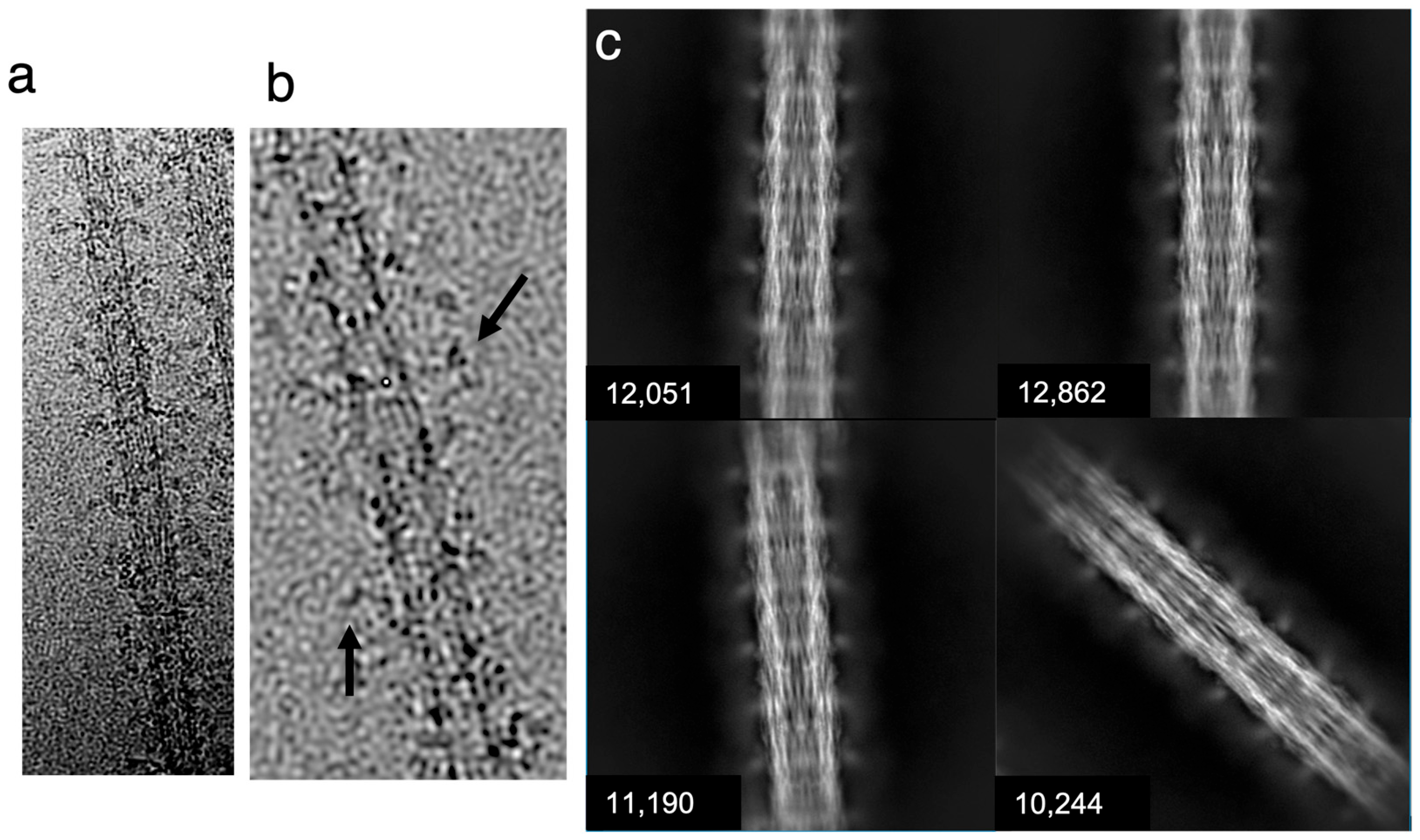
5.2. Electron Microscopy
5.3. Data Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hooper, S.L.; Hobbs, K.H.; Thuma, J.B. Invertebrate muscles: Thin and thick filament structure; molecular basis of contraction and its regulation, catch and asynchronous muscle. Prog. Neurobiol. 2008, 86, 72–127. [Google Scholar] [PubMed]
- Page, S.G.; Huxley, H.E. Filament Lengths in Striated Muscle. J. Cell Biol. 1963, 19, 369–390. [Google Scholar] [CrossRef]
- Al-Khayat, H.A.; Morris, E.P.; Kensler, R.W.; Squire, J.M. Myosin filament 3D structure in mammalian cardiac muscle. J. Struct. Biol. 2008, 163, 117–126. [Google Scholar] [CrossRef] [PubMed]
- Zoghbi, M.E.; Woodhead, J.L.; Moss, R.L.; Craig, R. Three-dimensional structure of vertebrate cardiac muscle myosin filaments. Proc. Natl. Acad. Sci. USA 2008, 105, 2386–2390. [Google Scholar] [CrossRef]
- Dutta, D.; Nguyen, V.; Campbell, K.; Padron, R.; Craig, R. Cryo-EM structure of the human cardiac myosin filament. bioRxiv 2023. bioRxiv:2023.04.11.536274. [Google Scholar]
- Tamborrini, D.; Wang, Z.; Wagner, T.; Tacke, S.; Stabrin, M.; Grange, M.; Kho, A.L.; Rees, M.; Bennett, P.; Gautel, M.; et al. In situ structures from relaxed cardiac myofibrils reveal the organization of the muscle thick filament. bioRxiv 2023. bioRxiv:2023.04.11.536387. [Google Scholar]
- Squire, J.M. General model for the structure of all myosin-containing filaments. Nature 1971, 233, 457–462. [Google Scholar] [CrossRef]
- Harrington, W.F.; Rodgers, M.E. Myosin. Annu. Rev. Biochem. 1984, 53, 35–73. [Google Scholar] [CrossRef]
- Tregear, R.T.; Hoyland, J.; Sayers, A.J. The Repeat Distance of Myosin in the Thick Filaments of Various Muscles. J. Mol. Biol. 1984, 176, 417–420. [Google Scholar] [CrossRef]
- Huxley, A.F. Muscular contraction. J. Physiol. 1974, 243, 1–43. [Google Scholar] [CrossRef]
- Reedy, M.K.; Holmes, K.C.; Tregear, R.T. Induced changes in orientation of the cross-bridges of glycerinated insect flight muscle. Nature 1965, 207, 1276–1280. [Google Scholar] [CrossRef] [PubMed]
- Stewart, M.; Kensler, R.W.; Levine, R.J. Structure of Limulus telson muscle thick filaments. J. Mol. Biol. 1981, 153, 781–790. [Google Scholar] [CrossRef]
- Crowther, R.A.; Padron, R.; Craig, R. Arrangement of the heads of myosin in relaxed thick filaments from tarantula muscle. J. Mol. Biol. 1985, 184, 429–439. [Google Scholar] [CrossRef] [PubMed]
- Huxley, H.E.; Brown, W. The low-angle x-ray diagram of vertebrate striated muscle and its behaviour during contraction and rigor. J. Mol. Biol. 1967, 30, 383–434. [Google Scholar] [CrossRef]
- Wendt, T.; Taylor, D.; Trybus, K.M.; Taylor, K. Three-dimensional image reconstruction of dephosphorylated smooth muscle heavy meromyosin reveals asymmetry in the interaction between myosin heads and placement of subfragment 2. Proc. Natl. Acad. Sci. USA 2001, 98, 4361–4366. [Google Scholar] [CrossRef]
- Sellers, J.R. Regulation of cytoplasmic and smooth muscle myosin. Curr. Opin. Cell Biol. 1991, 3, 98–104. [Google Scholar] [CrossRef]
- Liu, J.; Wendt, T.; Taylor, D.W.; Taylor, K.A. Refined model of the 10S conformation of smooth muscle myosin by cryo-electron microscopy 3D image reconstruction. J. Mol. Biol. 2003, 329, 963–972. [Google Scholar] [CrossRef] [PubMed]
- Burgess, S.A.; Yu, S.; Walker, M.L.; Hawkins, R.J.; Chalovich, J.M.; Knight, P.J. Structures of smooth muscle myosin and heavy meromyosin in the folded, shutdown state. J. Mol. Biol. 2007, 372, 1165–1178. [Google Scholar] [CrossRef] [PubMed]
- Scarff, C.A.; Carrington, G.; Casas-Mao, D.; Chalovich, J.M.; Knight, P.J.; Ranson, N.A.; Peckham, M. Structure of the shutdown state of myosin-2. Nature 2020, 588, 515–520. [Google Scholar] [CrossRef]
- Yang, S.; Tiwari, P.; Lee, K.H.; Sato, O.; Ikebe, M.; Padron, R.; Craig, R. Cryo-EM structure of the inhibited (10S) form of myosin II. Nature 2020, 588, 521–525. [Google Scholar] [CrossRef]
- Heissler, S.M.; Arora, A.S.; Billington, N.; Sellers, J.R.; Chinthalapudi, K. Cryo-EM structure of the autoinhibited state of myosin-2. Sci. Adv. 2021, 7, eabk3273. [Google Scholar] [CrossRef]
- Woodhead, J.L.; Zhao, F.Q.; Craig, R.; Egelman, E.H.; Alamo, L.; Padron, R. Atomic model of a myosin filament in the relaxed state. Nature 2005, 436, 1195–1199. [Google Scholar] [CrossRef]
- Jung, H.S.; Burgess, S.A.; Billington, N.; Colegrave, M.; Patel, H.; Chalovich, J.M.; Chantler, P.D.; Knight, P.J. Conservation of the regulated structure of folded myosin 2 in species separated by at least 600 million years of independent evolution. Proc. Natl. Acad. Sci. USA 2008, 105, 6022–6026. [Google Scholar] [CrossRef]
- Zhao, F.Q.; Craig, R.; Woodhead, J.L. Head-head interaction characterizes the relaxed state of Limulus muscle myosin filaments. J. Mol. Biol. 2009, 385, 423–431. [Google Scholar] [CrossRef]
- Sulbaran, G.; Alamo, L.; Pinto, A.; Marquez, G.; Mendez, F.; Padron, R.; Craig, R. An invertebrate smooth muscle with striated muscle myosin filaments. Proc. Natl. Acad. Sci. USA 2015, 112, E5660–E5668. [Google Scholar] [CrossRef]
- Jung, H.S.; Komatsu, S.; Ikebe, M.; Craig, R. Head-head and head-tail interaction: A general mechanism for switching off myosin II activity in cells. Mol. Biol. Cell 2008, 19, 3234–3242. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.H.; Sulbaran, G.; Yang, S.; Mun, J.Y.; Alamo, L.; Pinto, A.; Sato, O.; Ikebe, M.; Liu, X.; Korn, E.D.; et al. Interacting-heads motif has been conserved as a mechanism of myosin II inhibition since before the origin of animals. Proc. Natl. Acad. Sci. USA 2018, 115, E1991–E2000. [Google Scholar] [CrossRef]
- Naber, N.; Cooke, R.; Pate, E. Slow myosin ATP turnover in the super-relaxed state in tarantula muscle. J. Mol. Biol. 2011, 411, 943–950. [Google Scholar] [CrossRef]
- Trivedi, D.V.; Adhikari, A.S.; Sarkar, S.S.; Ruppel, K.M.; Spudich, J.A. Hypertrophic cardiomyopathy and the myosin mesa: Viewing an old disease in a new light. Biophys. Rev. 2018, 10, 27–48. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Taylor, D.W.; Reedy, M.K.; Edwards, R.J.; Taylor, K.A. Structure of myosin filaments from relaxed Lethocerus flight muscle by cryo-EM at 6 Å resolution. Sci. Adv. 2016, 2, e1600058. [Google Scholar] [CrossRef]
- Daneshparvar, N.; Taylor, D.W.; O’Leary, T.S.; Rahmani, H.; Abbasiyeganeh, F.; Previs, M.J.; Taylor, K.A. CryoEM structure of Drosophila flight muscle thick filaments at 7 A resolution. Life Sci. Alliance 2020, 3, e202000823. [Google Scholar] [CrossRef] [PubMed]
- Menetret, J.F.; Schroder, R.R.; Hofmann, W. Cryo-electron microscopic studies of relaxed striated muscle thick filaments. J. Muscle Res. Cell Motil. 1990, 11, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Farman, G.P.; Miller, M.S.; Reedy, M.C.; Soto-Adames, F.N.; Vigoreaux, J.O.; Maughan, D.W.; Irving, T.C. Phosphorylation and the N-terminal extension of the regulatory light chain help orient and align the myosin heads in Drosophila flight muscle. J. Struct. Biol. 2009, 168, 240–249. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Rahmani, H.; Abbasi Yeganeh, F.; Rastegarpouyani, H.; Taylor, D.W.; Wood, N.B.; Previs, M.J.; Iwamoto, H.; Taylor, K.A. Structure of the Flight Muscle Thick Filament from the Bumble Bee, Bombus ignitus, at 6 Å Resolution. Int. J. Mol. Sci. 2023, 24, 377. [Google Scholar] [CrossRef] [PubMed]
- Katzemich, A.; Kreiskother, N.; Alexandrovich, A.; Elliott, C.; Schock, F.; Leonard, K.; Sparrow, J.; Bullard, B. The function of the M-line protein obscurin in controlling the symmetry of the sarcomere in the flight muscle of Drosophila. J. Cell Sci. 2012, 125 Pt 14, 3367–3379. [Google Scholar] [CrossRef] [PubMed]
- Tonino, P.; Kiss, B.; Strom, J.; Methawasin, M.; Smith, J.E., 3rd; Kolb, J.; Labeit, S.; Granzier, H. The giant protein titin regulates the length of the striated muscle thick filament. Nat. Commun. 2017, 8, 1041. [Google Scholar] [CrossRef]
- Tskhovrebova, L.; Trinick, J. Roles of titin in the structure and elasticity of the sarcomere. J. Biomed. Biotechnol. 2010, 2010, 612482. [Google Scholar] [CrossRef]
- Hu, D.H.; Matsuno, A.; Terakado, K.; Matsuura, T.; Kimura, S.; Maruyama, K. Projectin is an invertebrate connectin (titin): Isolation from crayfish claw muscle and localization in crayfish claw muscle and insect flight muscle. J. Muscle Res. Cell Motil. 1990, 11, 497–511. [Google Scholar] [CrossRef]
- Lakey, A.; Ferguson, C.; Labeit, S.; Reedy, M.; Larkins, A.; Butcher, G.; Leonard, K.; Bullard, B. Identification and localization of high molecular weight proteins in insect flight and leg muscle. EMBO J. 1990, 9, 3459–3467. [Google Scholar] [CrossRef]
- Lakey, A.; Labeit, S.; Gautel, M.; Ferguson, C.; Barlow, D.P.; Leonard, K.; Bullard, B. Kettin, a large modular protein in the Z-disc of insect muscles. EMBO J. 1993, 12, 2863–2871. [Google Scholar] [CrossRef]
- Patel, S.R.; Saide, J.D. Stretchin-klp, a novel Drosophila indirect flight muscle protein, has both myosin dependent and independent isoforms. J. Muscle Res. Cell Motil. 2005, 26, 213–224. [Google Scholar] [CrossRef] [PubMed]
- Becker, K.D.; O’Donnell, P.T.; Heitz, J.M.; Vito, M.; Bernstein, S.I. Analysis of Drosophila paramyosin: Identification of a novel isoform which is restricted to a subset of adult muscles. J. Cell Biol. 1992, 116, 669–681. [Google Scholar] [CrossRef]
- Ayer, G.; Vigoreaux, J.O. Flightin is a myosin rod binding protein. Cell Biochem. Biophys. 2003, 38, 41–54. [Google Scholar] [CrossRef]
- Qiu, F.; Brendel, S.; Cunha, P.M.; Astola, N.; Song, B.; Furlong, E.E.; Leonard, K.R.; Bullard, B. Myofilin, a protein in the thick filaments of insect muscle. J. Cell Sci. 2005, 118 Pt 7, 1527–1536. [Google Scholar] [CrossRef]
- Beinbrech, G.; Ashton, F.T.; Pepe, F.A. Invertebrate myosin filament: Subfilament arrangement in the wall of tubular filaments of insect flight muscles. J. Mol. Biol. 1988, 201, 557–565. [Google Scholar] [CrossRef]
- He, S.; Scheres, S.H.W. Helical reconstruction in RELION. J. Struct. Biol. 2017, 198, 163–176. [Google Scholar] [CrossRef]
- Grant, T.; Rohou, A.; Grigorieff, N. cisTEM, user-friendly software for single-particle image processing. Elife 2018, 7, e35383. [Google Scholar] [CrossRef]
- Squire, J.M. General model of myosin filament structure. 3. Molecular packing arrangements in myosin filaments. J. Mol. Biol. 1973, 77, 291–323. [Google Scholar] [CrossRef]
- Dickinson, M.; Farman, G.; Frye, M.; Bekyarova, T.; Gore, D.; Maughan, D.; Irving, T. Molecular dynamics of cyclically contracting insect flight muscle in vivo. Nature 2005, 433, 330–334. [Google Scholar] [CrossRef] [PubMed]
- Rahmani, H.; Ma, W.; Hu, Z.; Daneshparvar, N.; Taylor, D.W.; McCammon, J.A.; Irving, T.C.; Edwards, R.J.; Taylor, K.A. The myosin II coiled-coil domain atomic structure in its native environment. Proc. Natl. Acad. Sci. USA 2021, 118, e202415111. [Google Scholar] [CrossRef] [PubMed]
- Daneshparvar, N.; Rahmani, H.; Taylor, K. Homology model of Drosophila melanogaster myosin filaments. Microsc. Microanal. 2021, 27, 1704–1706. [Google Scholar] [CrossRef]
- Crick, F.H.C. The packing of α-helices: Simple coiled coils. Acta Crystallogr. 1953, 6, 689–697. [Google Scholar] [CrossRef]
- Vigoreaux, J.O.; Saide, J.D.; Valgeirsdottir, K.; Pardue, M.L. Flightin, a novel myofibrillar protein of Drosophila stretch-activated muscles. J. Cell Biol. 1993, 121, 587–598. [Google Scholar] [CrossRef]
- Champagne, M.B.; Edwards, K.A.; Erickson, H.P.; Kiehart, D.P. Drosophila stretchin-MLCK is a novel member of the Titin/Myosin light chain kinase family. J. Mol. Biol. 2000, 300, 759–777. [Google Scholar] [CrossRef]
- Maroto, M.; Arredondo, J.; Goulding, D.; Marco, R.; Bullard, B.; Cervera, M. Drosophila paramyosin/miniparamyosin gene products show a large diversity in quantity, localization, and isoform pattern: A possible role in muscle maturation and function. J. Cell Biol. 1996, 134, 81–92. [Google Scholar] [CrossRef]
- Soto-Adames, F.N.; Alvarez-Ortiz, P.; Vigoreaux, J.O. An evolutionary analysis of flightin reveals a conserved motif unique and widespread in Pancrustacea. J. Mol. Evol. 2014, 78, 24–37. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Zidek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Torices, R.; Munoz-Pajares, A.J. Phenix: An R Package to Estimate a Size-Controlled Phenotypic Integration Index. Appl. Plant Sci. 2015, 3, apps.1400104. [Google Scholar] [CrossRef]
- Wingfield, P.T. N-Terminal Methionine Processing. Curr. Protoc. Protein Sci. 2017, 88, 6.14.1–6.14.3. [Google Scholar] [CrossRef]
- Tanner, B.C.; Miller, M.S.; Miller, B.M.; Lekkas, P.; Irving, T.C.; Maughan, D.W.; Vigoreaux, J.O. COOH-terminal truncation of flightin decreases myofilament lattice organization, cross-bridge binding, and power output in Drosophila indirect flight muscle. Am. J. Physiol. Cell. Physiol. 2011, 301, C383–C391. [Google Scholar] [CrossRef]
- Gasek, N.S.; Nyland, L.R.; Vigoreaux, J.O. The Contributions of the Amino and Carboxy Terminal Domains of Flightin to the Biomechanical Properties of Drosophila Flight Muscle Thick Filaments. Biology 2016, 5, 16. [Google Scholar] [CrossRef]
- Kronert, W.A.; O’Donnell, P.T.; Fieck, A.; Lawn, A.; Vigoreaux, J.O.; Sparrow, J.C.; Bernstein, S.I. Defects in the Drosophila myosin rod permit sarcomere assembly but cause flight muscle degeneration. J. Mol. Biol. 1995, 249, 111–125. [Google Scholar] [CrossRef]
- Parker, F.; Peckham, M. Disease mutations in striated muscle myosins. Biophys. Rev. 2020, 12, 887–894. [Google Scholar] [CrossRef]
- Knupp, C.; Luther, P.K.; Squire, J.M. Titin organisation and the 3D architecture of the vertebrate-striated muscle I-band. J. Mol. Biol. 2002, 322, 731–739. [Google Scholar] [CrossRef]
- Kolmerer, B.; Clayton, J.; Benes, V.; Allen, T.; Ferguson, C.; Leonard, K.; Weber, U.; Knekt, M.; Ansorge, W.; Labeit, S.; et al. Sequence and expression of the kettin gene in Drosophila melanogaster and Caenorhabditis elegans. J. Mol. Biol. 2000, 296, 435–448. [Google Scholar] [CrossRef]
- Ayme-Southgate, A.J.; Southgate, R.J.; Philipp, R.A.; Sotka, E.E.; Kramp, C. The myofibrillar protein, projectin, is highly conserved across insect evolution except for its PEVK domain. J. Mol. Evol. 2008, 67, 653–669. [Google Scholar] [CrossRef]
- Mogami, K.; O’Donnell, P.T.; Bernstein, S.I.; Wright, T.R.; Emerson, C.P., Jr. Mutations of the Drosophila myosin heavy-chain gene: Effects on transcription, myosin accumulation, and muscle function. Proc. Natl. Acad. Sci. USA 1986, 83, 1393–1397. [Google Scholar] [CrossRef]
- Craig, R. Molecular structure of muscle filaments determined by electron microscopy. Appl. Microsc. 2017, 47, 226–232. [Google Scholar] [CrossRef]
- Yeganeh, F.A.; Rastegarpouyani, H.; Taylor, K.A. High Resolution Cryo-EM Structure of Drosophila Thick Filaments. Microsc. Microanal. 2022, 28, 1102–1103. [Google Scholar] [CrossRef]
- Lowey, S.; Trybus, K.M. Common structural motifs for the regulation of divergent class II myosins. J. Biol. Chem. 2010, 285, 16403–16407. [Google Scholar] [CrossRef]
- Liu, J.; Wu, S.; Reedy, M.C.; Winkler, H.; Lucaveche, C.; Cheng, Y.; Reedy, M.K.; Taylor, K.A. Electron tomography of swollen rigor fibers of insect flight muscle reveals a short and variably angled S2 domain. J. Mol. Biol. 2006, 362, 844–860. [Google Scholar] [CrossRef]
- Hvidt, S.; Nestler, F.H.; Greaser, M.L.; Ferry, J.D. Flexibility of myosin rod determined from dilute solution viscoelastic measurements. Biochemistry 1982, 21, 4064–4073. [Google Scholar] [CrossRef]
- Taylor, K.A.; Reedy, M.C.; Cordova, L.; Reedy, M.K. Three-dimensional reconstruction of rigor insect flight muscle from tilted thin sections. Nature 1984, 310, 285–291. [Google Scholar] [CrossRef]
- White, D.C. Rigor contraction and the effect of various phosphate compounds on glycerinated insect flight and vertebrate muscle. J. Physiol. 1970, 208, 583–605. [Google Scholar] [CrossRef] [PubMed]
- Reedy, M.K.; Reedy, M.C. Rigor crossbridge structure in tilted single filament layers and flared-X formations from insect flight muscle. J. Mol. Biol. 1985, 185, 145–176. [Google Scholar] [CrossRef]
- Bullard, B.; Pastore, A. Through thick and thin: Dual regulation of insect flight muscle and cardiac muscle compared. J. Muscle Res. Cell Motil. 2019, 40, 99–110. [Google Scholar] [CrossRef] [PubMed]
- Misof, B.; Liu, S.; Meusemann, K.; Peters, R.S.; Donath, A.; Mayer, C.; Frandsen, P.B.; Ware, J.; Flouri, T.; Beutel, R.G.; et al. Phylogenomics resolves the timing and pattern of insect evolution. Science 2014, 346, 763–767. [Google Scholar] [CrossRef]
- McLachlan, A.D.; Karn, J. Periodic charge distributions in the myosin rod amino acid sequence match cross-bridge spacings in muscle. Nature 1982, 299, 226–231. [Google Scholar] [CrossRef]
- Zheng, S.Q.; Palovcak, E.; Armache, J.P.; Verba, K.A.; Cheng, Y.; Agard, D.A. MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 2017, 14, 331–332. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K. Gctf: Real-time CTF determination and correction. J. Struct. Biol. 2016, 193, 290–296. [Google Scholar] [CrossRef] [PubMed]
- Scheres, S.H. Processing of Structurally Heterogeneous Cryo-EM Data in RELION. Methods Enzymol. 2016, 579, 125–157. [Google Scholar] [PubMed]
- Punjani, A.; Rubinstein, J.L.; Fleet, D.J.; Brubaker, M.A. cryoSPARC: Algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 2017, 14, 290–296. [Google Scholar] [CrossRef] [PubMed]
- Vilas, J.L.; Gomez-Blanco, J.; Conesa, P.; Melero, R.; Miguel de la Rosa-Trevin, J.; Oton, J.; Cuenca, J.; Marabini, R.; Carazo, J.M.; Vargas, J.; et al. MonoRes: Automatic and Accurate Estimation of Local Resolution for Electron Microscopy Maps. Structure 2018, 26, 337–344.e4. [Google Scholar] [CrossRef] [PubMed]
- Ramirez-Aportela, E.; Vilas, J.L.; Glukhova, A.; Melero, R.; Conesa, P.; Martinez, M.; Maluenda, D.; Mota, J.; Jimenez, A.; Vargas, J.; et al. Automatic local resolution-based sharpening of cryo-EM maps. Bioinformatics 2020, 36, 765–772. [Google Scholar] [CrossRef]
- De la Rosa-Trevin, J.M.; Quintana, A.; del Cano, L.; Zaldivar, A.; Foche, I.; Gutierrez, J.; Gomez-Blanco, J.; Burguet-Castell, J.; Cuenca-Alba, J.; Abrishami, V.; et al. Scipion: A software framework toward integration, reproducibility and validation in 3D electron microscopy. J. Struct. Biol. 2016, 195, 93–99. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Emsley, P.; Cowtan, K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. Sect. D Biol. Crystallogr. 2004, 60, 2126–2132. [Google Scholar] [CrossRef] [PubMed]
- Afonine, P.V.; Poon, B.K.; Read, R.J.; Sobolev, O.V.; Terwilliger, T.C.; Urzhumtsev, A.; Adams, P.D. Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr. Sect. D Struct. Biol. 2018, 74, 531–544. [Google Scholar] [CrossRef]



| Domain | Residue Range | Domain Name | Domain Length | Linker Length |
|---|---|---|---|---|
| 1 | 456–543 | Ig-like | 88 | 11 |
| 2 | 554–643 | Ig-like | 90 | 67 |
| 3 | 710–798 | Ig-like | 89 | 17 |
| 4 | 815–904 | Ig-like | 89 | 61 |
| 5 | 965–1056 | Ig-like | 92 | 12 |
| 6 | 1068–1153 | Ig-like | 86 | 93 |
| 7 | 1246–1333 | Ig-like | 88 | 12 |
| 8 | 1345–1434 | Ig-like | 90 | 174 |
| 9 | 1608–1693 | Ig-like | 86 | 27 |
| 10 | 1720–1808 | Ig-like | 89 | 110 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abbasi Yeganeh, F.; Rastegarpouyani, H.; Li, J.; Taylor, K.A. Structure of the Drosophila melanogaster Flight Muscle Myosin Filament at 4.7 Å Resolution Reveals New Details of Non-Myosin Proteins. Int. J. Mol. Sci. 2023, 24, 14936. https://doi.org/10.3390/ijms241914936
Abbasi Yeganeh F, Rastegarpouyani H, Li J, Taylor KA. Structure of the Drosophila melanogaster Flight Muscle Myosin Filament at 4.7 Å Resolution Reveals New Details of Non-Myosin Proteins. International Journal of Molecular Sciences. 2023; 24(19):14936. https://doi.org/10.3390/ijms241914936
Chicago/Turabian StyleAbbasi Yeganeh, Fatemeh, Hosna Rastegarpouyani, Jiawei Li, and Kenneth A. Taylor. 2023. "Structure of the Drosophila melanogaster Flight Muscle Myosin Filament at 4.7 Å Resolution Reveals New Details of Non-Myosin Proteins" International Journal of Molecular Sciences 24, no. 19: 14936. https://doi.org/10.3390/ijms241914936
APA StyleAbbasi Yeganeh, F., Rastegarpouyani, H., Li, J., & Taylor, K. A. (2023). Structure of the Drosophila melanogaster Flight Muscle Myosin Filament at 4.7 Å Resolution Reveals New Details of Non-Myosin Proteins. International Journal of Molecular Sciences, 24(19), 14936. https://doi.org/10.3390/ijms241914936






