Proteomics Profiling of Bladder Cancer Tissues from Early to Advanced Stages Reveals NNMT and GALK1 as Biomarkers for Early Detection and Prognosis of BCa
Abstract
:1. Introduction
2. Results
2.1. Protein Identification
2.2. Proteins with Differential Abundance among Groups
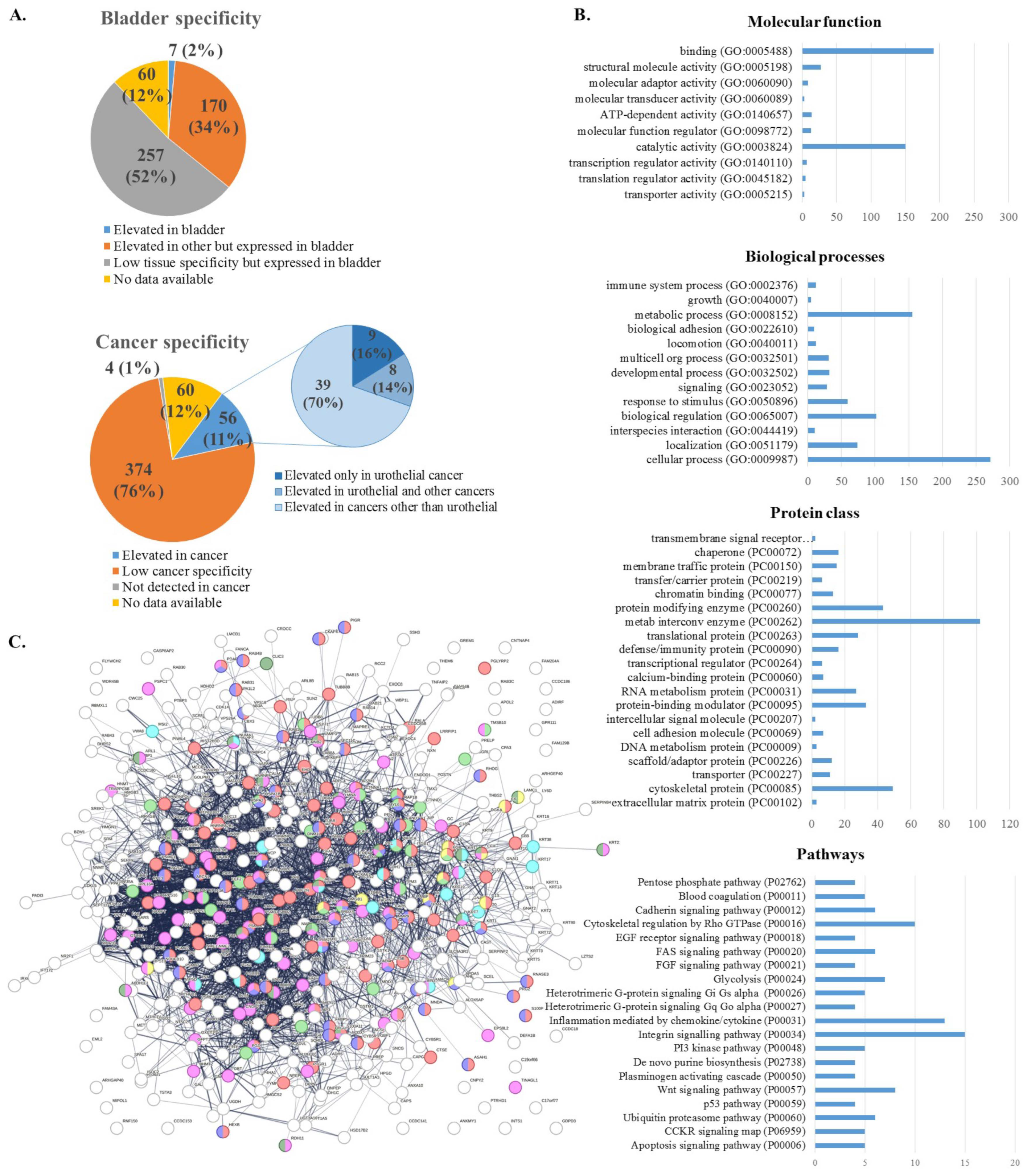
2.3. Functional Analysis
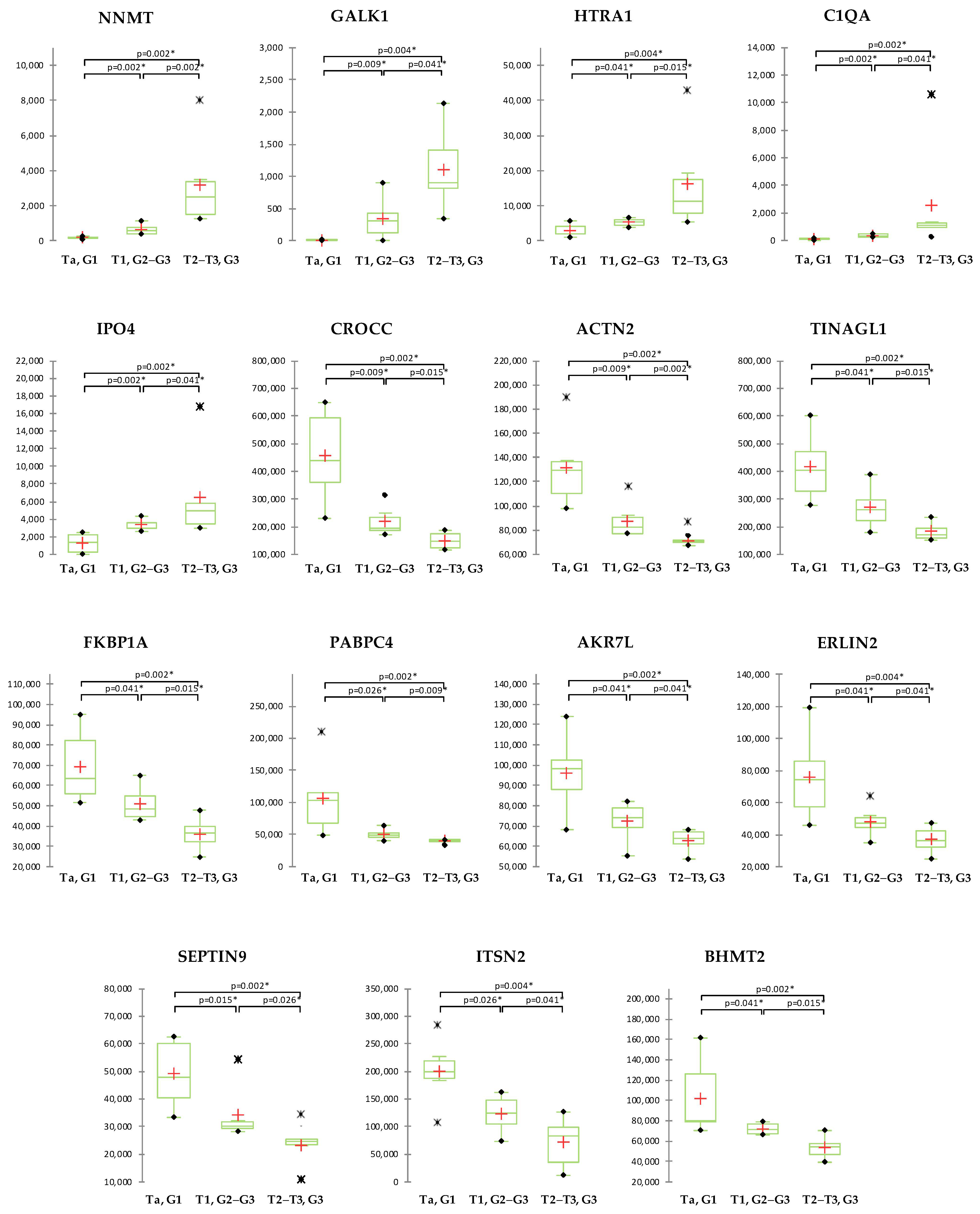
2.4. Correlation of Proteomics Findings with Clinical Parameters
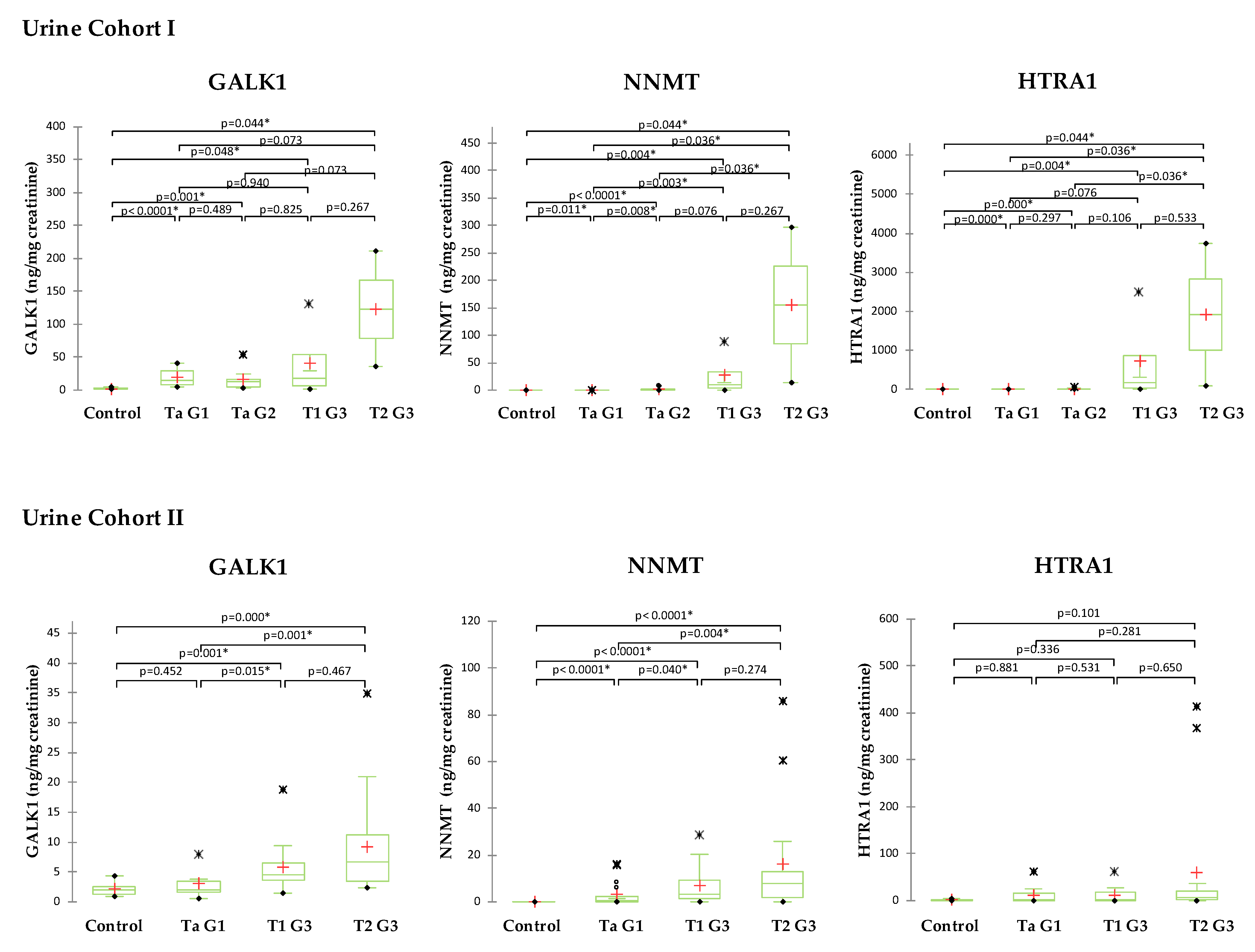
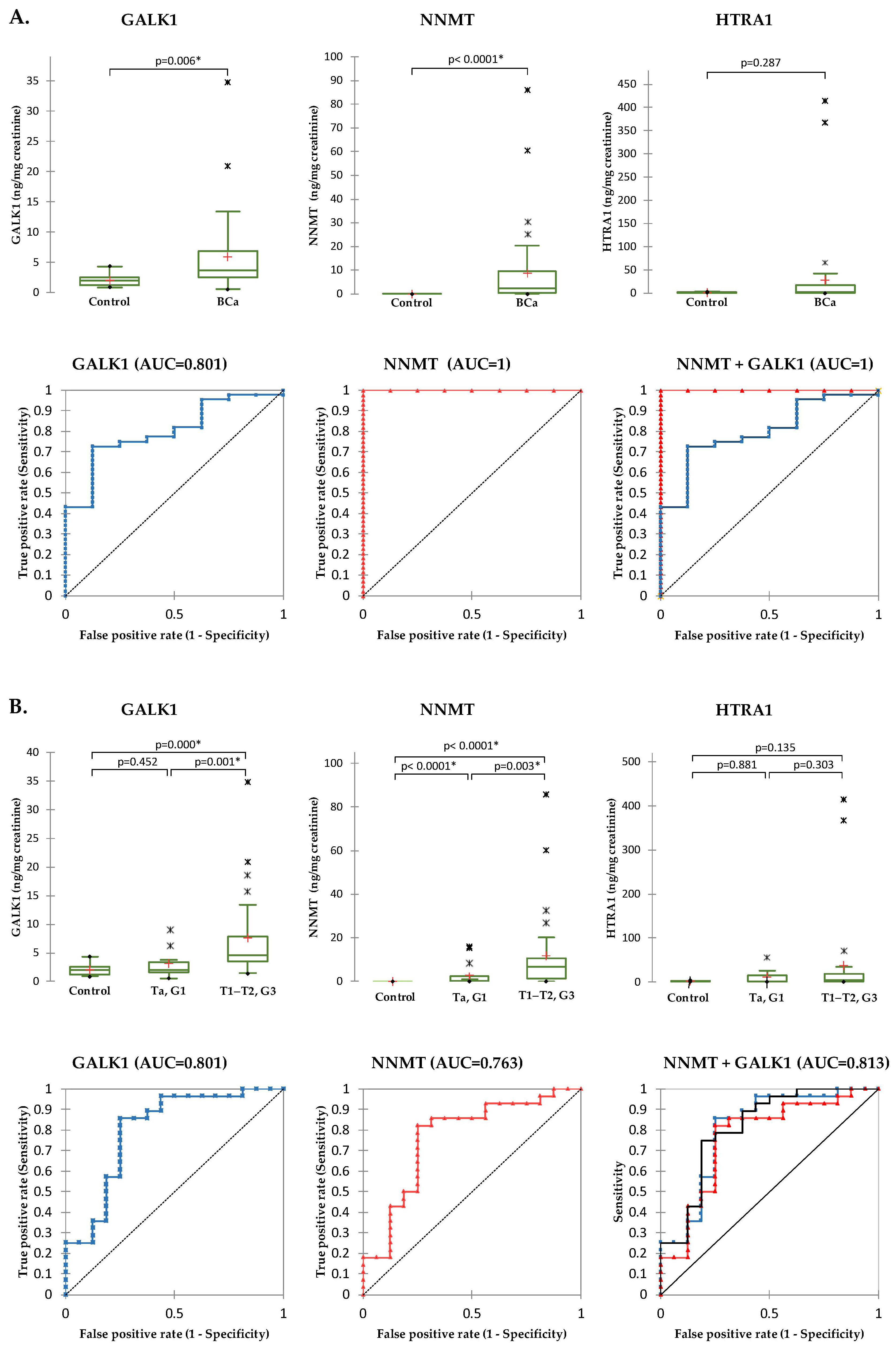
2.5. Validation of Selected Candidates in Urine
3. Discussion
4. Materials and Methods
4.1. Patients Samples
4.2. Sample Preparation
4.3. LC-MS/MS Data Acquisition
4.4. LC-MS/MS Data Processing
4.5. Quantitative Measurement of Candidate Proteins in Urine
4.6. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Nielsen, M.E.; Smith, A.B.; Meyer, A.M.; Kuo, T.M.; Tyree, S.; Kim, W.Y.; Milowsky, M.I.; Pruthi, R.S.; Millikan, R.C. Trends in stage-specific incidence rates for urothelial carcinoma of the bladder in the United States: 1988 to 2006. Cancer 2014, 120, 86–95. [Google Scholar] [CrossRef]
- Mak, R.H.; Hunt, D.; Shipley, W.U.; Efstathiou, J.A.; Tester, W.J.; Hagan, M.P.; Kaufman, D.S.; Heney, N.M.; Zietman, A.L. Long-term outcomes in patients with muscle-invasive bladder cancer after selective bladder-preserving combined-modality therapy: A pooled analysis of Radiation Therapy Oncology Group protocols 8802, 8903, 9506, 9706, 9906, and 0233. J. Clin. Oncol. 2014, 32, 3801–3809. [Google Scholar] [CrossRef]
- Babjuk, M.; Burger, M.; Zigeuner, R.; Shariat, S.F.; van Rhijn, B.W.; Comperat, E.; Sylvester, R.J.; Kaasinen, E.; Bohle, A.; Palou Redorta, J.; et al. EAU guidelines on non-muscle-invasive urothelial carcinoma of the bladder: Update 2013. Eur. Urol. 2013, 64, 639–653. [Google Scholar]
- Sylvester, R.J.; van der Meijden, A.P.; Oosterlinck, W.; Witjes, J.A.; Bouffioux, C.; Denis, L.; Newling, D.W.; Kurth, K. Predicting recurrence and progression in individual patients with stage Ta T1 bladder cancer using EORTC risk tables: A combined analysis of 2596 patients from seven EORTC trials. Eur. Urol. 2006, 49, 466–477. [Google Scholar] [CrossRef]
- Yafi, F.A.; Brimo, F.; Steinberg, J.; Aprikian, A.G.; Tanguay, S.; Kassouf, W. Prospective analysis of sensitivity and specificity of urinary cytology and other urinary biomarkers for bladder cancer. Urol. Oncol. 2015, 33, e25–e66.e31. [Google Scholar] [CrossRef]
- Fradet, Y.; Grossman, H.B.; Gomella, L.; Lerner, S.; Cookson, M.; Albala, D.; Droller, M.J. A comparison of hexaminolevulinate fluorescence cystoscopy and white light cystoscopy for the detection of carcinoma in situ in patients with bladder cancer: A phase III, multicenter study. J. Urol. 2007, 178, 68–73. [Google Scholar] [CrossRef]
- Grossman, H.B.; Gomella, L.; Fradet, Y.; Morales, A.; Presti, J.; Ritenour, C.; Nseyo, U.; Droller, M.J. A phase III, multicenter comparison of hexaminolevulinate fluorescence cystoscopy and white light cystoscopy for the detection of superficial papillary lesions in patients with bladder cancer. J. Urol. 2007, 178, 62–67. [Google Scholar] [CrossRef]
- Lee, C.S.; Yoon, C.Y.; Witjes, J.A. The past, present and future of cystoscopy: The fusion of cystoscopy and novel imaging technology. BJU Int. 2008, 102 Pt B, 1228–1233. [Google Scholar] [CrossRef]
- Soubra, A.; Risk, M.C. Diagnostics techniques in nonmuscle invasive bladder cancer. Indian J. Urol. 2015, 31, 283–288. [Google Scholar]
- Chakraborty, A.; Dasari, S.; Long, W.; Mohan, C. Urine protein biomarkers for the detection, surveillance, and treatment response prediction of bladder cancer. Am. J. Cancer Res. 2019, 9, 1104–1117. [Google Scholar]
- Frantzi, M.; Latosinska, A.; Fluhe, L.; Hupe, M.C.; Critselis, E.; Kramer, M.W.; Merseburger, A.S.; Mischak, H.; Vlahou, A. Developing proteomic biomarkers for bladder cancer: Towards clinical application. Nat. Rev. Urol. 2015, 12, 317–330. [Google Scholar] [CrossRef]
- Hong, M.; He, G.; Goh, S.; Low, A.W.X.; Tay, K.J.; Lim, T.K.H.; Yeong, J.; Khor, L.Y.; Lim, T.S. Biomarkers for Precision Urothelial Carcinoma Diagnosis: Current Approaches and the Application of Single-Cell Technologies. Cancers 2021, 13, 260. [Google Scholar] [CrossRef]
- Latosinska, A.; Frantzi, M.; Vlahou, A.; Merseburger, A.S.; Mischak, H. Clinical Proteomics for Precision Medicine: The Bladder Cancer Case. Proteom. Clin. Appl. 2018, 12, 1700074. [Google Scholar] [CrossRef]
- Frantzi, M.; Vlahou, A. Ten Years of Proteomics in Bladder Cancer: Progress and Future Directions. Bladder Cancer 2017, 3, 1–18. [Google Scholar] [CrossRef]
- Cooper, J.; Giancotti, F.G. Integrin Signaling in Cancer: Mechanotransduction, Stemness, Epithelial Plasticity, and Therapeutic Resistance. Cancer Cell 2019, 35, 347–367. [Google Scholar] [CrossRef]
- Hembruff, S.L.; Cheng, N. Chemokine signaling in cancer: Implications on the tumor microenvironment and therapeutic targeting. Cancer Ther. 2009, 7, 254–267. [Google Scholar]
- Vega, F.M.; Ridley, A.J. Rho GTPases in cancer cell biology. FEBS Lett. 2008, 582, 2093–2101. [Google Scholar] [CrossRef]
- Wong, R.S. Apoptosis in cancer: From pathogenesis to treatment. J. Exp. Clin. Cancer Res. 2011, 30, 87. [Google Scholar] [CrossRef]
- Houston, A.; O’Connell, J. The Fas signaling pathway and its role in the pathogenesis of cancer. Curr. Opin. Pharm. 2004, 4, 321–326. [Google Scholar] [CrossRef]
- Vazquez, A.; Bond, E.E.; Levine, A.J.; Bond, G.L. The genetics of the p53 pathway, apoptosis and cancer therapy. Nat. Rev. Drug Discov. 2008, 7, 979–987. [Google Scholar] [CrossRef]
- Normanno, N.; De Luca, A.; Bianco, C.; Strizzi, L.; Mancino, M.; Maiello, M.R.; Carotenuto, A.; De Feo, G.; Caponigro, F.; Salomon, D.S. Epidermal growth factor receptor (EGFR) signaling in cancer. Gene 2006, 366, 2–16. [Google Scholar] [CrossRef]
- Fresno Vara, J.A.; Casado, E.; de Castro, J.; Cejas, P.; Belda-Iniesta, C.; Gonzalez-Baron, M. PI3K/Akt signalling pathway and cancer. Cancer Treat. Rev. 2004, 30, 193–204. [Google Scholar] [CrossRef]
- Yu, W.; Yang, L.; Li, T.; Zhang, Y. Cadherin Signaling in Cancer: Its Functions and Role as a Therapeutic Target. Front. Oncol. 2019, 9, 989. [Google Scholar] [CrossRef]
- Zhan, T.; Rindtorff, N.; Boutros, M. Wnt signaling in cancer. Oncogene 2017, 36, 1461–1473. [Google Scholar] [CrossRef]
- Schwartz, A.L.; Ciechanover, A. The ubiquitin-proteasome pathway and pathogenesis of human diseases. Annu. Rev. Med. 1999, 50, 57–74. [Google Scholar] [CrossRef]
- Patra, K.C.; Hay, N. The pentose phosphate pathway and cancer. Trends Biochem. Sci. 2014, 39, 347–354. [Google Scholar] [CrossRef]
- Zhou, D.; Duan, Z.; Li, Z.; Ge, F.; Wei, R.; Kong, L. The significance of glycolysis in tumor progression and its relationship with the tumor microenvironment. Front. Pharmacol. 2022, 13, 1091779. [Google Scholar] [CrossRef]
- Arang, N.; Gutkind, J.S. G Protein-Coupled receptors and heterotrimeric G proteins as cancer drivers. FEBS Lett. 2020, 594, 4201–4232. [Google Scholar] [CrossRef]
- Kalafati, L.; Mitroulis, I.; Verginis, P.; Chavakis, T.; Kourtzelis, I. Neutrophils as Orchestrators in Tumor Development and Metastasis Formation. Front. Oncol. 2020, 10, 581457. [Google Scholar] [CrossRef]
- Claesson-Welsh, L.; Welsh, M. VEGFA and tumor angiogenesis. J. Intern. Med. 2013, 273, 114–127. [Google Scholar] [CrossRef]
- Revel, M.; Daugan, M.V.; Sautes-Fridman, C.; Fridman, W.H.; Roumenina, L.T. Complement System: Promoter or Suppressor of Cancer Progression? Antibodies 2020, 9, 57. [Google Scholar] [CrossRef]
- Niu, H.T.; Zhang, Y.B.; Jiang, H.P.; Cheng, B.; Sun, G.; Wang, Y.; Jun, Y.E.; de Pang, Q.; Chang, J.W. Differences in shotgun protein expression profile between superficial bladder transitional cell carcinoma and normal urothelium. Urol. Oncol. 2009, 27, 400–406. [Google Scholar] [CrossRef]
- Liu, P.F.; Wang, Y.H.; Cao, Y.W.; Jiang, H.P.; Yang, X.C.; Wang, X.S.; Niu, H.T. Far from resolved: Stromal cell-based iTRAQ research of muscle-invasive bladder cancer regarding heterogeneity. Oncol. Rep. 2014, 32, 1489–1496. [Google Scholar] [CrossRef]
- Fristrup, N.; Ulhoi, B.P.; Birkenkamp-Demtroder, K.; Mansilla, F.; Sanchez-Carbayo, M.; Segersten, U.; Malmstrom, P.U.; Hartmann, A.; Palou, J.; Alvarez-Mugica, M.; et al. Cathepsin E, maspin, Plk1, and survivin are promising prognostic protein markers for progression in non-muscle invasive bladder cancer. Am. J. Pathol. 2012, 180, 1824–1834. [Google Scholar] [CrossRef]
- Sheng, K.H.; Yao, Y.C.; Chuang, S.S.; Wu, H.; Wu, T.F. Search for the tumor-related proteins of transition cell carcinoma in Taiwan by proteomic analysis. Proteomics 2006, 6, 1058–1065. [Google Scholar] [CrossRef]
- Barboro, P.; Rubagotti, A.; Orecchia, P.; Spina, B.; Truini, M.; Repaci, E.; Carmignani, G.; Romagnoli, A.; Introini, C.; Boccardo, F.; et al. Differential proteomic analysis of nuclear matrix in muscle-invasive bladder cancer: Potential to improve diagnosis and prognosis. Cell. Oncol. 2008, 30, 13–26. [Google Scholar] [CrossRef]
- Soloway, M.S.; Briggman, V.; Carpinito, G.A.; Chodak, G.W.; Church, P.A.; Lamm, D.L.; Lange, P.; Messing, E.; Pasciak, R.M.; Reservitz, G.B.; et al. Use of a new tumor marker, urinary NMP22, in the detection of occult or rapidly recurring transitional cell carcinoma of the urinary tract following surgical treatment. J. Urol. 1996, 156 Pt 1, 363–367. [Google Scholar] [CrossRef]
- Rini, J.; Szumlanski, C.; Guerciolini, R.; Weinshilboum, R.M. Human liver nicotinamide N-methyltransferase: Ion-pairing radiochemical assay, biochemical properties and individual variation. Clin. Chim. Acta 1990, 186, 359–374. [Google Scholar] [CrossRef]
- Campagna, R.; Pozzi, V.; Spinelli, G.; Sartini, D.; Milanese, G.; Galosi, A.B.; Emanuelli, M. The Utility of Nicotinamide N-Methyltransferase as a Potential Biomarker to Predict the Oncological Outcomes for Urological Cancers: An Update. Biomolecules 2021, 11, 1214. [Google Scholar] [CrossRef]
- Wang, W.; Yang, C.; Wang, T.; Deng, H. Complex roles of nicotinamide N-methyltransferase in cancer progression. Cell. Death Dis 2022, 13, 267. [Google Scholar] [CrossRef]
- Eckert, M.A.; Coscia, F.; Chryplewicz, A.; Chang, J.W.; Hernandez, K.M.; Pan, S.; Tienda, S.M.; Nahotko, D.A.; Li, G.; Blazenovic, I.; et al. Proteomics reveals NNMT as a master metabolic regulator of cancer-associated fibroblasts. Nature 2019, 569, 723–728. [Google Scholar] [CrossRef]
- Gao, Y.; van Haren, M.J.; Buijs, N.; Innocenti, P.; Zhang, Y.; Sartini, D.; Campagna, R.; Emanuelli, M.; Parsons, R.B.; Jespers, W.; et al. Potent Inhibition of Nicotinamide N-Methyltransferase by Alkene-Linked Bisubstrate Mimics Bearing Electron Deficient Aromatics. J. Med. Chem. 2021, 64, 12938–12963. [Google Scholar] [CrossRef]
- van Haren, M.J.; Gao, Y.; Buijs, N.; Campagna, R.; Sartini, D.; Emanuelli, M.; Mateuszuk, L.; Kij, A.; Chlopicki, S.; Escude Martinez de Castilla, P.; et al. Esterase-Sensitive Prodrugs of a Potent Bisubstrate Inhibitor of Nicotinamide N-Methyltransferase (NNMT) Display Cellular Activity. Biomolecules 2021, 11, 1357. [Google Scholar] [CrossRef]
- van Haren, M.J.; Zhang, Y.; Thijssen, V.; Buijs, N.; Gao, Y.; Mateuszuk, L.; Fedak, F.A.; Kij, A.; Campagna, R.; Sartini, D.; et al. Macrocyclic peptides as allosteric inhibitors of nicotinamide N-methyltransferase (NNMT). RSC Chem. Biol. 2021, 2, 1546–1555. [Google Scholar] [CrossRef]
- Riester, M.; Taylor, J.M.; Feifer, A.; Koppie, T.; Rosenberg, J.E.; Downey, R.J.; Bochner, B.H.; Michor, F. Combination of a novel gene expression signature with a clinical nomogram improves the prediction of survival in high-risk bladder cancer. Clin. Cancer Res. 2012, 18, 1323–1333. [Google Scholar] [CrossRef]
- Sartini, D.; Muzzonigro, G.; Milanese, G.; Pozzi, V.; Vici, A.; Morganti, S.; Rossi, V.; Mazzucchelli, R.; Montironi, R.; Emanuelli, M. Upregulation of tissue and urinary nicotinamide N-methyltransferase in bladder cancer: Potential for the development of a urine-based diagnostic test. Cell Biochem. Biophys. 2013, 65, 473–483. [Google Scholar] [CrossRef]
- Pozzi, V.; Di Ruscio, G.; Sartini, D.; Campagna, R.; Seta, R.; Fulvi, P.; Vici, A.; Milanese, G.; Brandoni, G.; Galosi, A.B.; et al. Clinical performance and utility of a NNMT-based urine test for bladder cancer. Int. J. Biol. Markers 2018, 33, 94–101. [Google Scholar] [CrossRef]
- Barretina, J.; Caponigro, G.; Stransky, N.; Venkatesan, K.; Margolin, A.A.; Kim, S.; Wilson, C.J.; Lehar, J.; Kryukov, G.V.; Sonkin, D.; et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 2012, 483, 603–607. [Google Scholar] [CrossRef]
- Tang, M.; Etokidem, E.; Lai, K. The Leloir Pathway of Galactose Metabolism—A Novel Therapeutic Target for Hepatocellular Carcinoma. Anticancer. Res. 2016, 36, 6265–6271. [Google Scholar] [CrossRef]
- Wu, Z.; Wen, Z.; Li, Z.; Yu, M.; Ye, G. Identification and prognostic value of a glycolysis-related gene signature in patients with bladder cancer. Medicine 2021, 100, e23836. [Google Scholar] [CrossRef]
- Qiu, T.; Chen, Y.; Meng, L.; Xu, T.; Zhang, H. Identification of a metabolism-related gene signature predicting overall survival for bladder cancer. Genomics 2022, 114, 110402. [Google Scholar] [CrossRef]
- Huang, C.; Li, Y.; Ling, Q.; Wei, C.; Fang, B.; Mao, X.; Yang, R.; Zhang, L.; Huang, S.; Cheng, J.; et al. Establishment of a risk score model for bladder urothelial carcinoma based on energy metabolism-related genes and their relationships with immune infiltration. FEBS Open Bio 2023, 13, 736–750. [Google Scholar] [CrossRef]
- Shridhar, V.; Sen, A.; Chien, J.; Staub, J.; Avula, R.; Kovats, S.; Lee, J.; Lillie, J.; Smith, D.I. Identification of underexpressed genes in early- and late-stage primary ovarian tumors by suppression subtraction hybridization. Cancer Res. 2002, 62, 262–270. [Google Scholar]
- Esposito, V.; Campioni, M.; De Luca, A.; Spugnini, E.P.; Baldi, F.; Cassandro, R.; Mancini, A.; Vincenzi, B.; Groeger, A.; Caputi, M.; et al. Analysis of HtrA1 serine protease expression in human lung cancer. Anticancer. Res. 2006, 26, 3455–3459. [Google Scholar]
- Baldi, A.; De Luca, A.; Morini, M.; Battista, T.; Felsani, A.; Baldi, F.; Catricala, C.; Amantea, A.; Noonan, D.M.; Albini, A.; et al. The HtrA1 serine protease is down-regulated during human melanoma progression and represses growth of metastatic melanoma cells. Oncogene 2002, 21, 6684–6688. [Google Scholar] [CrossRef]
- Lorenzi, T.; Lorenzi, M.; Altobelli, E.; Marzioni, D.; Mensa, E.; Quaranta, A.; Paolinelli, F.; Morroni, M.; Mazzucchelli, R.; De Luca, A.; et al. HtrA1 in human urothelial bladder cancer: A secreted protein and a potential novel biomarker. Int. J. Cancer 2013, 133, 2650–2661. [Google Scholar] [CrossRef]
- Klose, R.; Adam, M.G.; Weis, E.M.; Moll, I.; Wustehube-Lausch, J.; Tetzlaff, F.; Oka, C.; Ehrmann, M.; Fischer, A. Inactivation of the serine protease HTRA1 inhibits tumor growth by deregulating angiogenesis. Oncogene 2018, 37, 4260–4272. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Yu, Y.Q.; Gilar, M.; Lee, P.J.; Bouvier, E.S.; Gebler, J.C. Enzyme-friendly, mass spectrometry-compatible surfactant for in-solution enzymatic digestion of proteins. Anal. Chem. 2003, 75, 6023–6028. [Google Scholar] [CrossRef]
- Davalieva, K.; Kiprijanovska, S.; Dimovski, A.; Rosoklija, G.; Dwork, A.J. Comparative evaluation of two methods for LC-MS/MS proteomic analysis of formalin fixed and paraffin embedded tissues. J. Proteom. 2021, 235, 104117. [Google Scholar] [CrossRef]
- Distler, U.; Kuharev, J.; Navarro, P.; Levin, Y.; Schild, H.; Tenzer, S. Drift time-specific collision energies enable deep-coverage data-independent acquisition proteomics. Nat. Methods 2014, 11, 167–170. [Google Scholar] [CrossRef]
- Distler, U.; Kuharev, J.; Navarro, P.; Tenzer, S. Label-free quantification in ion mobility-enhanced data-independent acquisition proteomics. Nat. Protoc. 2016, 11, 795–812. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Society. Ser. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Mi, H.; Ebert, D.; Muruganujan, A.; Mills, C.; Albou, L.-P.; Mushayamaha, T.; Thomas, P.D. PANTHER version 16: A revised family classification, tree-based classification tool, enhancer regions and extensive API. Nucleic Acids Res. 2020, 49, D394–D403. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef]
- Uhlen, M.; Fagerberg, L.; Hallstrom, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, A.; Kampf, C.; Sjostedt, E.; Asplund, A.; et al. Proteomics. Tissue-Based Map of the Human Proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef]


| Gr1 vs. Gr2 (Ta vs. T1) | Gr2 vs. Gr3 (T1 vs. T2/3) | Gr1 vs. GR3 (Ta vs. T2/3) | |
|---|---|---|---|
| Differentially expressed (Mann–Whitney p ≤ 0.05) | 340 | 95 | 482 |
| Differentially expressed (B-H p ≤ 0.05) | 81 | 0 | 195 |
| Up-regulated (B-H p ≤ 0.05) | 35 | / | 75 |
| Down-regulated (B-H p ≤ 0.05) | 46 | / | 121 |
| Variables | Age | Stage | GALK1 | NNMT | HTRA1 |
|---|---|---|---|---|---|
| Age | 1 | 0.412 | 0.360 | 0.351 | 0.188 |
| Stage | 0.412 | 1 | 0.566 | 0.627 | 0.206 |
| GALK1 | 0.360 | 0.566 | 1 | 0.634 | 0.565 |
| NNMT | 0.351 | 0.627 | 0.634 | 1 | 0.598 |
| HTRA1 | 0.188 | 0.206 | 0.565 | 0.598 | 1 |
| Group | Diagnosis | Patients Per Group | Age (Mean ± SD) | Age (Median) | TNM Classification | Grade | ||
|---|---|---|---|---|---|---|---|---|
| T | N | M | ||||||
| Tissue samples for the discovery of proteomics | ||||||||
| Group 1 (Ta, G1) | Non-invasive low-grade papillary urothelial carcinoma | 6 | 58.5 ± 9.8 | 62 | Ta | N0 | M0 | I |
| Group 2 (T1, G2–G3) | Invasive low/high-grade papillary urothelial carcinoma | 6 | 62.2 ± 6.7 | 62 | T1 | N0 | M0 | II–III |
| Group 3 (T2–T3, G3) | Invasive/Infiltrative high-grade papillary urothelial carcinoma | 6 | 72.2 ± 8.0 | 73.5 | T2–T3 | Nx | Mx | III |
| Urine cohort I for ELISA validation | ||||||||
| Group 1 (Ta, G1) | Non-invasive low-grade papillary urothelial carcinoma | 9 | 58.2 ± 12.0 | 63 | Ta | N0 | M0 | I |
| Group 2 (Ta, G2) | Non-invasive low-grade papillary urothelial carcinoma | 9 | 69.9 ± 6.6 | 68 | Ta | N0 | M0 | II |
| Group 3 (T1, G3) | Invasive high-grade papillary urothelial carcinoma | 4 | 69.5 ± 1.3 | 69.5 | T1 | N0 | M0 | III |
| Group 4 (T2, G2–G3) | Infiltrative high-grade papillary urothelial carcinoma | 2 | 76.0 ± 7.1 | 76 | T2 | Nx | Mx | II–III |
| Urine cohort II for ELISA validation | ||||||||
| Group 1 (Ta, G1) | Non-invasive low-grade papillary urothelial carcinoma | 16 | 64.6 ± 14.3 | 65 | Ta | N0 | M0 | I |
| Group 2 (T1, G2–G3) | Invasive low/high-grade papillary urothelial carcinoma | 13 | 64.6 ± 7.8 | 64 | T1 | N0 | M0 | II–III |
| Group 3 (T2, G3) | Invasive/Infiltrative high-grade papillary urothelial carcinoma | 15 | 68.9 ± 10.3 | 66 | T2 | Nx | Mx | III |
| Control group | / | 8 | 48.9 ± 9.8 | 47 | / | / | / | / |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Davalieva, K.; Kiprijanovska, S.; Ivanovski, O.; Trifunovski, A.; Saidi, S.; Dimovski, A.; Popov, Z. Proteomics Profiling of Bladder Cancer Tissues from Early to Advanced Stages Reveals NNMT and GALK1 as Biomarkers for Early Detection and Prognosis of BCa. Int. J. Mol. Sci. 2023, 24, 14938. https://doi.org/10.3390/ijms241914938
Davalieva K, Kiprijanovska S, Ivanovski O, Trifunovski A, Saidi S, Dimovski A, Popov Z. Proteomics Profiling of Bladder Cancer Tissues from Early to Advanced Stages Reveals NNMT and GALK1 as Biomarkers for Early Detection and Prognosis of BCa. International Journal of Molecular Sciences. 2023; 24(19):14938. https://doi.org/10.3390/ijms241914938
Chicago/Turabian StyleDavalieva, Katarina, Sanja Kiprijanovska, Ognen Ivanovski, Aleksandar Trifunovski, Skender Saidi, Aleksandar Dimovski, and Zivko Popov. 2023. "Proteomics Profiling of Bladder Cancer Tissues from Early to Advanced Stages Reveals NNMT and GALK1 as Biomarkers for Early Detection and Prognosis of BCa" International Journal of Molecular Sciences 24, no. 19: 14938. https://doi.org/10.3390/ijms241914938







