Multi-Omics Profiling of Human Endothelial Cells from the Coronary Artery and Internal Thoracic Artery Reveals Molecular but Not Functional Heterogeneity
Abstract
:1. Introduction
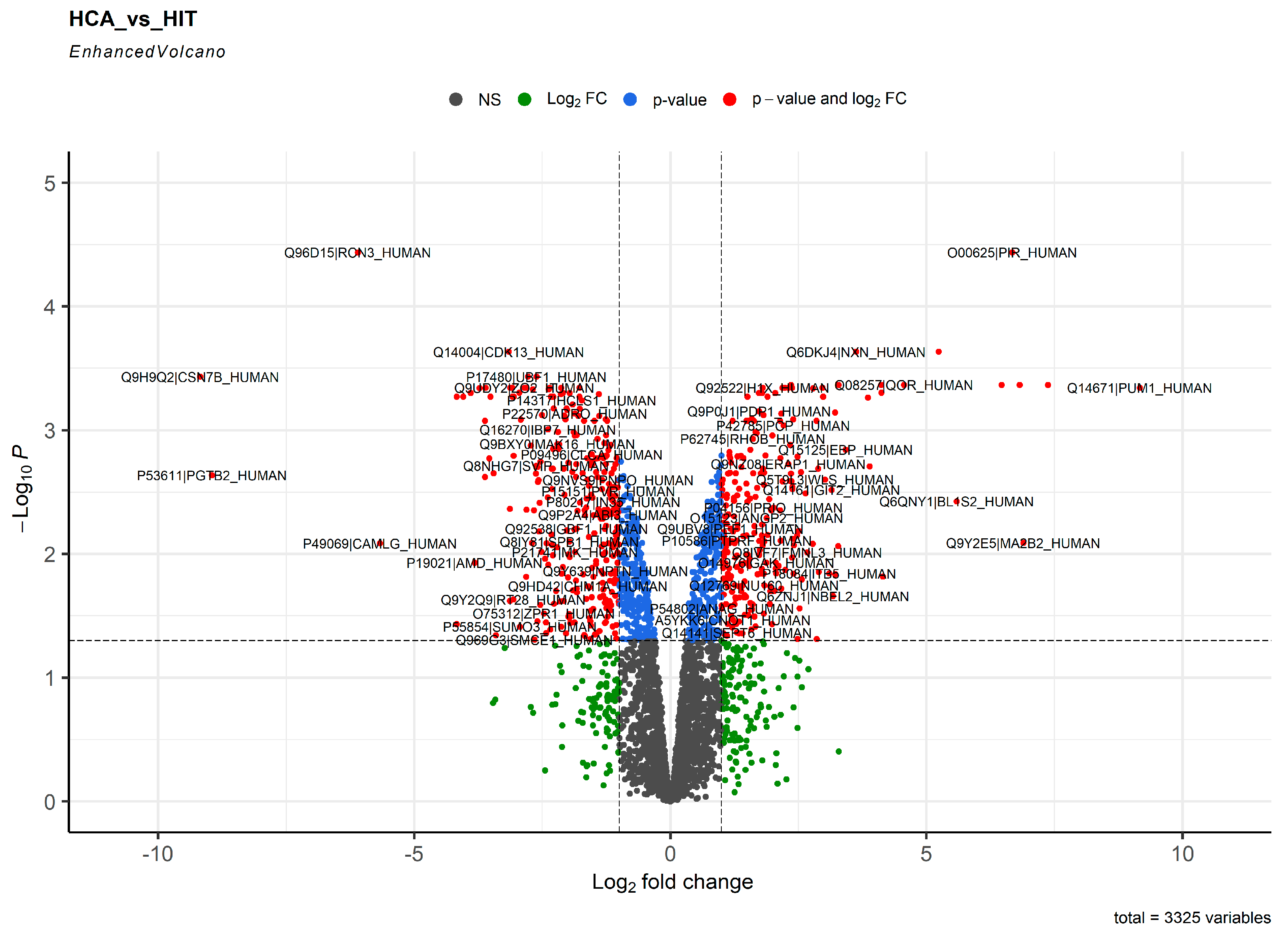
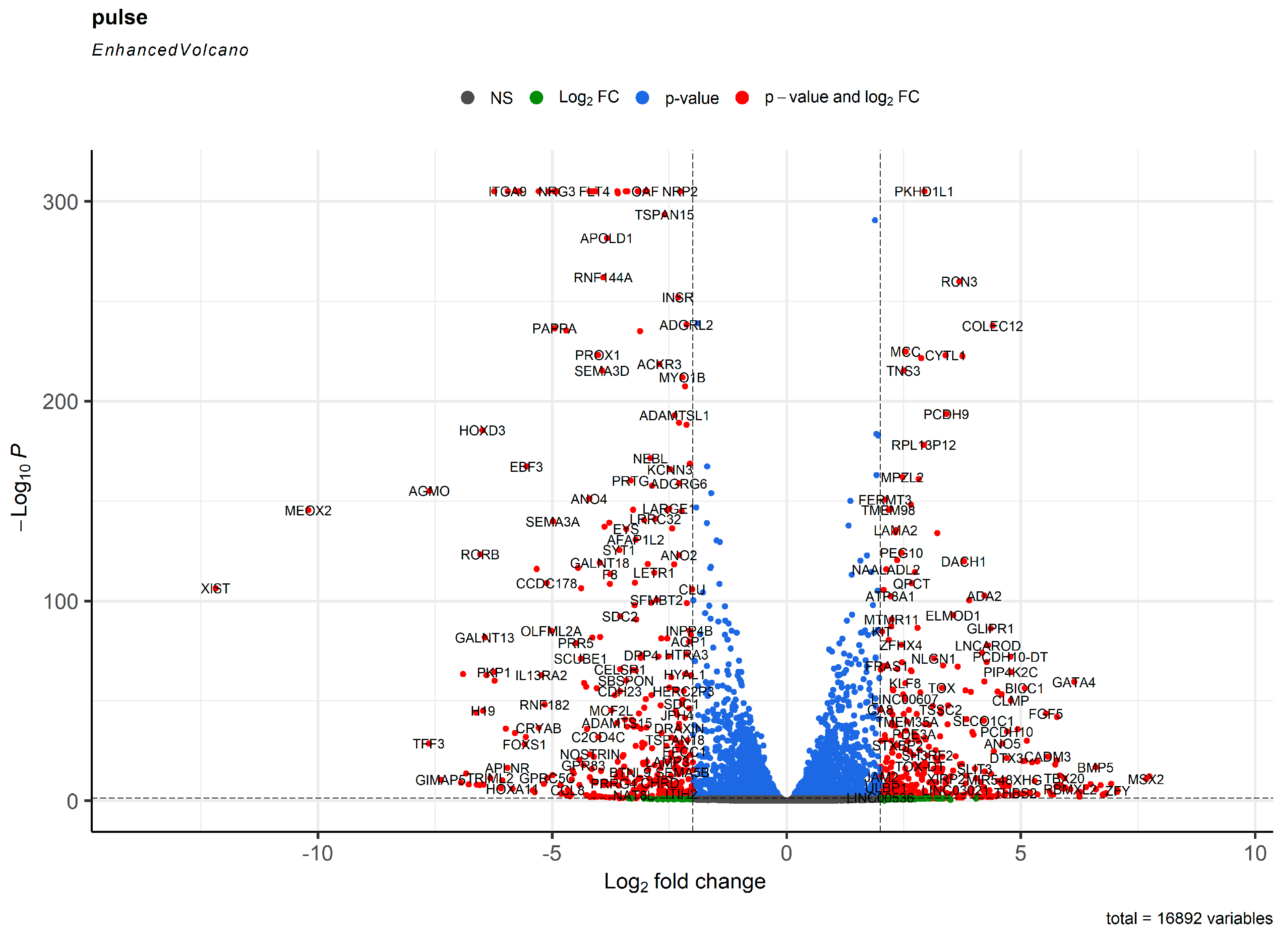
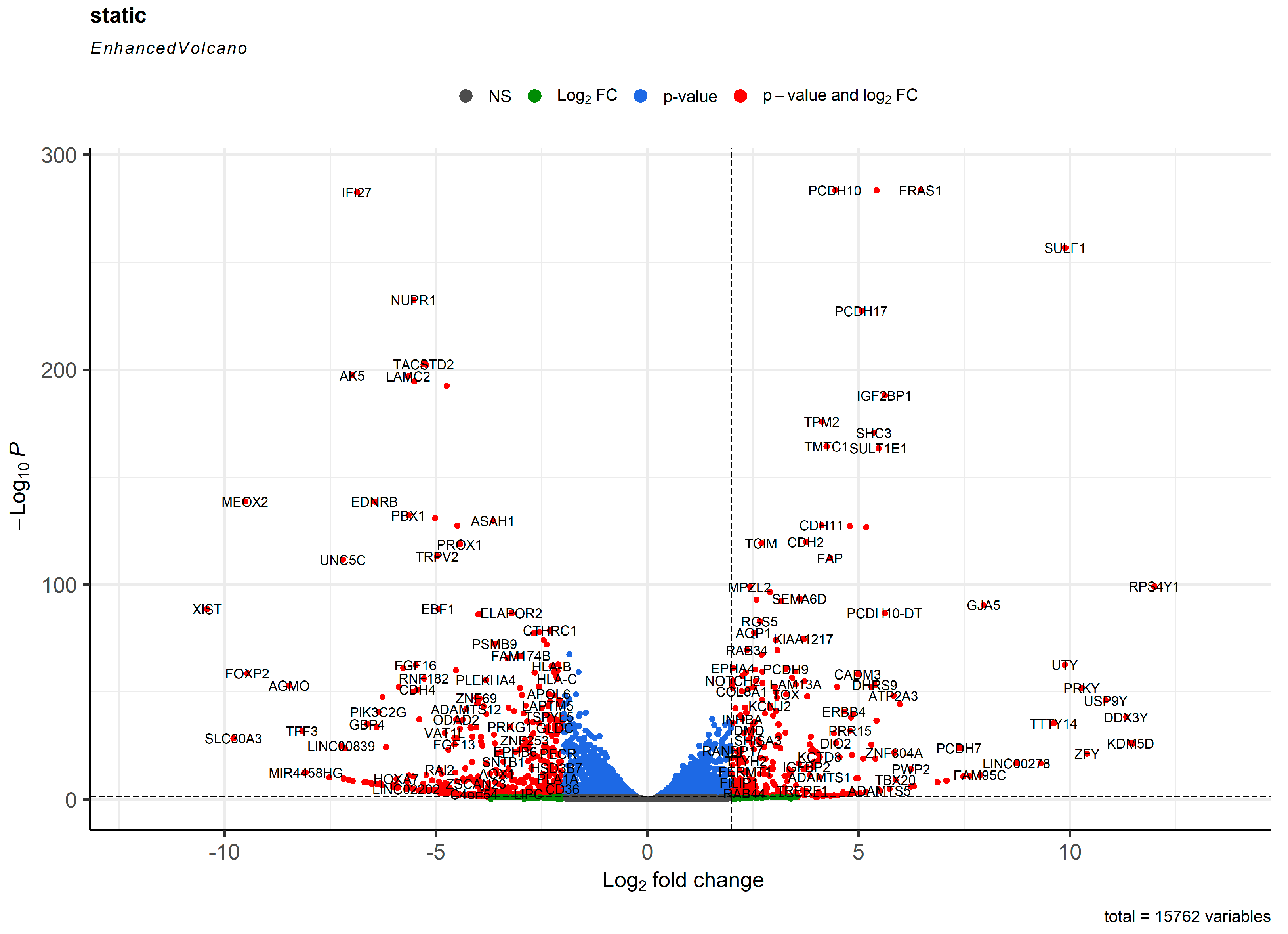
2. Results
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Whole-Transcriptome Analysis
4.3. Proteomic Profiling
4.4. Bioinformatic Analysis of the HCAEC/HITAEC Interactome
4.5. RT-qPCR Analysis
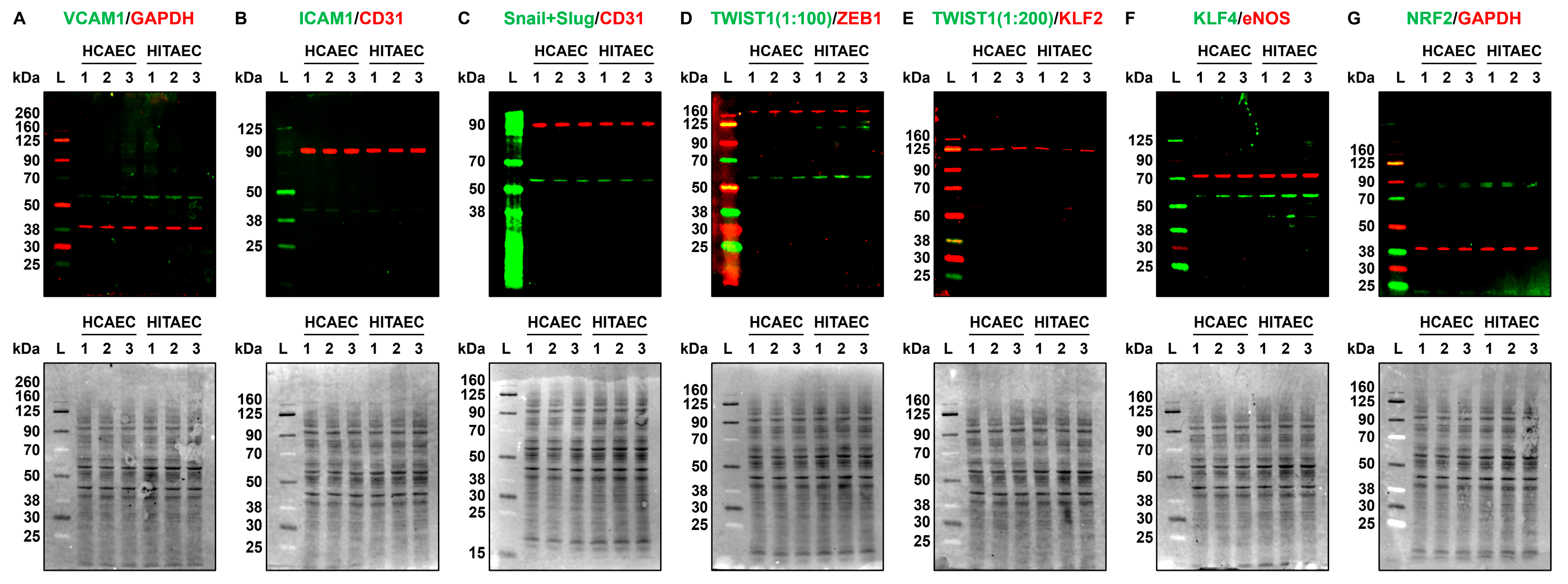
4.6. Western Blotting
4.7. Analysis of Bypass Dysfunctions after CABG Surgery at 10 Years of Follow-Up
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Rafii, S.; Butler, J.M.; Ding, B.S. Angiocrine Functions of Organ-Specific Endothelial Cells. Nature 2016, 529, 316–325. [Google Scholar] [CrossRef]
- Pasquier, J.; Ghiabi, P.; Chouchane, L.; Razzouk, K.; Rafii, S.; Rafii, A. Angiocrine Endothelium: From Physiology to Cancer. J. Transl. Med. 2020, 18, 52. [Google Scholar] [CrossRef] [PubMed]
- Gomez-Salinero, J.M.; Itkin, T.; Rafii, S. Developmental Angiocrine Diversification of Endothelial Cells for Organotypic Regeneration. Dev. Cell 2021, 56, 3042–3051. [Google Scholar] [CrossRef] [PubMed]
- Kutikhin, A.G.; Shishkova, D.K.; Velikanova, E.A.; Sinitsky, M.Y.; Sinitskaya, A.V.; Markova, V.E. Endothelial Dysfunction in the Context of Blood-Brain Barrier Modeling. J. Evol. Biochem. Physiol. 2022, 58, 781–806. [Google Scholar] [CrossRef] [PubMed]
- Bishop, D.; Schwarz, Q.; Wiszniak, S. Endothelial-Derived Angiocrine Factors as Instructors of Embryonic Development. Front. Cell Dev. Biol. 2023, 11, 1172114. [Google Scholar] [CrossRef]
- Feng, W.; Chen, L.; Nguyen, P.K.; Wu, S.M.; Li, G. Single Cell Analysis of Endothelial Cells Identified Organ-Specific Molecular Signatures and Heart-Specific Cell Populations and Molecular Features. Front. Cardiovasc. Med. 2019, 6, 165. [Google Scholar] [CrossRef]
- Paik, D.T.; Tian, L.; Williams, I.M.; Rhee, S.; Zhang, H.; Liu, C.; Mishra, R.; Wu, S.M.; Red-Horse, K.; Wu, J.C. Single-Cell RNA Sequencing Unveils Unique Transcriptomic Signatures of Organ-Specific Endothelial Cells. Circulation 2020, 142, 1848–1862. [Google Scholar] [CrossRef]
- Stone, O.A.; Zhou, B.; Red-Horse, K.; Stainier, D.Y.R. Endothelial Ontogeny and the Establishment of Vascular Heterogeneity. Bioessays 2021, 43, e2100036. [Google Scholar] [CrossRef]
- Jambusaria, A.; Hong, Z.; Zhang, L.; Srivastava, S.; Jana, A.; Toth, P.T.; Dai, Y.; Malik, A.B.; Rehman, J. Endothelial Heterogeneity across Distinct Vascular Beds during Homeostasis and Inflammation. Elife 2020, 9, e51413. [Google Scholar] [CrossRef]
- Dejana, E.; Hirschi, K.K.; Simons, M. The Molecular Basis of Endothelial Cell Plasticity. Nat. Commun. 2017, 8, 14361. [Google Scholar] [CrossRef]
- Jakab, M.; Augustin, H.G. Understanding Angiodiversity: Insights from Single Cell Biology. Development 2020, 147, dev146621. [Google Scholar] [CrossRef] [PubMed]
- Greenspan, L.J.; Weinstein, B.M. To Be or Not to Be: Endothelial Cell Plasticity in Development, Repair, and Disease. Angiogenesis 2021, 24, 251–269. [Google Scholar] [CrossRef]
- Naiche, L.A.; Villa, S.R.; Kitajewski, J.K. Endothelial Cell Fate Determination: A Top Notch Job in Vascular Decision-Making. Cold Spring Harb. Perspect. Med. 2022, 12, a041183. [Google Scholar] [CrossRef]
- Becker, L.M.; Chen, S.H.; Rodor, J.; de Rooij, L.P.M.H.; Baker, A.H.; Carmeliet, P. Deciphering Endothelial Heterogeneity in Health and Disease at Single-Cell Resolution: Progress and Perspectives. Cardiovasc. Res. 2023, 119, 6–27. [Google Scholar] [CrossRef]
- Trimm, E.; Red-Horse, K. Vascular Endothelial Cell Development and Diversity. Nat. Rev. Cardiol. 2023, 20, 197–210. [Google Scholar] [CrossRef] [PubMed]
- Norrmén, C.; Tammela, T.; Petrova, T.V.; Alitalo, K. Biological Basis of Therapeutic Lymphangiogenesis. Circulation 2011, 123, 1335–1351. [Google Scholar] [CrossRef]
- Campbell, K.T.; Silva, E.A. Biomaterial-Based Strategies for Engineering New Lymphatic Vasculature. Adv. Healthc. Mater. 2020, 9, e2000895. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, Y.; Che, Y.; Murohara, T. Therapeutic Lymphangiogenesis Is a Promising Strategy for Secondary Lymphedema. Int. J. Mol. Sci. 2023, 24, 7774. [Google Scholar] [CrossRef]
- Elliott, M.B.; Ginn, B.; Fukunishi, T.; Bedja, D.; Suresh, A.; Chen, T.; Inoue, T.; Dietz, H.C.; Santhanam, L.; Mao, H.Q.; et al. Regenerative and Durable Small-Diameter Graft as an Arterial Conduit. Proc. Natl. Acad. Sci. USA 2019, 116, 12710–12719. [Google Scholar] [CrossRef]
- Kutikhin, A.G.; Tupikin, A.E.; Matveeva, V.G.; Shishkova, D.K.; Antonova, L.V.; Kabilov, M.R.; Velikanova, E.A. Human Peripheral Blood-Derived Endothelial Colony-Forming Cells Are Highly Similar to Mature Vascular Endothelial Cells yet Demonstrate a Transitional Transcriptomic Signature. Cells 2020, 9, 876. [Google Scholar] [CrossRef]
- Seiffert, N.; Tang, P.; Keshi, E.; Reutzel-Selke, A.; Moosburner, S.; Everwien, H.; Wulsten, D.; Napierala, H.; Pratschke, J.; Sauer, I.M.; et al. In Vitro Recellularization of Decellularized Bovine Carotid Arteries Using Human Endothelial Colony Forming Cells. J. Biol. Eng. 2021, 15, 15. [Google Scholar] [CrossRef]
- Velikanova, E.A.; Sinitsky, M.Y.; Sinitskaya, A.V.; Matveeva, V.G.; Khanova, M.Y.; Antonova, L.V. Evaluation of the Feasibility of Endothelial Colony-Forming Cells to Develop Tissue-Engineered Vascular Grafts Based on the Gene Expression Profile Analysis. Sovrem. Tekhnol. Med. 2022, 14, 15–19. [Google Scholar] [CrossRef] [PubMed]
- Kraus, X.; van de Flierdt, E.; Renzelmann, J.; Thoms, S.; Witt, M.; Scheper, T.; Blume, C. Peripheral Blood Derived Endothelial Colony Forming Cells as Suitable Cell Source for Pre-Endothelialization of Arterial Vascular Grafts under Dynamic Flow Conditions. Microvasc. Res. 2022, 143, 104402. [Google Scholar] [CrossRef] [PubMed]
- Melly, L.; Torregrossa, G.; Lee, T.; Jansens, J.L.; Puskas, J.D. Fifty Years of Coronary Artery Bypass Grafting. J. Thorac. Dis. 2018, 10, 1960–1967. [Google Scholar] [CrossRef] [PubMed]
- Squiers, J.J.; Mack, M.J. Coronary Artery Bypass Grafting-Fifty Years of Quality Initiatives Since Favaloro. Ann. Cardiothorac. Surg. 2018, 7, 516–520. [Google Scholar] [CrossRef] [PubMed]
- Beerkens, F.J.; Claessen, B.E.; Mahan, M.; Gaudino, M.F.L.; Tam, D.Y.; Henriques, J.P.S.; Mehran, R.; Dangas, G.D. Contemporary Coronary Artery Bypass Graft Surgery and Subsequent Percutaneous Revascularization. Nat. Rev. Cardiol. 2022, 19, 195–208. [Google Scholar] [CrossRef]
- Welt, F.G.P. CABG versus PCI End of the Debate? N. Engl. J. Med. 2022, 386, 185–187. [Google Scholar] [CrossRef]
- Serruys, P.W.; Morice, M.C.; Kappetein, A.P.; Colombo, A.; Holmes, D.R.; Mack, M.J.; Ståhle, E.; Feldman, T.E.; van den Brand, M.; Bass, E.J.; et al. Percutaneous Coronary Intervention versus Coronary-Artery Bypass Grafting for Severe Coronary Artery Disease. N. Engl. J. Med. 2009, 360, 961–972. [Google Scholar] [CrossRef]
- Mohr, F.W.; Morice, M.C.; Kappetein, A.P.; Feldman, T.E.; Ståhle, E.; Colombo, A.; Mack, M.J.; Holmes, D.R.; Morel, M.A.; Van Dyck, N.; et al. Coronary Artery Bypass Graft Surgery versus Percutaneous Coronary Intervention in Patients with Three-Vessel Disease and Left Main Coronary Disease: 5-Year Follow-Up of the Randomised, Clinical SYNTAX Trial. Lancet 2013, 381, 629–638. [Google Scholar] [CrossRef]
- Head, S.J.; Milojevic, M.; Daemen, J.; Ahn, J.M.; Boersma, E.; Christiansen, E.H.; Domanski, M.J.; Farkouh, M.E.; Flather, M.; Fuster, V.; et al. Mortality after Coronary Artery Bypass Grafting versus Percutaneous Coronary Intervention with Stenting for Coronary Artery Disease: A Pooled Analysis of Individual Patient Data. Lancet 2018, 391, 939–948. [Google Scholar] [CrossRef]
- Thuijs, D.J.F.M.; Kappetein, A.P.; Serruys, P.W.; Mohr, F.W.; Morice, M.C.; Mack, M.J.; Holmes, D.R., Jr.; Curzen, N.; Davierwala, P.; Noack, T.; et al. Percutaneous coronary intervention versus coronary artery bypass grafting in patients with three-vessel or left main coronary artery disease: 10-year follow-up of the multicentre randomised controlled SYNTAX trial. Lancet 2019, 394, 1325–1334. [Google Scholar] [CrossRef] [PubMed]
- Gaudino, M.; Hameed, I.; Farkouh, M.E.; Rahouma, M.; Naik, A.; Robinson, N.B.; Ruan, Y.; Demetres, M.; Biondi-Zoccai, G.; Angiolillo, D.J.; et al. Overall and Cause-Specific Mortality in Randomized Clinical Trials Comparing Percutaneous Interventions With Coronary Bypass Surgery: A Meta-analysis. JAMA Intern. Med. 2020, 180, 1638–1646. [Google Scholar] [CrossRef] [PubMed]
- Taggart, D.P.; Altman, D.G.; Flather, M.; Gerry, S.; Gray, A.; Lees, B.; Benedetto, U.; ART (Arterial Revascularization Trial) Investigators. Associations Between Adding a Radial Artery Graft to Single and Bilateral Internal Thoracic Artery Grafts and Outcomes: Insights From the Arterial Revascularization Trial. Circulation 2017, 136, 454–463. [Google Scholar] [CrossRef] [PubMed]
- Rocha, R.V.; Tam, D.Y.; Karkhanis, R.; Nedadur, R.; Fang, J.; Tu, J.V.; Gaudino, M.; Royse, A.; Fremes, S.E. Multiple Arterial Grafting Is Associated With Better Outcomes for Coronary Artery Bypass Grafting Patients. Circulation 2018, 138, 2081–2090. [Google Scholar] [CrossRef] [PubMed]
- Gaudino, M.; Rahouma, M.; Abouarab, A.; Tam, D.Y.; Di Franco, A.; Leonard, J.; Benedetto, U.; Iannaccone, M.; D’Ascenzo, F.; Biondi-Zoccai, G.; et al. Meta-analysis Comparing Outcomes of Drug Eluting Stents Versus Single and Multiarterial Coronary Artery Bypass Grafting. Am. J. Cardiol. 2018, 122, 2018–2025. [Google Scholar] [CrossRef] [PubMed]
- Samadashvili, Z.; Sundt, T.M., 3rd; Wechsler, A.; Chikwe, J.; Adams, D.H.; Smith, C.R.; Jordan, D.; Girardi, L.; Lahey, S.J.; Gold, J.P.; et al. Multiple Versus Single Arterial Coronary Bypass Graft Surgery for Multivessel Disease. J. Am. Coll. Cardiol. 2019, 74, 1275–1285. [Google Scholar] [CrossRef]
- Gaudino, M.; Lorusso, R.; Rahouma, M.; Abouarab, A.; Tam, D.Y.; Spadaccio, C.; Saint-Hilary, G.; Leonard, J.; Iannaccone, M.; D’Ascenzo, F.; et al. Radial Artery Versus Right Internal Thoracic Artery Versus Saphenous Vein as the Second Conduit for Coronary Artery Bypass Surgery: A Network Meta-Analysis of Clinical Outcomes. J. Am. Heart Assoc. 2019, 8, e010839. [Google Scholar] [CrossRef]
- Taggart, D.P.; Audisio, K.; Gerry, S.; Robinson, N.B.; Rahouma, M.; Soletti, G.J.; Cancelli, G.; Benedetto, U.; Lees, B.; Gray, A.; et al. Effect of Total Arterial Grafting in the Arterial Revascularization Trial. J. Thorac. Cardiovasc. Surg. 2022, 163, 1002–1009.e6. [Google Scholar] [CrossRef]
- Saraiva, F.A.; Leite-Moreira, J.P.; Barros, A.S.; Lourenço, A.P.; Benedetto, U.; Leite-Moreira, A.F. Multiple versus Single Arterial Grafting in Coronary Artery Bypass Grafting: A Meta-Analysis of Randomized Controlled Trials and Propensity Score Studies. Int. J. Cardiol. 2020, 320, 55–63. [Google Scholar] [CrossRef]
- Gaudino, M.; Di Franco, A.; Flather, M.; Gerry, S.; Bagiella, E.; Gray, A.; Pearcey, L.; Saw, T.H.; Lees, B.; Benedetto, U.; et al. Association of Age with 10-Year Outcomes After Coronary Surgery in the Arterial Revascularization Trial. J. Am. Coll. Cardiol. 2021, 77, 18–26. [Google Scholar] [CrossRef]
- Saraiva, F.A.; Moreira, R.; Cerqueira, R.J.; Mancio, J.; Barros, A.S.; Lourenço, A.P.; Leite-Moreira, A.F. Multiple versus single arterial grafting in the elderly: A meta-analysis of randomized controlled trials and propensity score studies. J. Cardiovasc. Surg. 2022, 63, 169–178. [Google Scholar] [CrossRef] [PubMed]
- Taggart, D.P.; Audisio, K.; Gerry, S.; Robinson, N.B.; Rahouma, M.; Soletti, G.J.; Cancelli, G.; Benedetto, U.; Lees, B.; Gray, A.; et al. Single versus multiple arterial grafting in diabetic patients at 10 years: The Arterial Revascularization Trial. Eur. Heart J. 2022, 43, 4644–4652. [Google Scholar] [CrossRef] [PubMed]
- Thuijs, D.J.F.M.; Davierwala, P.; Milojevic, M.; Deo, S.V.; Noack, T.; Kappetein, A.P.; Serruys, P.W.; Mohr, F.W.; Morice, M.C.; Mack, M.J.; et al. Long-term survival after coronary bypass surgery with multiple versus single arterial grafts. Eur. J. Cardiothorac. Surg. 2022, 61, 925–933. [Google Scholar] [CrossRef]
- Davierwala, P.M.; Gao, C.; Thuijs, D.J.F.M.; Wang, R.; Hara, H.; Ono, M.; Noack, T.; Garg, S.; O’leary, N.; Milojevic, M.; et al. Single or multiple arterial bypass graft surgery vs. percutaneous coronary intervention in patients with three-vessel or left main coronary artery disease. Eur. Heart J. 2022, 43, 1334–1344. [Google Scholar] [CrossRef]
- Taggart, D.P.; Altman, D.G.; Gray, A.M.; Lees, B.; Gerry, S.; Benedetto, U.; Flather, M.; ART Investigators. Randomized Trial of Bilateral versus Single Internal-Thoracic-Artery Grafts. N. Engl. J. Med. 2016, 375, 2540–2549. [Google Scholar] [CrossRef] [PubMed]
- Rubino, A.S.; Gatti, G.; Reichart, D.; Tauriainen, T.; De Feo, M.; Onorati, F.; Pappalardo, A.; Chocron, S.; Gulbins, H.; Dalén, M.; et al. Early Outcome of Bilateral Versus Single Internal Mammary Artery Grafting in the Elderly. Ann. Thorac. Surg. 2018, 105, 1717–1723. [Google Scholar] [CrossRef] [PubMed]
- Iribarne, A.; Schmoker, J.D.; Malenka, D.J.; Leavitt, B.J.; McCullough, J.N.; Weldner, P.W.; DeSimone, J.P.; Westbrook, B.M.; Quinn, R.D.; Klemperer, J.D.; et al. Does Use of Bilateral Internal Mammary Artery Grafting Reduce Long-Term Risk of Repeat Coronary Revascularization? A Multicenter Analysis. Circulation 2017, 136, 1676–1685. [Google Scholar] [CrossRef]
- Buttar, S.N.; Yan, T.D.; Taggart, D.P.; Tian, D.H. Long-term and short-term outcomes of using bilateral internal mammary artery grafting versus left internal mammary artery grafting: A meta-analysis. Heart 2017, 103, 1419–1426. [Google Scholar] [CrossRef]
- Little, M.; Gray, A.M.; Altman, D.G.; Benedetto, U.; Flather, M.; Gerry, S.; Lees, B.; Murphy, J.; Gaudino, M.; Taggart, D.P.; et al. Cost-effectiveness of bilateral vs. single internal thoracic artery grafts at 10 years. Eur. Heart J. Qual. Care Clin. Outcomes 2022, 8, 324–332. [Google Scholar] [CrossRef]
- Taggart, D.P.; Benedetto, U.; Gerry, S.; Altman, D.G.; Gray, A.M.; Lees, B.; Gaudino, M.; Zamvar, V.; Bochenek, A.; Buxton, B.; et al. Bilateral versus Single Internal-Thoracic-Artery Grafts at 10 Years. N. Engl. J. Med. 2019, 380, 437–446. [Google Scholar] [CrossRef]
- Pevni, D.; Nesher, N.; Kramer, A.; Paz, Y.; Farkash, A.; Ben-Gal, Y. Does bilateral versus single thoracic artery grafting provide survival benefit in female patients? Interact. Cardiovasc. Thorac. Surg. 2019, 28, 860–867. [Google Scholar] [CrossRef] [PubMed]
- Gaudino, M.; Alexander, J.H.; Bakaeen, F.G.; Ballman, K.; Barili, F.; Calafiore, A.M.; Davierwala, P.; Goldman, S.; Kappetein, P.; Lorusso, R.; et al. Randomized comparison of the clinical outcome of single versus multiple arterial grafts: The ROMA trial-rationale and study protocol. Eur. J. Cardiothorac. Surg. 2017, 52, 1031–1040. [Google Scholar] [CrossRef] [PubMed]
- Gaudino, M.F.L.; Taggart, D.P.; Fremes, S.E. The ROMA trial: Why it is needed. Curr. Opin. Cardiol. 2018, 33, 622–626. [Google Scholar] [CrossRef] [PubMed]
- Gaudino, M.; Bakaeen, F.G.; Benedetto, U.; Di Franco, A.; Fremes, S.; Glineur, D.; Girardi, L.N.; Grau, J.; Puskas, J.D.; Ruel, M.; et al. Arterial Grafts for Coronary Bypass: A Critical Review After the Publication of ART and RADIAL. Circulation 2019, 140, 1273–1284. [Google Scholar] [CrossRef] [PubMed]
- Gaudino, M.; Audisio, K.; Rahouma, M.; Chadow, D.; Cancelli, G.; Soletti, G.J.; Gray, A.; Lees, B.; Gerry, S.; Benedetto, U.; et al. Comparison of Long-term Clinical Outcomes of Skeletonized vs Pedicled Internal Thoracic Artery Harvesting Techniques in the Arterial Revascularization Trial. JAMA Cardiol. 2021, 6, 1380–1386. [Google Scholar] [CrossRef]
- Kusu-Orkar, T.E.; Kermali, M.; Masharani, K.; Noshirwani, A.; MacCarthy-Ofosu, B.; Oguamanam, N.; Bin Saeid, J.; Muir, A.D.; Harky, A. Skeletonized or Pedicled Harvesting of Left Internal Mammary Artery: A Systematic Review and Meta-analysis. Semin. Thorac. Cardiovasc. Surg. 2021, 33, 10–18. [Google Scholar] [CrossRef]
- Iddawela, S.; Mellor, S.L.; Zahra, S.A.; Khare, Y.; Harky, A. Pedicled or skeletonized bilateral internal mammary artery harvesting a meta-analysis and trial sequential analysis. Expert Rev. Cardiovasc. Ther. 2021, 19, 647–654. [Google Scholar] [CrossRef]
- Dreifaldt, M.; Samano, N.; Geijer, H.; Lidén, M.; Bodin, L.; Souza, D. Pedicled versus skeletonized internal thoracic artery grafts: A randomized trial. Asian Cardiovasc. Thorac. Ann. 2021, 29, 490–497. [Google Scholar] [CrossRef]
- Sá, M.P.; Cavalcanti, P.E.; de Andrade Costa Santos, H.J.; Soares, A.F.; Albuquerque Miranda, R.G.; Araújo, M.L.; Lima, R.C. Skeletonized versus pedicled bilateral internal mammary artery grafting: Outcomes and concerns analyzed through a meta-analytical approach. Int. J. Surg. 2015, 16, 146–152. [Google Scholar] [CrossRef]
- Otsuka, F.; Yahagi, K.; Sakakura, K.; Virmani, R. Why is the mammary artery so special and what protects it from atherosclerosis? Ann. Cardiothorac. Surg. 2013, 2, 519–526. [Google Scholar] [CrossRef]
- Martínez-González, B.; Reyes-Hernández, C.G.; Quiroga-Garza, A.; Rodríguez-Rodríguez, V.E.; Esparza-Hernández, C.N.; Elizondo-Omaña, R.E.; Guzmán-López, S. Conduits Used in Coronary Artery Bypass Grafting: A Review of Morphological Studies. Ann. Thorac. Cardiovasc. Surg. 2017, 23, 55–65. [Google Scholar] [CrossRef] [PubMed]
- Gharibeh, L.; Ferrari, G.; Ouimet, M.; Grau, J.B. Conduits’ Biology Regulates the Outcomes of Coronary Artery Bypass Grafting. JACC Basic Transl. Sci. 2021, 6, 388–396. [Google Scholar] [CrossRef] [PubMed]
- Shadrin, I.Y.; Holmes, D.R.; Behfar, A. Left Internal Mammary Artery as an Endocrine Organ: Insights Into Graft Biology and Long-term Impact Following Coronary Artery Bypass Grafting. Mayo Clin. Proc. 2023, 98, 150–162. [Google Scholar] [CrossRef] [PubMed]
- Canver, C.C.; Ricotta, J.J.; Bhayana, J.N.; Fiedler, R.C.; Mentzer, R.M., Jr. Use of duplex imaging to assess suitability of the internal mammary artery for coronary artery surgery. J. Vasc. Surg. 1991, 13, 294–300. [Google Scholar] [CrossRef]
- Dodge, J.T., Jr.; Brown, B.G.; Bolson, E.L.; Dodge, H.T. Lumen diameter of normal human coronary arteries. Influence of age, sex, anatomic variation, and left ventricular hypertrophy or dilation. Circulation 1992, 86, 232–246. [Google Scholar] [CrossRef]
- Waller, B.F.; Orr, C.M.; Slack, J.D.; Pinkerton, C.A.; Van Tassel, J.; Peters, T. Anatomy, histology, and pathology of coronary arteries: A review relevant to new interventional and imaging techniques—Part I. Clin. Cardiol. 1992, 15, 451–457. [Google Scholar] [CrossRef]
- Canham, P.B.; Finlay, H.M.; Boughner, D.R. Contrasting structure of the saphenous vein and internal mammary artery used as coronary bypass vessels. Cardiovasc. Res. 1997, 34, 557–567. [Google Scholar] [CrossRef]
- Jung, Y.; Ahn, B.H.; Kim, G.S.; Jeong, I.S.; Lee, K.S.; Song, S.Y.; Na, K.J.; Oh, S.G. Change in luminal diameter of the left internal thoracic artery anastomosed to the totally occluded left anterior descending coronary artery. J. Cardiothorac. Surg. 2016, 11, 157. [Google Scholar] [CrossRef]
- Gopal, D.; Singh, N.G.; Jagadeesh, A.M.; Ture, A.; Thimmarayappa, A. Comparison of left internal mammary artery diameter before and after left stellate ganglion block. Ann. Card Anaesth. 2013, 16, 238–242. [Google Scholar] [CrossRef]
- Ylä-Herttuala, S.; Sumuvuori, H.; Karkola, K.; Möttönen, M.; Nikkari, T. Glycosaminoglycans in normal and atherosclerotic human coronary arteries. Lab Invest. 1986, 54, 402–407. [Google Scholar]
- Sisto, T.; Ylä-Herttuala, S.; Luoma, J.; Riekkinen, H.; Nikkari, T. Biochemical composition of human internal mammary artery and saphenous vein. J. Vasc. Surg. 1990, 11, 418–422. [Google Scholar] [CrossRef] [PubMed]
- Merrilees, M.J.; Beaumont, B.; Scott, L.J. Comparison of deposits of versican, biglycan and decorin in saphenous vein and internal thoracic, radial and coronary arteries: Correlation to patency. Coron. Artery Dis. 2001, 12, 7–16. [Google Scholar] [CrossRef]
- Anstadt, M.P.; Franga, D.L.; Portik-Dobos, V.; Pennathur, A.; Bannan, M.; Mawulawde, K.; Ergul, A. Native matrix metalloproteinase characteristics may influence early stenosis of venous versus arterial coronary artery bypass grafting conduits. Chest 2004, 125, 1853–1858. [Google Scholar] [CrossRef] [PubMed]
- Predel, H.G.; Yang, Z.; von Segesser, L.; Turina, M.; Bühler, F.R.; Lüscher, T.F. Implications of pulsatile stretch on growth of saphenous vein and mammary artery smooth muscle. Lancet 1992, 340, 878–879. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Oemar, B.S.; Carrel, T.; Kipfer, B.; Julmy, F.; Lüscher, T.F. Different proliferative properties of smooth muscle cells of human arterial and venous bypass vessels: Role of PDGF receptors, mitogen-activated protein kinase, and cyclin-dependent kinase inhibitors. Circulation 1998, 97, 181–187. [Google Scholar] [CrossRef]
- Del Rizzo, D.F.; Yurkova, N.; Moon, M.C.; Litchie, B.; Zahradka, P. Platelet-derived growth factor-induced expression of c-fos in human vascular smooth muscle cells: Implications for long-term graft patency. Ann. Thorac. Surg. 2002, 74, 90–95. [Google Scholar] [CrossRef] [PubMed]
- Friedl, R.; Li, J.; Schumacher, B.; Hanke, H.; Waltenberger, J.; Hannekum, A.; Stracke, S. Intimal hyperplasia and expression of transforming growth factor-beta1 in saphenous veins and internal mammary arteries before coronary artery surgery. Ann. Thorac. Surg. 2004, 78, 1312–1318. [Google Scholar] [CrossRef]
- Hata, J.A.; Petrofski, J.A.; Schroder, J.N.; Williams, M.L.; Timberlake, S.H.; Pippen, A.; Corwin, M.T.; Solan, A.K.; Jakoi, A.; Gehrig, T.R.; et al. Modulation of phosphatidylinositol 3-kinase signaling reduces intimal hyperplasia in aortocoronary saphenous vein grafts. J. Thorac. Cardiovasc. Surg. 2005, 129, 1405–1413. [Google Scholar] [CrossRef]
- Frischknecht, K.; Greutert, H.; Weisshaupt, C.; Kaspar, M.; Yang, Z.; Luscher, T.F.; Carrel, T.P.; Tanner, F.C. Different vascular smooth muscle cell apoptosis in the human internal mammary artery and the saphenous vein. Implications for bypass graft disease. J. Vasc. Res. 2006, 43, 338–346. [Google Scholar] [CrossRef]
- Mekontso-Dessap, A.; Kirsch, M.; Guignambert, C.; Zadigue, P.; Adnot, S.; Loisance, D.; Eddahibi, S. Vascular-wall remodeling of 3 human bypass vessels: Organ culture and smooth muscle cell properties. J. Thorac. Cardiovasc. Surg. 2006, 131, 651–658. [Google Scholar] [CrossRef]
- Turner, N.A.; Ho, S.; Warburton, P.; O’Regan, D.J.; Porter, K.E. Smooth muscle cells cultured from human saphenous vein exhibit increased proliferation, invasion, and mitogen-activated protein kinase activation in vitro compared with paired internal mammary artery cells. J. Vasc. Surg. 2007, 45, 1022–1028. [Google Scholar] [CrossRef] [PubMed]
- Mitra, A.K.; Jia, G.; Gangahar, D.M.; Agrawal, D.K. Temporal PTEN inactivation causes proliferation of saphenous vein smooth muscle cells of human CABG conduits. J. Cell. Mol. Med. 2009, 13, 177–187. [Google Scholar] [CrossRef] [PubMed]
- Zhu, T.X.; Lan, B.; Meng, L.Y.; Yang, Y.L.; Li, R.X.; Li, E.M.; Zheng, S.Y.; Xu, L.Y. ECM-related gene expression profile in vascular smooth muscle cells from human saphenous vein and internal thoracic artery. J. Cardiothorac. Surg. 2013, 8, 155. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.H.; Diederich, D.; Schneider, K.; Siebenmann, R.; Stulz, P.; von Segesser, L.; Turina, M.; Bühler, F.R.; Lüscher, T.F. Endothelium-derived relaxing factor and protection against contractions induced by histamine and serotonin in the human internal mammary artery and in the saphenous vein. Circulation 1989, 80, 1041–1048. [Google Scholar] [CrossRef]
- Yang, Z.H.; von Segesser, L.; Bauer, E.; Stulz, P.; Turina, M.; Lüscher, T.F. Different activation of the endothelial L-arginine and cyclooxygenase pathway in the human internal mammary artery and saphenous vein. Circ. Res. 1991, 68, 52–60. [Google Scholar] [CrossRef]
- Tadjkarimi, S.; O’Neil, G.S.; Luu, T.N.; Allen, S.P.; Schyns, C.J.; Chester, A.H.; Yacoub, M.H. Comparison of cyclic GMP in human internal mammary artery and saphenous vein: Implications for coronary artery bypass graft patency. Cardiovasc. Res. 1992, 26, 297–300. [Google Scholar] [CrossRef]
- Thorin-Trescases, N.; Hamilton, C.A.; Reid, J.L.; McPherson, K.L.; Jardine, E.; Berg, G.; Bohr, D.; Dominiczak, A.F. Inducible L-arginine/nitric oxide pathway in human internal mammary artery and saphenous vein. Am. J. Physiol. 1995, 268, H1122–H1132. [Google Scholar] [CrossRef]
- Liu, Z.G.; Ge, Z.D.; He, G.W. Difference in endothelium-derived hyperpolarizing factor-mediated hyperpolarization and nitric oxide release between human internal mammary artery and saphenous vein. Circulation 2000, 102, III296–III301. [Google Scholar] [CrossRef]
- Broeders, M.A.; Doevendans, P.A.; Maessen, J.G.; van Gorsel, E.; Egbrink, M.G.; Daemen, M.J.; Tangelder, G.J.; Reneman, R.S.; van der Zee, R. The human internal thoracic artery releases more nitric oxide in response to vascular endothelial growth factor than the human saphenous vein. J. Thorac. Cardiovasc. Surg. 2001, 122, 305–309. [Google Scholar] [CrossRef]
- Gaudino, M.; Toesca, A.; Maggiano, N.; Pragliola, C.; Possati, G. Localization of nitric oxide synthase type III in the internal thoracic and radial arteries and the great saphenous vein: A comparative immunohistochemical study. J. Thorac. Cardiovasc. Surg. 2003, 125, 1510–1515. [Google Scholar] [CrossRef]
- Shishkova, D.; Markova, V.; Sinitsky, M.; Tsepokina, A.; Frolov, A.; Zagorodnikov, N.; Bogdanov, L.; Kutikhin, A. Co-Culture of Primary Human Coronary Artery and Internal Thoracic Artery Endothelial Cells Results in Mutually Beneficial Paracrine Interactions. Int. J. Mol. Sci. 2020, 21, 8032. [Google Scholar] [CrossRef]
- Schmalfuss, C.M.; Chen, L.Y.; Bott, J.N.; Staples, E.D.; Mehta, J.L. Superoxide Anion Generation, Superoxide Dismutase Activity, and Nitric Oxide Release in Human Internal Mammary Artery and Saphenous Vein Segments. J. Cardiovasc. Pharmacol. Ther. 1999, 4, 249–257. [Google Scholar] [CrossRef] [PubMed]
- Shapira, O.M.; Xu, A.; Aldea, G.S.; Vita, J.A.; Shemin, R.J.; Keaney, J.F., Jr. Enhanced nitric oxide-mediated vascular relaxation in radial artery compared with internal mammary artery or saphenous vein. Circulation 1999, 100, II322–II327. [Google Scholar] [CrossRef] [PubMed]
- Mangoush, O.; Athanasiou, T.; Nakamura, K.; Johnson, P.; Smoienski, R.; Sarathchandra, P.; Oury, T.; Chester, A.H.; Amrani, M. Anti-oxidant properties of the internal thoracic artery and the radial artery. Heart Lung Circ. 2008, 17, 40–47. [Google Scholar] [CrossRef] [PubMed]
- Uydeş-Doğan, B.S.; Nebigil, M.; Aslamaci, S.; Onuk, E.; Kanzik, I.; Akar, F. The comparison of vascular reactivities of arterial and venous grafts to vasodilators: Management of graft spasm. Int. J. Cardiol. 1996, 53, 137–145. [Google Scholar] [CrossRef]
- Hammerer-Lercher, A.; Fersterer, J.; Holzmann, S.; Bonatti, J.; Ruttmann, E.; Hoefer, D.; Mair, J.; Puschendorf, B. Direct comparison of relaxation and cGMP production in human coronary bypass grafts in response to stimulation with natriuretic peptides and a nitric oxide donor. Clin. Sci. 2006, 111, 225–231. [Google Scholar] [CrossRef]
- Mirkhani, H.; Shafa, M.; Khazraei, H. Comparison of the effects of levosimendan and papaverine on human internal mammary artery and saphenous vein. Cardiovasc. Drugs Ther. 2009, 23, 355–359. [Google Scholar] [CrossRef]
- Nguyen, H.G.; Korach, A.; Collura, C.; Eskenazi, B.R.; Vita, J.A.; Shapira, O.M. Differential effects of natriuretic peptides on arterial and venous coronary artery bypass conduits. Ann. Thorac. Surg. 2009, 87, 748–756. [Google Scholar] [CrossRef]
- Nishioka, H.; Kitamura, S.; Kameda, Y.; Taniguchi, S.; Kawata, T.; Mizuguchi, K. Difference in acetylcholine-induced nitric oxide release of arterial and venous grafts in patients after coronary bypass operations. J. Thorac. Cardiovasc. Surg. 1998, 116, 454–459. [Google Scholar] [CrossRef]
- Augustin, H.G.; Koh, G.Y. Organotypic vasculature: From descriptive heterogeneity to functional pathophysiology. Science 2017, 357, eaal2379. [Google Scholar] [CrossRef]
- Payeli, S.K.; Latini, R.; Gebhard, C.; Patrignani, A.; Wagner, U.; Lüscher, T.F.; Tanner, F.C. Prothrombotic gene expression profile in vascular smooth muscle cells of human saphenous vein, but not internal mammary artery. Arterioscler. Thromb. Vasc. Biol. 2008, 28, 705–710. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Shi, G.P. Vascular wall extracellular matrix proteins and vascular diseases. Biochim. Biophys. Acta 2014, 1842, 2106–2119. [Google Scholar] [CrossRef] [PubMed]
- Del Monte-Nieto, G.; Fischer, J.W.; Gorski, D.J.; Harvey, R.P.; Kovacic, J.C. Basic Biology of Extracellular Matrix in the Cardiovascular System, Part 1/4: JACC Focus Seminar. J. Am. Coll. Cardiol. 2020, 75, 2169–2188. [Google Scholar] [CrossRef] [PubMed]
- Barallobre-Barreiro, J.; Loeys, B.; Mayr, M.; Rienks, M.; Verstraeten, A.; Kovacic, J.C. Extracellular Matrix in Vascular Disease, Part 2/4: JACC Focus Seminar. J. Am. Coll. Cardiol. 2020, 75, 2189–2203. [Google Scholar] [CrossRef] [PubMed]
- Cai, Z.; Gong, Z.; Li, Z.; Li, L.; Kong, W. Vascular Extracellular Matrix Remodeling and Hypertension. Antioxid. Redox Signal. 2021, 34, 765–783. [Google Scholar] [CrossRef]
- Mammoto, A.; Matus, K.; Mammoto, T. Extracellular Matrix in Aging Aorta. Front. Cell Dev. Biol. 2022, 10, 822561. [Google Scholar] [CrossRef]
- Komarova, Y.A.; Kruse, K.; Mehta, D.; Malik, A.B. Protein Interactions at Endothelial Junctions and Signaling Mechanisms Regulating Endothelial Permeability. Circ. Res. 2017, 120, 179–206. [Google Scholar] [CrossRef]
- Wettschureck, N.; Strilic, B.; Offermanns, S. Passing the Vascular Barrier: Endothelial Signaling Processes Controlling Extravasation. Physiol. Rev. 2019, 99, 1467–1525. [Google Scholar] [CrossRef]
- Duong, C.N.; Vestweber, D. Mechanisms Ensuring Endothelial Junction Integrity Beyond VE-Cadherin. Front. Physiol. 2020, 11, 519. [Google Scholar] [CrossRef]
- Claesson-Welsh, L.; Dejana, E.; McDonald, D.M. Permeability of the Endothelial Barrier: Identifying and Reconciling Controversies. Trends Mol. Med. 2021, 27, 314–331. [Google Scholar] [CrossRef]
- Arora, S.; Yim, E.K.F.; Toh, Y.C. Environmental Specification of Pluripotent Stem Cell Derived Endothelial Cells Toward Arterial and Venous Subtypes. Front. Bioeng. Biotechnol. 2019, 7, 143. [Google Scholar] [CrossRef] [PubMed]
- Gifre-Renom, L.; Daems, M.; Luttun, A.; Jones, E.A.V. Organ-Specific Endothelial Cell Differentiation and Impact of Microenvironmental Cues on Endothelial Heterogeneity. Int. J. Mol. Sci. 2022, 23, 1477. [Google Scholar] [CrossRef] [PubMed]
- Di Russo, J.; Luik, A.L.; Yousif, L.; Budny, S.; Oberleithner, H.; Hofschröer, V.; Klingauf, J.; van Bavel, E.; Bakker, E.N.; Hellstrand, P.; et al. Endothelial basement membrane laminin 511 is essential for shear stress response. EMBO J. 2017, 36, 183–201. [Google Scholar] [CrossRef]
- Yamamoto, K.; Nogimori, Y.; Imamura, H.; Ando, J. Shear stress activates mitochondrial oxidative phosphorylation by reducing plasma membrane cholesterol in vascular endothelial cells. Proc. Natl. Acad. Sci. USA 2020, 117, 33660–33667. [Google Scholar] [CrossRef] [PubMed]
- Moore, K.H.; Murphy, H.A.; George, E.M. The glycocalyx: A central regulator of vascular function. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2021, 320, R508–R518. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, A.; Tomita, H.; Okada, H. Form follows function: The endothelial glycocalyx. Transl. Res. 2022, 247, 158–167. [Google Scholar] [CrossRef]
- Villalba, N.; Baby, S.; Yuan, S.Y. The Endothelial Glycocalyx as a Double-Edged Sword in Microvascular Homeostasis and Pathogenesis. Front. Cell Dev. Biol. 2021, 9, 711003. [Google Scholar] [CrossRef]
- Hu, Z.; Cano, I.; D’Amore, P.A. Update on the Role of the Endothelial Glycocalyx in Angiogenesis and Vascular Inflammation. Front. Cell Dev. Biol. 2021, 9, 734276. [Google Scholar] [CrossRef]
- Antonova, L.; Kutikhin, A.; Sevostianova, V.; Lobov, A.; Repkin, E.; Krivkina, E.; Velikanova, E.; Mironov, A.; Mukhamadiyarov, R.; Senokosova, E.; et al. Controlled and Synchronised Vascular Regeneration upon the Implantation of Iloprost- and Cationic Amphiphilic Drugs-Conjugated Tissue-Engineered Vascular Grafts into the Ovine Carotid Artery: A Proteomics-Empowered Study. Polymers 2022, 14, 5149. [Google Scholar] [CrossRef]
- Antonova, L.V.; Sevostianova, V.V.; Silnikov, V.N.; Krivkina, E.O.; Velikanova, E.A.; Mironov, A.V.; Shabaev, A.R.; Senokosova, E.A.; Khanova, M.Y.; Glushkova, T.V.; et al. Comparison of the Patency and Regenerative Potential of Biodegradable Vascular Prostheses of Different Polymer Compositions in an Ovine Model. Int. J. Mol. Sci. 2023, 24, 8540. [Google Scholar] [CrossRef]
- Eelen, G.; de Zeeuw, P.; Treps, L.; Harjes, U.; Wong, B.W.; Carmeliet, P. Endothelial Cell Metabolism. Physiol. Rev. 2018, 98, 3–58. [Google Scholar] [CrossRef]
- Li, X.; Sun, X.; Carmeliet, P. Hallmarks of Endothelial Cell Metabolism in Health and Disease. Cell Metab. 2019, 30, 414–433. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Kumar, A.; Carmeliet, P. Metabolic Pathways Fueling the Endothelial Cell Drive. Annu. Rev. Physiol. 2019, 81, 483–503. [Google Scholar] [CrossRef] [PubMed]
- Peng, H.; Wang, X.; Du, J.; Cui, Q.; Huang, Y.; Jin, H. Metabolic Reprogramming of Vascular Endothelial Cells: Basic Research and Clinical Applications. Front. Cell Dev. Biol. 2021, 9, 626047. [Google Scholar] [CrossRef] [PubMed]
- Donato, A.J.; Machin, D.R.; Lesniewski, L.A. Mechanisms of Dysfunction in the Aging Vasculature and Role in Age-Related Disease. Circ. Res. 2018, 123, 825–848. [Google Scholar] [CrossRef] [PubMed]
- Ungvari, Z.; Tarantini, S.; Kiss, T.; Wren, J.D.; Giles, C.B.; Griffin, C.T.; Murfee, W.L.; Pacher, P.; Csiszar, A. Endothelial dysfunction and angiogenesis impairment in the ageing vasculature. Nat. Rev. Cardiol. 2018, 15, 555–565. [Google Scholar] [CrossRef]
- Ungvari, Z.; Tarantini, S.; Sorond, F.; Merkely, B.; Csiszar, A. Mechanisms of Vascular Aging, A Geroscience Perspective: JACC Focus Seminar. J. Am. Coll. Cardiol. 2020, 75, 931–941. [Google Scholar] [CrossRef]
- Sinitsky, M.Y.; Kutikhin, A.G.; Tsepokina, A.V.; Shishkova, D.K.; Asanov, M.A.; Yuzhalin, A.E.; Minina, V.I.; Ponasenko, A.V. Mitomycin C induced genotoxic stress in endothelial cells is associated with differential expression of proinflammatory cytokines. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 2020, 858–860, 503252. [Google Scholar] [CrossRef]
- Sinitsky, M.; Sinitskaya, A.; Shishkova, D.; Tupikin, A.; Asanov, M.; Khutornaya, M.; Kabilov, M.; Ponasenko, A. Identification of Key Genes and Pathways in Genotoxic Stress Induced Endothelial Dysfunction: Results of Whole Transcriptome Sequencing. Biomedicines 2022, 10, 2067. [Google Scholar] [CrossRef]
- Sinitsky, M.; Asanov, M.; Sinitskaya, A.; Shishkova, D.; Khutornaya, M.; Minina, V.; Ponasenko, A. Atorvastatin Can Modulate DNA Damage Repair in Endothelial Cells Exposed to Mitomycin C. Int. J. Mol. Sci. 2023, 24, 6783. [Google Scholar] [CrossRef]
- Bautista-Niño, P.K.; Portilla-Fernandez, E.; Rubio-Beltrán, E.; van der Linden, J.J.; de Vries, R.; van Veghel, R.; de Boer, M.; Durik, M.; Ridwan, Y.; Brandt, R.; et al. Local endothelial DNA repair deficiency causes aging-resembling endothelial-specific dysfunction. Clin. Sci. 2020, 134, 727–746. [Google Scholar] [CrossRef] [PubMed]
- Mameli, E.; Martello, A.; Caporali, A. Autophagy at the interface of endothelial cell homeostasis and vascular disease. FEBS J. 2022, 289, 2976–2991. [Google Scholar] [CrossRef] [PubMed]
- Pacinella, G.; Ciaccio, A.M.; Tuttolomondo, A. Endothelial Dysfunction and Chronic Inflammation: The Cornerstones of Vascular Alterations in Age-Related Diseases. Int. J. Mol. Sci. 2022, 23, 15722. [Google Scholar] [CrossRef] [PubMed]
- Marzoog, B.A. Endothelial cell autophagy in the context of disease development. Anat. Cell Biol. 2023, 56, 16–24. [Google Scholar] [CrossRef] [PubMed]
- Cai, J.; Culley, M.K.; Zhao, Y.; Zhao, J. The role of ubiquitination and deubiquitination in the regulation of cell junctions. Protein Cell 2018, 9, 754–769. [Google Scholar] [CrossRef]
- Totland, M.Z.; Rasmussen, N.L.; Knudsen, L.M.; Leithe, E. Regulation of gap junction intercellular communication by connexin ubiquitination: Physiological and pathophysiological implications. Cell Mol. Life Sci. 2020, 77, 573–591. [Google Scholar] [CrossRef] [PubMed]
- Yadid, M.; Lind, J.U.; Ardoña, H.A.M.; Sheehy, S.P.; Dickinson, L.E.; Eweje, F.; Bastings, M.M.C.; Pope, B.; O’Connor, B.B.; Straubhaar, J.R.; et al. Endothelial extracellular vesicles contain protective proteins and rescue ischemia-reperfusion injury in a human heart-on-chip. Sci. Transl. Med. 2020, 12, eaax8005. [Google Scholar] [CrossRef]
- Li, T.; Wang, B.; Ding, H.; Chen, S.; Cheng, W.; Li, Y.; Wu, X.; Wang, L.; Jiang, Y.; Lu, Z.; et al. Effect of Extracellular Vesicles From Multiple Cells on Vascular Smooth Muscle Cells in Atherosclerosis. Front. Pharmacol. 2022, 13, 857331. [Google Scholar] [CrossRef]
- He, Y.; Li, Q.; Feng, F.; Gao, R.; Li, H.; Chu, Y.; Li, S.; Wang, Y.; Mao, R.; Ji, Z.; et al. Extracellular vesicles produced by human-induced pluripotent stem cell-derived endothelial cells can prevent arterial stenosis in mice via autophagy regulation. Front. Cardiovasc. Med. 2022, 9, 922790. [Google Scholar] [CrossRef]
- Abel, F.; Murke, F.; Gaida, M.; Garnier, N.; Ochsenfarth, C.; Theiss, C.; Thielmann, M.; Kleinbongard, P.; Giebel, B.; Peters, J.; et al. Extracellular vesicles isolated from patients undergoing remote ischemic preconditioning decrease hypoxia-evoked apoptosis of cardiomyoblasts after isoflurane but not propofol exposure. PLoS ONE 2020, 15, e0228948. [Google Scholar] [CrossRef]
- Desgres, M.; Lima Correa, B.; Petrusca, L.; Autret, G.; Pezzana, C.; Marigny, C.; Guillas, C.; Bellamy, V.; Vilar, J.; Perier, M.C.; et al. Therapeutic potential of extracellular vesicles derived from cardiac progenitor cells in rodent models of chemotherapy-induced cardiomyopathy. Front. Cardiovasc. Med. 2023, 10, 1206279. [Google Scholar] [CrossRef]
- Alfì, E.; Thairi, C.; Femminò, S.; Alloatti, G.; Moccia, F.; Brizzi, M.F.; Pagliaro, P.; Penna, C. Extracellular vesicles (EVs) in ischemic conditioning and angiogenesis: Focus on endothelial derived EVs. Vascul. Pharmacol. 2021, 140, 106873. [Google Scholar] [CrossRef]
- Markova, V.; Bogdanov, L.; Velikanova, E.; Kanonykina, A.; Frolov, A.; Shishkova, D.; Lazebnaya, A.; Kutikhin, A. Endothelial Cell Markers Are Inferior to Vascular Smooth Muscle Cells Markers in Staining Vasa Vasorum and Are Non-Specific for Distinct Endothelial Cell Lineages in Clinical Samples. Int. J. Mol. Sci. 2023, 24, 1959. [Google Scholar] [CrossRef] [PubMed]
- Bogdanov, L.A.; Velikanova, E.A.; Kanonykina, A.Y.; Frolov, A.V.; Shishkova, D.K.; Lazebnaya, A.I.; Kutikhin, A.G. Vascular smooth muscle cell contractile proteins as universal markers of vessels of microcirculatory bed. Compl. Iss. Cardiovasc. Dis. 2022, 11, 162–176. [Google Scholar] [CrossRef]
- Shishkova, D.; Lobov, A.; Zainullina, B.; Matveeva, V.; Markova, V.; Sinitskaya, A.; Velikanova, E.; Sinitsky, M.; Kanonykina, A.; Dyleva, Y.; et al. Calciprotein Particles Cause Physiologically Significant Pro-Inflammatory Response in Endothelial Cells and Systemic Circulation. Int. J. Mol. Sci. 2022, 23, 14941. [Google Scholar] [CrossRef]
- Shishkova, D.; Velikanova, E.; Bogdanov, L.; Sinitsky, M.; Kostyunin, A.; Tsepokina, A.; Gruzdeva, O.; Mironov, A.; Mukhamadiyarov, R.; Glushkova, T.; et al. Calciprotein Particles Link Disturbed Mineral Homeostasis with Cardiovascular Disease by Causing Endothelial Dysfunction and Vascular Inflammation. Int. J. Mol. Sci. 2021, 22, 12458. [Google Scholar] [CrossRef] [PubMed]
- Shishkova, D.; Markova, V.; Sinitsky, M.; Tsepokina, A.; Velikanova, E.; Bogdanov, L.; Glushkova, T.; Kutikhin, A. Calciprotein Particles Cause Endothelial Dysfunction under Flow. Int. J. Mol. Sci. 2020, 21, 8802. [Google Scholar] [CrossRef] [PubMed]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [PubMed]
- Gene Ontology Consortium. The Gene Ontology resource: Enriching a GOld mine. Nucleic Acids Res. 2021, 49, D325–D334. [Google Scholar] [CrossRef]
- UniProt Consortium. UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Res. 2021, 49, D480–D489. [Google Scholar] [CrossRef] [PubMed]
- Gillespie, M.; Jassal, B.; Stephan, R.; Milacic, M.; Rothfels, K.; Senff-Ribeiro, A.; Griss, J.; Sevilla, C.; Matthews, L.; Gong, C.; et al. The reactome pathway knowledgebase 2022. Nucleic Acids Res. 2022, 50, D687–D692. [Google Scholar] [CrossRef] [PubMed]
- Griss, J.; Viteri, G.; Sidiropoulos, K.; Nguyen, V.; Fabregat, A.; Hermjakob, H. ReactomeGSA Efficient Multi-Omics Comparative Pathway Analysis. Mol. Cell. Proteomics 2020, 19, 2115–2125. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Furumichi, M.; Sato, Y.; Ishiguro-Watanabe, M.; Tanabe, M. KEGG: Integrating viruses and cellular organisms. Nucleic Acids Res. 2021, 49, D545–D551. [Google Scholar] [CrossRef]
- Chen, H.; Boutros, P.C. VennDiagram: A package for the generation of highly-customizable Venn and Euler diagrams in R. BMC Bioinform. 2011, 12, 35. [Google Scholar] [CrossRef]
- Dixon, P. VEGAN, A Package of R Functions for Community Ecology. J. Veget. Sci. 2003, 14, 927–930. [Google Scholar] [CrossRef]
- Rohart, F.; Gautier, B.; Singh, A.; Lê Cao, K.A. mixOmics: An R package for ’omics feature selection and multiple data integration. PLoS Comput. Biol. 2017, 13, e1005752. [Google Scholar] [CrossRef]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef] [PubMed]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016. [Google Scholar]
- Blighe, K.; Rana, S.; Lewis, M. EnhancedVolcano: Publication-Ready Volcano Plots with Enhanced Colouring and Labeling, R package version 1.14.0; R Core Team: Vienna, Austria, 2022. [Google Scholar]
- Perez-Riverol, Y.; Bai, J.; Bandla, C.; García-Seisdedos, D.; Hewapathirana, S.; Kamatchinathan, S.; Kundu, D.J.; Prakash, A.; Frericks-Zipper, A.; Eisenacher, M.; et al. The PRIDE database resources in 2022: A hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res. 2022, 50, D543–D552. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
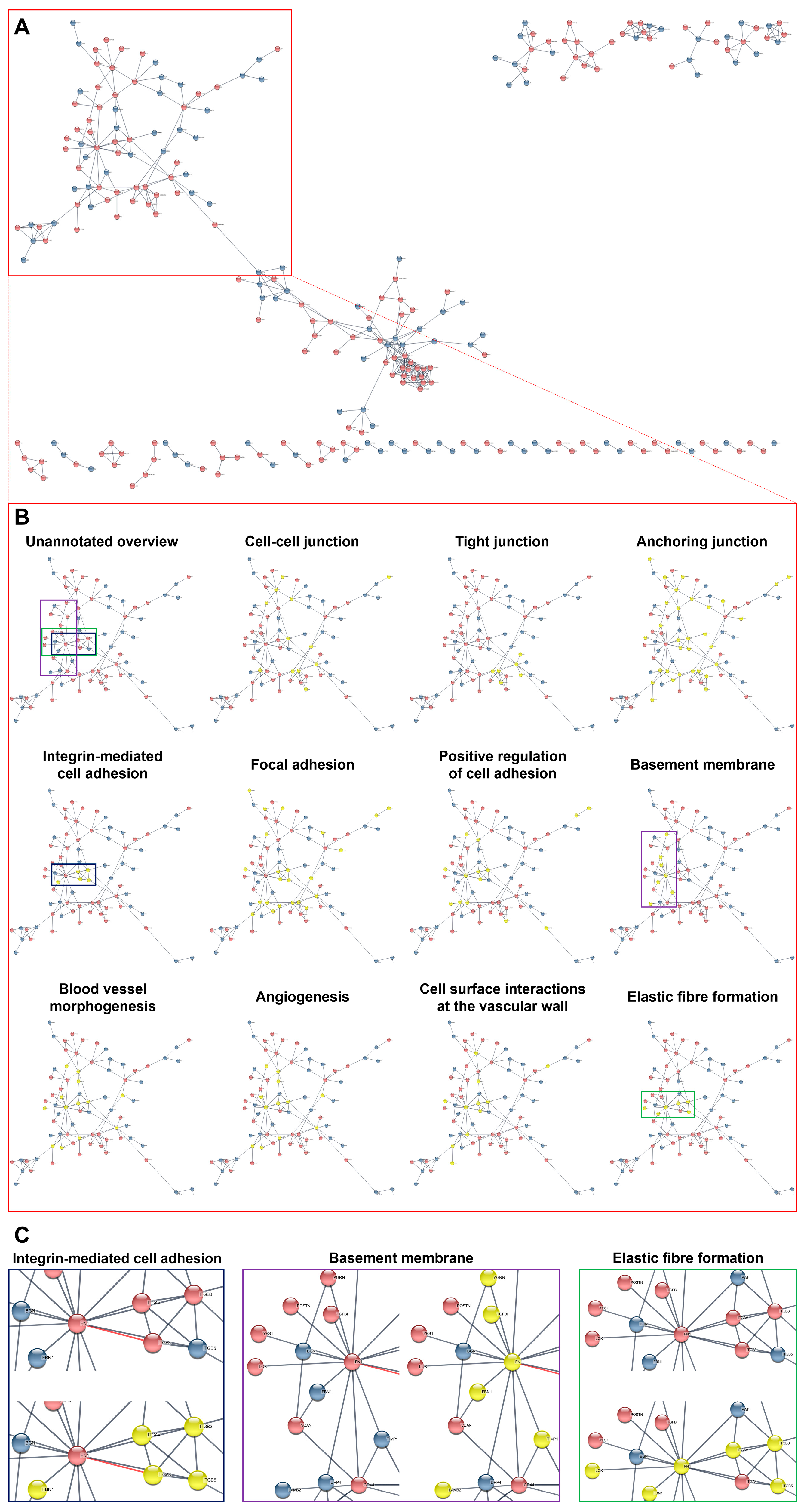
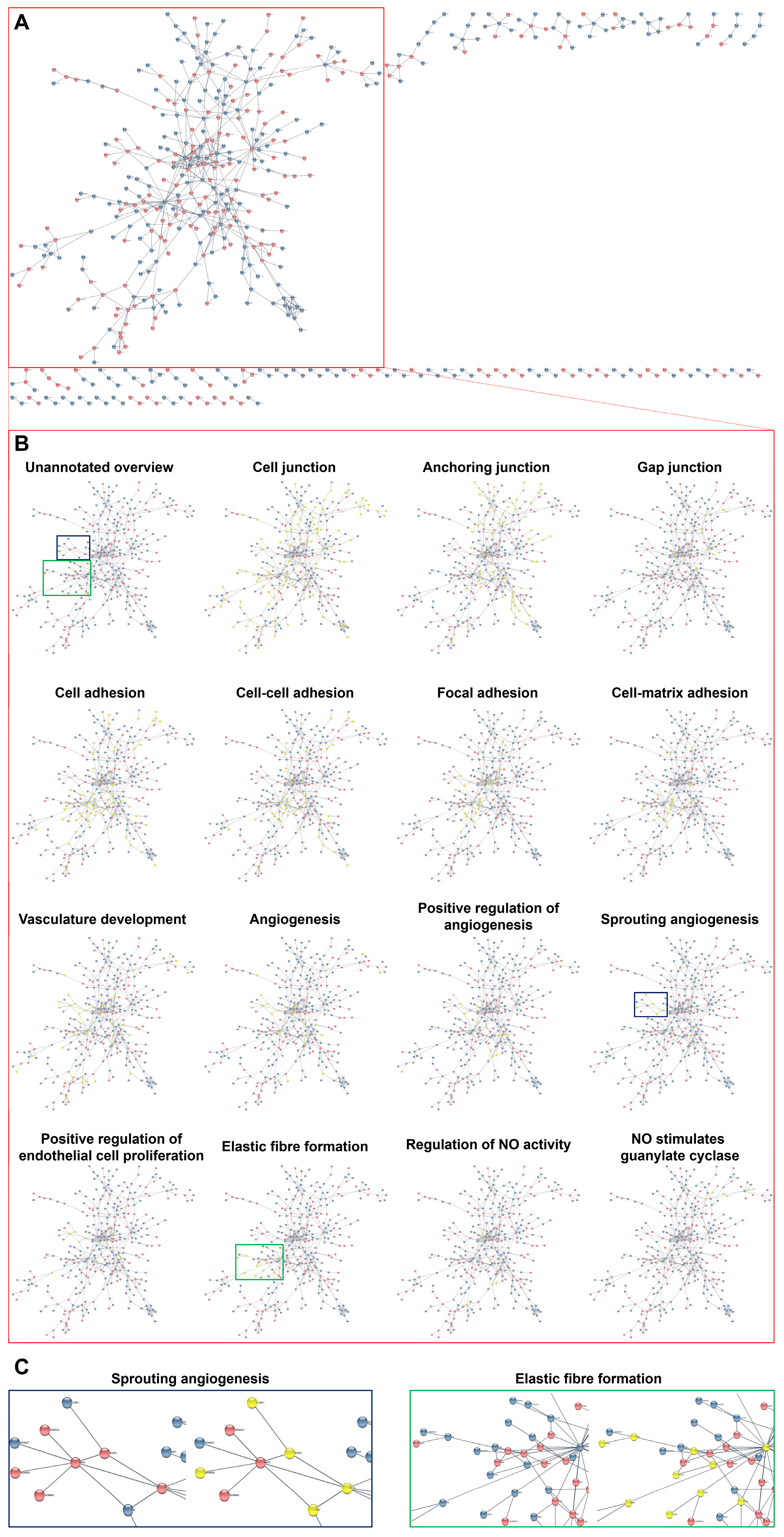
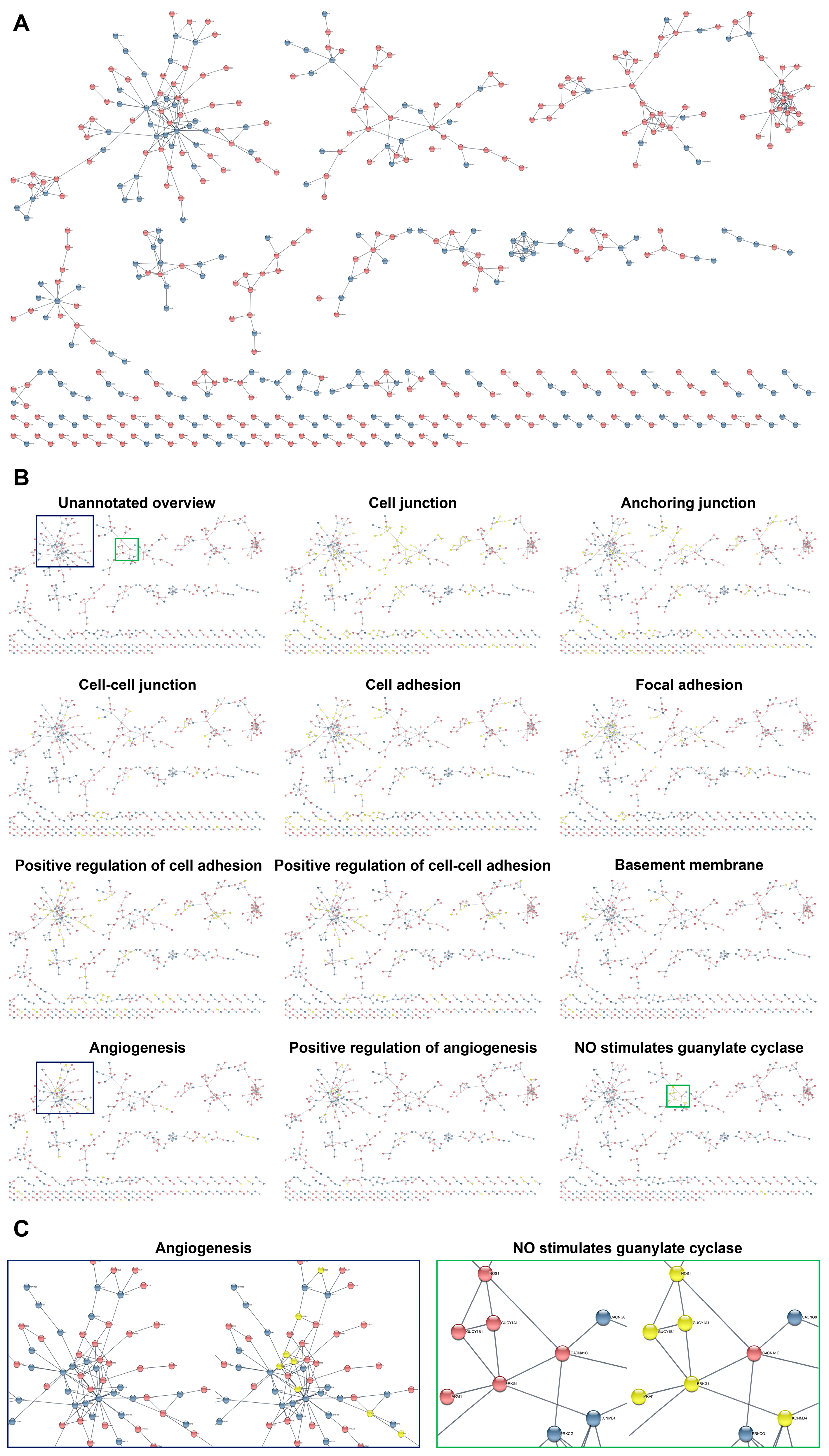
| Gene | Flow Culture Chambers (µ-Slide y-Shaped) | Static Conditions Experiment #1 | Static Conditions Experiment #2 | Static Conditions Experiment #3 | Static Conditions Experiment #4 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HCAECs, ΔCt (Mean ± Standard Deviation) | HITAECs, ΔCt (Mean ± Standard Deviation) | Fold Change (HCAECs to HITAECs) | HCAECs, ΔCt (Mean ± Standard Deviation) | HITAECs, ΔCt (Mean ± Standard Deviation) | Fold Change (HCAECs to HITAECs) | HCAECs, ΔCt (Mean ± Standard Deviation) | HITAECs, ΔCt (Mean ± Standard Deviation) | Fold Change (HCAECs to HITAECs) | HCAECs, ΔCt (Mean ± Standard Deviation) | HITAECs, ΔCt (Mean ± Standard Deviation) | Fold Change (HCAECs to HITAECs) | HCAECs, ΔCt (Mean ± Standard Deviation) | HITAECs, ΔCt (Mean ± Standard Deviation) | Fold Change (HCAECs to HITAECs) | |
| VCAM1 | 0.0051 ± 0.0004 | 0.0053 ± 0.0006 | 0.962 | 0.0015 ± 0.0004 | 0.0098 ± 0.0009 | 0.153 | N/D | N/D | N/D | 0.0013 ± 0.0002 | 0.0955 ± 0.0039 | 0.014 | 0.0051 ± 0.0002 | 0.0455 ± 0.0041 | 0.112 |
| ICAM1 | 0.0331 ± 0.0021 | 0.1659 ± 0.0121 | 0.200 | 0.0050 ± 0.0003 | 0.0407 ± 0.0027 | 0.123 | 0.0279 ± 0.0132 | 0.0559 ± 0.0231 | 0.499 | 0.0163 ± 0.0079 | 2.0781 ± 0.7478 | 0.008 | 0.0313 ± 0.0096 | 0.2675 ± 0.1376 | 0.117 |
| SELE | 0.0012 ± 0.0001 | 0.0265 ± 0.0067 | 0.045 | 0.0011 ± 0.0002 | 0.0481 ± 0.0039 | 0.023 | 0.0002 ± 0.00002 | 0.0219 ± 0.0016 | 0.009 | 0.0004 ± 0.0001 | 0.0942 ± 0.0110 | 0.004 | 0.0029 ± 0.0002 | 0.0939 ± 0.0057 | 0.031 |
| SELP | 0.0010 ± 0.0001 | 0.0184 ± 0.0028 | 0.054 | 0.0025 ± 0.0009 | 0.0044 ± 0.0011 | 0.568 | 0.0899 ± 0.0035 | 0.0080 ± 0.0037 | 11.238 | 0.0127 ± 0.0016 | 0.0555 ± 0.0099 | 0.229 | 0.0321 ± 0.0022 | 0.0203 ± 0.0011 | 1.581 |
| IL6 | 0.0044 ± 0.0004 | 0.0199 ± 0.0016 | 0.221 | 0.0029 ± 0.0007 | 0.0007 ± 0.0003 | 4.143 | 0.0042 ± 0.0003 | 0.0211 ± 0.0019 | 0.199 | 0.0029 ± 0.0002 | 0.0095 ± 0.0020 | 0.305 | 0.0084 ± 0.0009 | 0.0050 ± 0.0003 | 1.680 |
| CXCL8 | 0.0337 ± 0.0009 | 0.1774 ± 0.0129 | 0.190 | 0.0701 ± 0.0045 | 0.0557 ± 0.0051 | 1.259 | 0.0256 ± 0.0029 | 0.3286 ± 0.0285 | 0.078 | 0.0301 ± 0.0028 | 0.5886 ± 0.0732 | 0.051 | 0.1107 ± 0.0063 | 0.4469 ± 0.0335 | 0.248 |
| CCL2 | 0.0112 ± 0.0009 | 0.2993 ± 0.0247 | 0.037 | 0.0279 ± 0.0091 | 0.2563 ± 0.0137 | 0.109 | 0.0614 ± 0.0038 | 1.0850 ± 0.1059 | 0.057 | 0.0543 ± 0.0026 | 4.3406 ± 0.9546 | 0.013 | 0.0920 ± 0.0048 | 0.9923 ± 0.0732 | 0.093 |
| CXCL1 | N/D | N/D | N/D | 0.0763 ± 0.0080 | 0.0651 ± 0.0061 | 1.172 | 0.0984 ± 0.0274 | 0.2571 ± 0.0556 | 0.383 | 0.1074 ± 0.0154 | 1.6440 ± 0.4153 | 0.065 | 0.2617 ± 0.0185 | 0.2715 ± 0.0439 | 0.964 |
| MIF | N/D | N/D | N/D | 0.2152 ± 0.0231 | 0.0812 ± 0.0074 | 2.650 | 0.4353 ± 0.0956 | 2.6439 ± 0.1977 | 0.165 | 0.1903 ± 0.0117 | 4.0451 ± 0.5167 | 0.047 | 0.4234 ± 0.0139 | 0.6183 ± 0.0129 | 0.685 |
| KLF2 | 0.0047 ± 0.0006 | 0.0021 ± 0.0002 | 2.238 | 0.0018 ± 0.0009 | 0.0004 ± 0.0001 | 4.500 | N/D | N/D | N/D | N/D | N/D | N/D | N/D | N/D | N/D |
| KLF4 | 0.0239 ± 0.0012 | 0.1268 ± 0.0178 | 0.188 | 0.0010 ± 0.0002 | 0.0005 ± 0.0001 | 2.000 | N/D | N/D | N/D | N/D | N/D | N/D | N/D | N/D | N/D |
| NFE2L2 | 0.0618 ± 0.0041 | 0.0293 ± 0.0010 | 2.109 | 0.0866 ± 0.088 | 0.0523 ± 0.0026 | 1.656 | N/D | N/D | N/D | N/D | N/D | N/D | N/D | N/D | N/D |
| SNAI1 | 0.0312 ± 0.0011 | 0.3193 ± 0.0632 | 0.098 | 0.0071 ± 0.0004 | 0.0121 ± 0.0017 | 0.587 | 0.0286 ± 0.0031 | 0.0381 ± 0.0065 | 0.751 | 0.0011 ± 0.0001 | 0.1209 ± 0.0140 | 0.009 | 0.0236 ± 0.0011 | 0.0237 ± 0.0031 | 0.996 |
| SNAI2 | 0.0003 ± 0.00002 | 0.0002 ± 0.00004 | 1.500 | 0.0002 ± 0.00001 | 0.0001 ± 0.00001 | 2.000 | 0.0371 ± 0.0045 | 0.0025 ± 0.0007 | 14.840 | 0.00006 ± 0.00002 | 0.0043 ± 0.0015 | 0.014 | 0.0034 ± 0.0002 | 0.0001 ± 0.00005 | 34.000 |
| TWIST1 | 0.0005 ± 0.0001 | 0.0001 ± 0.00001 | 5.000 | 0.0004 ± 0.00001 | 0.00001 ± 0.00001 | 40.000 | 0.0010 ± 0.0004 | 0.0007 ± 0.0002 | 1.429 | 0.00008 ± 0.00002 | 0.00014 ± 0.0001 | 0.571 | 0.0002 ± 0.00004 | 0.00004 ± 0.00002 | 5.000 |
| ZEB1 | 0.0845 ± 0.0096 | 0.0886 ± 0.0056 | 0.954 | 0.0350 ± 0.0057 | 0.0343 ± 0.0034 | 1.020 | 0.0818 ± 0.0037 | 0.1628 ± 0.0196 | 0.502 | 0.0068 ± 0.0004 | 0.0655 ± 0.0094 | 0.104 | 0.0486 ± 0.0024 | 0.0552 ± 0.0083 | 0.880 |
| CDH5 | 0.6792 ± 0.0132 | 3.0981 ± 0.3207 | 0.219 | 0.4247 ± 0.0985 | 0.5115 ± 0.0189 | 0.830 | 0.3888 ± 0.0087 | 0.3852 ± 0.0165 | 1.009 | 0.9632 ± 0.0871 | 9.4160 ± 2.8673 | 0.102 | 1.1429 ± 0.1359 | 2.1643 ± 0.0577 | 0.528 |
| CDH2 | 0.0088 ± 0.0013 | 0.0103 ± 0.0015 | 0.854 | 0.0108 ± 0.0015 | 0.0028 ± 0.0009 | 3.857 | 0.0670 ± 0.0056 | 0.0001 ± 0.0002 | 670.000 | 0.0479 ± 0.0032 | 0.0146 ± 0.0039 | 3.281 | 0.1393 ± 0.0119 | 0.0079 ± 0.0003 | 17.633 |
| NOS3 | 0.0450 ± 0.0008 | 0.1536 ± 0.0102 | 0.293 | 0.0066 ± 0.00011 | 0.0020 ± 0.0007 | 3.300 | 0.0015 ± 0.0004 | 0.0009 ± 0.0007 | 1.667 | 0.0057 ± 0.0005 | 0.2375 ± 0.0387 | 0.024 | 0.0262 ± 0.0026 | 0.0125 ± 0.0003 | 2.096 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Frolov, A.; Lobov, A.; Kabilov, M.; Zainullina, B.; Tupikin, A.; Shishkova, D.; Markova, V.; Sinitskaya, A.; Grigoriev, E.; Markova, Y.; et al. Multi-Omics Profiling of Human Endothelial Cells from the Coronary Artery and Internal Thoracic Artery Reveals Molecular but Not Functional Heterogeneity. Int. J. Mol. Sci. 2023, 24, 15032. https://doi.org/10.3390/ijms241915032
Frolov A, Lobov A, Kabilov M, Zainullina B, Tupikin A, Shishkova D, Markova V, Sinitskaya A, Grigoriev E, Markova Y, et al. Multi-Omics Profiling of Human Endothelial Cells from the Coronary Artery and Internal Thoracic Artery Reveals Molecular but Not Functional Heterogeneity. International Journal of Molecular Sciences. 2023; 24(19):15032. https://doi.org/10.3390/ijms241915032
Chicago/Turabian StyleFrolov, Alexey, Arseniy Lobov, Marsel Kabilov, Bozhana Zainullina, Alexey Tupikin, Daria Shishkova, Victoria Markova, Anna Sinitskaya, Evgeny Grigoriev, Yulia Markova, and et al. 2023. "Multi-Omics Profiling of Human Endothelial Cells from the Coronary Artery and Internal Thoracic Artery Reveals Molecular but Not Functional Heterogeneity" International Journal of Molecular Sciences 24, no. 19: 15032. https://doi.org/10.3390/ijms241915032
APA StyleFrolov, A., Lobov, A., Kabilov, M., Zainullina, B., Tupikin, A., Shishkova, D., Markova, V., Sinitskaya, A., Grigoriev, E., Markova, Y., & Kutikhin, A. (2023). Multi-Omics Profiling of Human Endothelial Cells from the Coronary Artery and Internal Thoracic Artery Reveals Molecular but Not Functional Heterogeneity. International Journal of Molecular Sciences, 24(19), 15032. https://doi.org/10.3390/ijms241915032






