Alternative Splicing of TaHsfA2-7 Is Involved in the Improvement of Thermotolerance in Wheat
Abstract
:1. Introduction
2. Results
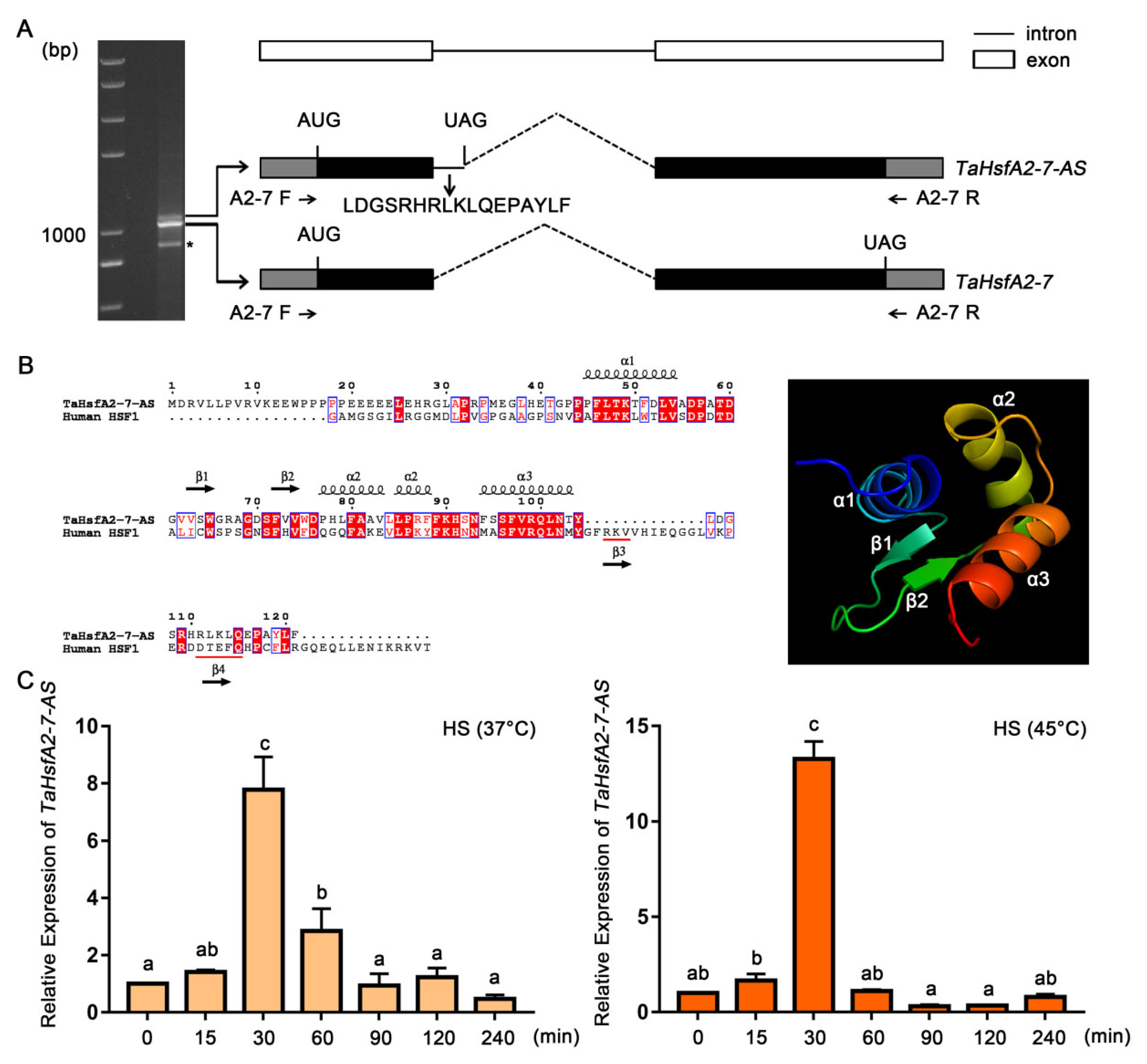
2.1. Characteristics of a New Splice Variant of TaHsfA2-7
2.2. Expression Patterns of TaHsfA2-7-AS
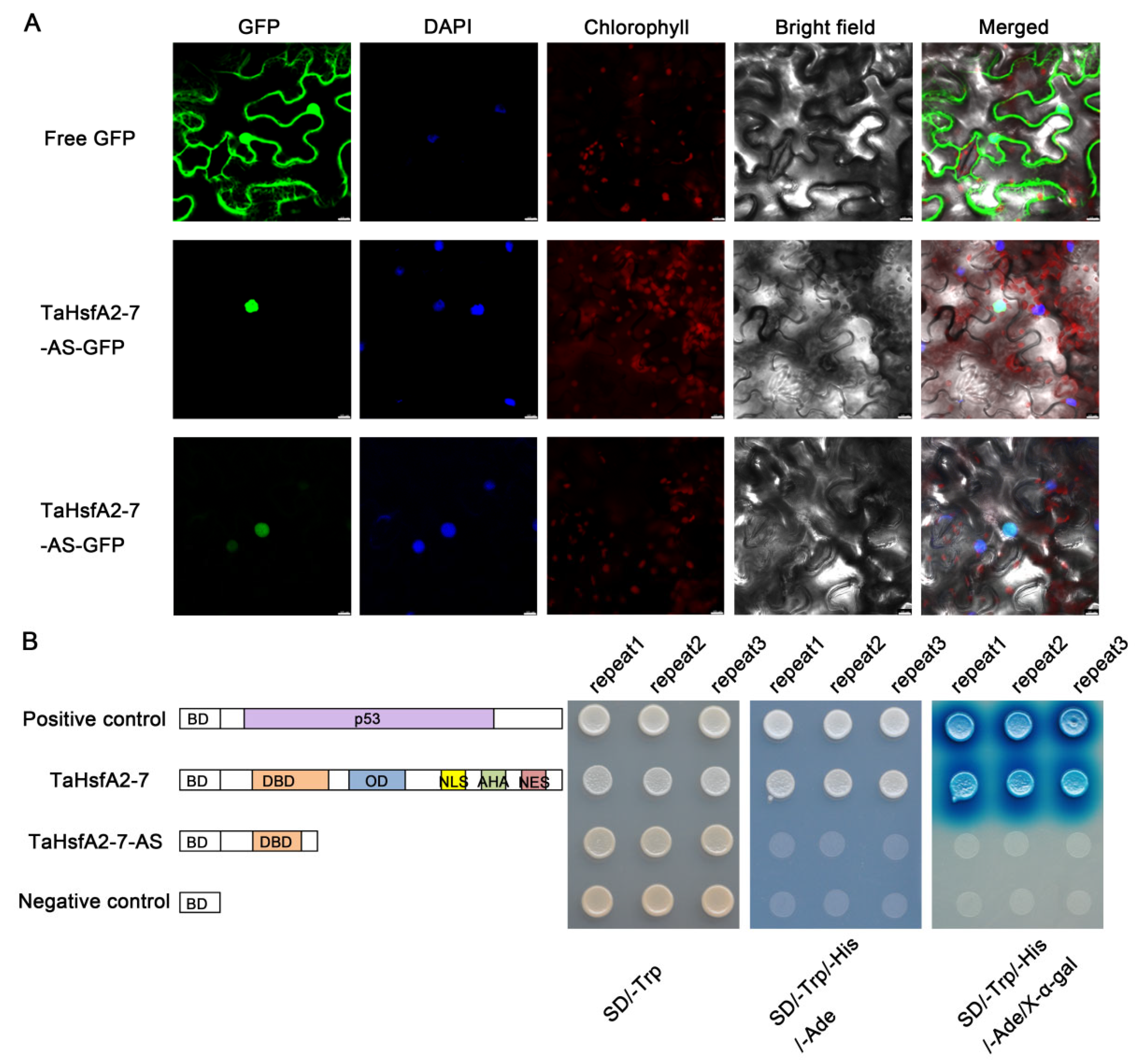
2.3. TaHsfA2-7-AS Is Localized to Nucleus with No Transcriptional Activity
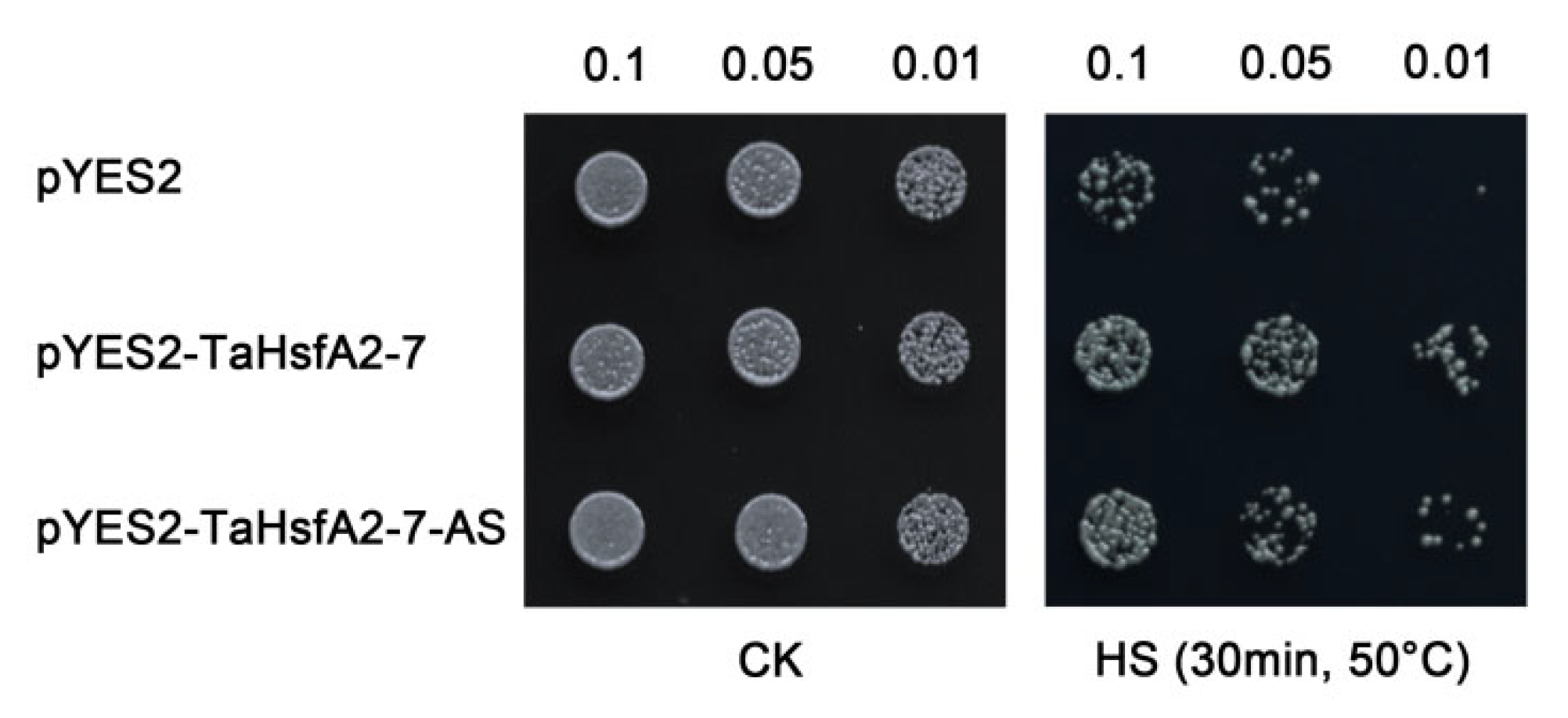
2.4. Ectopic Expression of TaHsfA2-7-AS Improves Thermotolerance in Yeast
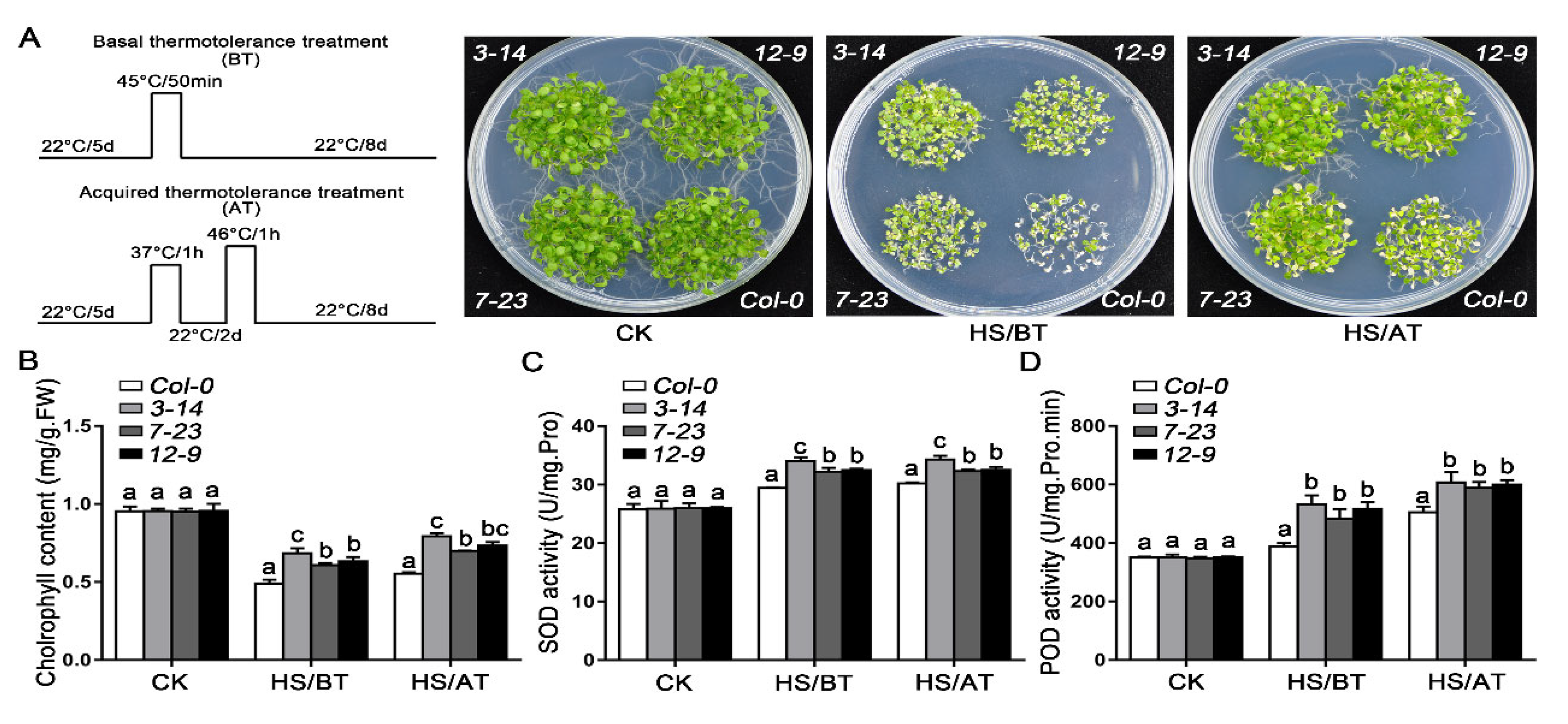
2.5. The Overexpression of TaHsfA2-7-AS Enhanced the Tolerance of Transgenic Arabidopsis to Heat Stress
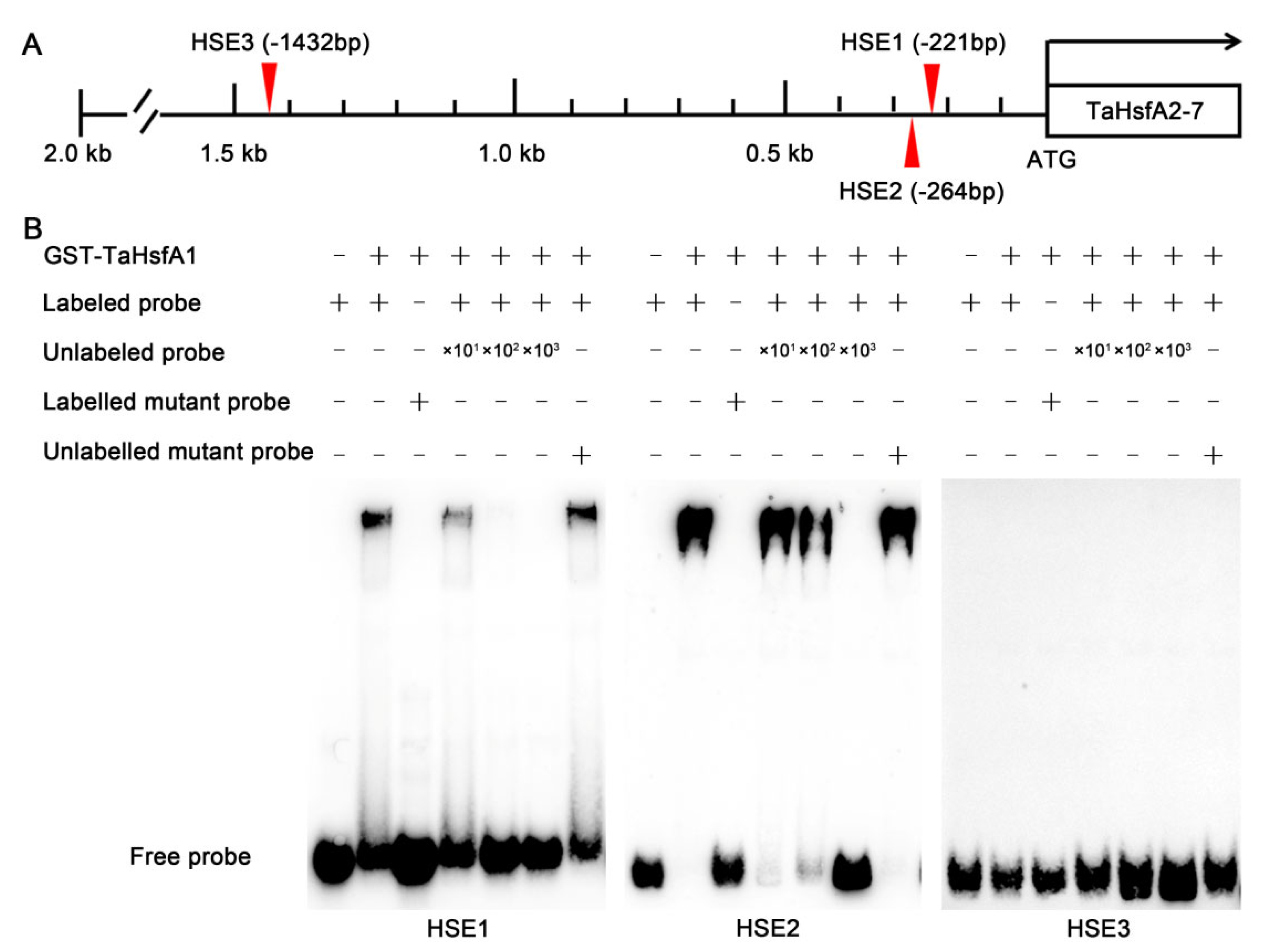
2.6. TaHsfA1 Acts as an Upstream Transcriptional Regulator of TaHsfA2-7 and TaHsfA2-7-AS
3. Discussion
3.1. TaHsfA2-7-AS Is a HS-Induced Splice Variant of TaHsfA2-7
3.2. TaHsfA2-7-AS Enhances Tolerance to Heat Stress
3.3. The Transcription of TaHsfA2-7 and TaHSFA2-7-AS Is Regulated by TaHsfA1
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Isolation of Total RNA and Identification of Splice Variants
4.3. Expression Analysis Using RT-qPCR
4.4. Subcellular Localization
4.5. Transactivation Analysis in Yeast
4.6. Thermotolerance Assay in Yeast
4.7. Generation of Transgenic Plants
4.8. Protein Extraction and Western Blotting
4.9. HS Treatment in Arabidopsis
4.10. Measurements of Physiological Indexes
4.11. EMSA
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Bita, C.E.; Gerats, T. Plant tolerance to high temperature in a changing environment: Scientifific fundamentals and production of heat stress-tolerant crops. Front. Plant Sci. 2013, 4, 273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, C.; Liu, B.; Piao, S.; Wang, X.; Lobell, D.B.; Huang, Y.; Huang, M.; Yao, Y.; Bassu, S.; Ciais, P.; et al. Temperature increase reduces global yields of majorcrops in four independent estimates. Proc. Natl. Acad. Sci. USA 2017, 114, 9326–9331. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, B.; Liu, L.; Tian, L.; Cao, W.; Zhu, Y.; Asseng, S. Post-heading heat stress and yield impact in winter wheat of China. Glob. Chang. Biol. 2014, 20, 372–381. [Google Scholar] [CrossRef] [PubMed]
- Fragkostefanakis, S.; Roth, S.; Schleiff, E.; Scharf, K.D. Prospects of engineering thermotolerance in crops through modulation of heat stress transcription factor and heat shock protein networks. Plant Cell Environ. 2015, 38, 1881–1895. [Google Scholar] [CrossRef] [PubMed]
- Andrasi, N.; Pettko-Szandtner, A.; Szabados, L. Diversity of plant heat shock factors: Regulation, interactions, and functions. J. Exp. Bot. 2021, 72, 1558–1575. [Google Scholar] [CrossRef] [PubMed]
- Guo, M.; Liu, J.H.; Ma, X.; Luo, D.X.; Gong, Z.H.; Lu, M.H. The plant heat stress transcription factors (HSFs): Structure, regulation, and function in response to abiotic stresses. Front. Plant Sci. 2016, 7, 114. [Google Scholar] [CrossRef] [Green Version]
- Hahn, A.; Bublak, D.; Schleiff, E.; Scharf, K.D. Crosstalk between Hsp90 and Hsp70 chaperones and heat stress transcription factors in tomato. Plant Cell 2011, 23, 741–755. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ogawa, D.; Yamaguchi, K.; Nishiuchi, T. High-level overexpression of the Arabidopsis HsfA2 gene confers not only increased themotolerance but also salt/osmotic stress tolerance and enhanced callus growth. J. Exp. Bot. 2007, 58, 3373–3383. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Li, Y.; Xing, D.; Gao, C. Characterization of mitochondrial dynamics and subcellular localization of ROS reveal that HsfA2 alleviates oxidative damage caused by heat stress in Arabidopsis. J. Exp. Bot. 2009, 60, 2073–2091. [Google Scholar] [CrossRef] [Green Version]
- Ikeda, M.; Mitsuda, N.; Ohme-Takagi, M. Arabidopsis HsfB1 and HsfB2b act as repressors of the expression of heat-inducible Hsfs but positively regulate the acquired thermotolerance. Plant Physiol. 2011, 157, 1243–1254. [Google Scholar] [CrossRef]
- Liu, Z.; Qin, J.; Tian, X.; Xu, S.; Wang, Y.; Li, H.; Wang, X.; Peng, H.; Yao, Y.; Hu, Z.; et al. Global profifiling of alternative splicing landscape responsive to drought, heat and their combination in wheat (Triticum aestivum L.). Plant Biotechnol. J. 2018, 16, 714–726. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- John, S.; Olas, J.J.; Mueller-Roeber, B. Regulation of alternative splicing in response to temperature variation in plants. J. Exp. Bot. 2021, 72, 6150–6163. [Google Scholar] [CrossRef] [PubMed]
- Kelemen, O.; Convertini, P.; Zhang, Z.; Wen, Y.; Shen, M.; Falaleeva, M.; Stamm, S. Function of alternative splicing. Gene 2013, 514, 1–20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barbazuk, W.B.; Fu, Y.; McGinnis, K.M. Genome-wide analyses of alternative splicing in plants: Opportunities and challenges. Genome Res. 2008, 18, 1381–1392. [Google Scholar] [CrossRef] [Green Version]
- Nilsen, T.W.; Graveley, B.R. Expansion of the eukaryotic proteome by alternative splicing. Nature 2010, 463, 457–463. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simpson, C.G.; Lewandowska, D.; Fuller, J.; Maronova, M.; Kalyna, M.; Davidson, D.; McNicol, J.; Raczynska, D.; Jarmolowski, A.; Barta, A.; et al. Alternative splicing in plants. Biochem. Soc. Trans. 2008, 36, 508–510. [Google Scholar] [CrossRef] [Green Version]
- Lam, P.Y.; Wang, L.; Lo, C.; Zhu, F.Y. Alternative splicing and its roles in plant metabolism. Int. J. Mol. Sci. 2022, 23, 7355. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Yang, J.; Shang, X.; Lv, W.; Xia, C.; Wang, C.; Feng, J.; Cao, Y.; He, H.; Li, L.; et al. SKIP regulates environmental fitness and floral transition by forming two distinct complexes in Arabidopsis. New Phytol. 2019, 224, 321–335. [Google Scholar] [CrossRef]
- Jiang, J.; Liu, X.; Liu, C.; Liu, G.; Li, S.; Wang, L. Integrating omics and alternative splicing reveals insights into grape response to high temperature. Plant Physiol. 2017, 173, 1502–1518. [Google Scholar] [CrossRef] [Green Version]
- Gu, J.; Xia, Z.; Luo, Y.; Jiang, X.; Qian, B.; Xie, H.; Zhu, J.; Xiong, L.; Zhu, J.; Wang, Z. Spliceosomal protein U1A is involved in alternative splicing and salt stress tolerance in Arabidopsis thaliana. Nucleic Acids Res. 2018, 46, 1777–1792. [Google Scholar] [CrossRef]
- Calixto, C.P.G.; Guo, W.; James, A.B.; Tzioutziou, N.A.; Entizne, J.C.; Panter, P.E.; Knight, H.; Nimmo, H.G.; Zhang, R.; Brown, J.W.S. Rapid and dynamic alternative splicing impacts the Arabidopsis cold response transcriptome. Plant Cell 2018, 30, 1424–1444. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tian, L.; Zhao, X.; Liu, H.; Ku, L.; Wang, S.; Han, Z.; Wu, L.; Shi, Y.; Song, X.; Chen, Y. Alternative splicing of ZmCCA1 mediates drought response in tropical maize. PLoS ONE 2019, 14, e0211623. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petrillo, E.; Kalyna, M.; Mandadi, K.K.; Tu, S.L.; Simpson, C.G. Editorial: Alternative splicing regulation in plants. Front. Plant Sci. 2020, 11, 913. [Google Scholar] [CrossRef] [PubMed]
- Filichkin, S.A.; Priest, H.D.; Givan, S.A.; Shen, R.; Bryant, D.W.; Fox, S.E.; Wong, W.-K.; Mockler, T.C. Genome-wide mapping of alternative splicing in Arabidopsis thaliana. Genome Res. 2010, 20, 45–58. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, T.; Lu, G.; Fan, D.; Zhu, C.; Li, W.; Zhao, Q.; Feng, Q.; Zhao, Y.; Guo, Y.; Li, W.; et al. Function annotation of the rice transcriptome at single-nucleotide resolution by RNA-seq. Genome Res. 2010, 20, 1238–1249. [Google Scholar] [CrossRef] [Green Version]
- Yang, S.; Tang, F.; Zhu, H. Alternative splicing in plant immunity. Int. J. Mol. Sci. 2014, 15, 10424–10445. [Google Scholar] [CrossRef] [Green Version]
- Reddy, A.S.N. Alternative splicing of pre-messenger RNAs in plants in the genomic era. Annu. Rev. Plant Biol. 2007, 58, 267–294. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Zhou, J. Splicing and alternative splicing in rice and humans. BMB Rep. 2013, 46, 439–447. [Google Scholar]
- Drechsel, G.; Kahles, A.; Kesarwani, A.K.; Stauffer, E.; Behr, J.; Drewe, P.; Rätsch, G.; Wachter, A. Nonsense-mediated decay of alternative precursor mRNA splicing variants is a major determinant of the Arabidopsis steady state transcriptome. Plant Cell 2013, 25, 3726–3742. [Google Scholar] [CrossRef] [Green Version]
- Kalyna, M.; Simpson, C.G.; Syed, N.H.; Lewandowska, D.; Marquez, Y.; Kusenda, B.; Marshall, J.; Fuller, J.; Cardle, L.; McNicol, J.; et al. Alternative splicing and nonsense-mediated decay modulate expression of important regulatory genes in Arabidopsis. Nucleic Acids Res. 2012, 40, 2454–2469. [Google Scholar] [CrossRef] [Green Version]
- Hamada, S.; Ito, H.; Hiraga, S.; Inagaki, K.; Nozaki, K.; Isono, N.; Yoshimoto, Y.; Takeda, Y.; Matsui, H. Differential characteristics and subcellular localization of two starch-branching enzyme isoforms encoded by a single gene in Phaseolus vulgaris L. J. Biol. Chem. 2002, 277, 16538–16546. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Chen, X.; Liang, X.; Zhou, X.; Yang, F.; Liu, J.; He, S.Y.; Guo, Z. Alternative splicing of rice WRKY62 and WRKY76 transcription factor genes in pathogen defense. Plant Physiol. 2016, 171, 1427–1442. [Google Scholar] [CrossRef] [Green Version]
- Capovilla, G.; Pajoro, A.; Immink, R.G.H.; Schmid, M. Role of alternative pre-mRNA splicing in temperature signaling. Curr. Opin. Plant Biol. 2015, 27, 97–103. [Google Scholar] [CrossRef] [PubMed]
- Keller, M.; Hu, Y.; Mesihovic, A.; Fragkostefanakis, S.; Schleiff, E.; Simm, S. Alternative splicing in tomato pollen in response to heat stress. Dna Res. 2017, 24, 205–217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, Z.; Liang, J.; Wang, C.; Ding, L.; Zhao, X.; Cao, X.; Xu, S.; Teng, N.; Yi, M. Alternative splicing provides a mechanism to regulate LlHSFA3 function in response to heat stress in lily. Plant Physiol. 2019, 181, 1651–1667. [Google Scholar] [CrossRef]
- Deng, Y.; Humbert, S.; Liu, J.; Srivastava, R.; Rothstein, S.J.; Howell, S.H. Heat induces the splicing by IRE1 of a mRNA encoding a transcription factor involved in the unfolded protein response in Arabidopsis. Proc. Natl. Acad. Sci. USA 2011, 108, 7247–7252. [Google Scholar] [CrossRef] [Green Version]
- Ling, Y.; Serrano, N.; Gao, G.; Atia, M.; Mokhtar, M.; Woo, Y.H.; Bazin, J.; Veluchamy, A.; Benhamed, M.; Crespi, M.; et al. Thermopriming triggers splicing memory in Arabidopsis. J. Exp. Bot. 2018, 69, 2659–2675. [Google Scholar] [CrossRef] [Green Version]
- Duan, S.; Liu, B.; Zhang, Y.; Li, G.; Guo, X. Genome-wide identification and abiotic stress-responsive pattern of heat shock transcription factor family in Triticum aestivum L. BMC Genom. 2019, 20, 257. [Google Scholar] [CrossRef] [Green Version]
- Neudegger, T.; Verghese, J.; Hayer-Hartl, M.; Hartl, F.U.; Bracher, A. Structure of human heat-shock transcription factor 1 in complex with DNA. Nat. Struct. Mol. Biol. 2016, 23, 140–146. [Google Scholar] [CrossRef]
- Ni, Z.; Li, H.; Zhao, Y.; Peng, H.; Hu, Z.; Xin, M.; Sun, Q. Genetic improvement of heat tolerance in wheat: Recent progress in understanding the underlying molecular mechanisms. Crop J. 2018, 6, 32–41. [Google Scholar] [CrossRef]
- Zhang, Y.J.; Zhang, Y.Y.; Zhang, H.N.; Qin, N.; Li, G.L.; Guo, X.L. Characterization and regulatory roles in thermotolerance of wheat heat shock transcription factor gene TaHsfA2e. Acta Agron. Sin. 2018, 44, 1818–1828. [Google Scholar] [CrossRef]
- Yoshida, T.; Ohama, N.; Nakajima, J.; Kidokoro, S.; Mizoi, J.; Nakashima, K.; Maruyama, K.; Kim, J.M.; Seki, M.; Todaka, D.; et al. Arabidopsis HsfA1 transcription factors function as the main positive regulators in heat shock-responsive gene expression. Mol. Genet. Genom. 2011, 286, 321–332. [Google Scholar] [CrossRef] [PubMed]
- Ohama, N.; Sato, H.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Transcriptional regulatory network of plant heat stress response. Trends Plant Sci. 2017, 22, 53–65. [Google Scholar] [CrossRef]
- Gong, Z.; Xiong, L.; Shi, H.; Yang, S.; Herrera-Estrella, L.R.; Xu, G.; Chao, D.; Li, J.; Wang, P.; Qin, F.; et al. Plant abiotic stress response and nutrient use efficiency. Sci. China Life Sci. 2020, 63, 635–674. [Google Scholar] [CrossRef]
- Liu, H.C.; Liao, H.T.; Charng, Y.Y. The role of class A1 heat shock factors (HSFA1s) in response to heat and other stresses in Arabidopsis. Plant Cell Environ. 2011, 34, 738–751. [Google Scholar] [CrossRef]
- Chang, C.Y.; Lin, W.D.; Tu, S.L. Genome-wide analysis of heat-sensitive alternative splicing in Physcomitrella patens. Plant Physiol. 2014, 165, 826–840. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Sun, N.; Liu, M.; Liu, J.; Du, B.; Wang, X.; Qi, X. An autoregulatory loop controlling Arabidopsis HsfA2 expression: Role of heat shock-induced alternative splicing. Plant Physiol. 2013, 162, 512–521. [Google Scholar] [CrossRef] [Green Version]
- Cheng, Q.; Zhou, Y.; Liu, Z.; Zhang, L.; Song, G.; Guo, Z.; Wang, W.; Qu, X.; Zhu, Y.; Yang, D. An alternatively spliced heat shock transcription factor, OsHSFA2dI, functions in the heat stress-induced unfolded protein response in rice. Plant Biol. 2015, 17, 419–429. [Google Scholar] [CrossRef]
- Nishizawa-Yokoi, A.; Nosaka, R.; Hayashi, H.; Tainaka, H.; Maruta, T.; Tamoi, M.; Ikeda, M.; Ohme-Takagi, M.; Yoshimura, K.; Yabuta, Y.; et al. HsfA1d and HsfA1e involved in the transcriptional regulation of HsfA2 function as key regulators for the Hsf signaling network in response to environmental stress. Plant Cell Physiol. 2011, 52, 933–945. [Google Scholar] [CrossRef] [Green Version]
- Xue, G.P.; Sadat, S.; Drenth, J.; Mclntyre, C.L. The heat shock factor family from Triticum aestivum in response to heat and other major abiotic stresses and their role in regulation of heat shock protein genes. J. Exp. Bot. 2014, 65, 539–557. [Google Scholar] [CrossRef] [Green Version]
- Titz, B.; Thomas, S.; Rajagopala, S.V.; Chiba, T.; Ito, T.; Uetz, P. Transcriptional activitors in yeast. Nucleic Acids Res. 2006, 34, 955–967. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Li, G.; Zhang, H.; Zhang, Y.; Zhang, Y.; Duan, S.; Sheteiwy, M.S.A.; Zhang, H.; Shao, H.; Guo, X. TaHsfA2-1, a new gene for thermotolerance in wheat seedlings: Characterization and functional roles. J Plant Physiol. 2020, 246–247, 153135. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Zhang, H.; Shao, H.; Wang, G.; Zhang, Y.; Zhang, Y.; Zhao, L.; Guo, X.; Sheteiwy, M.S.A. ZmHsf05, a new heat shock transcription factor from Zea mays L. improves thermotolerance in Arabidopsis thaliana and rescues thermotolerance defects of the athsfa2mutant. Plant Sci. 2019, 283, 375–384. [Google Scholar] [CrossRef] [PubMed]
- Porra, R.J.; Thompson, W.A.; Kriedemann, P.E. Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: Verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim. Biophys. Acta 1989, 975, 384–394. [Google Scholar] [CrossRef]
- Li, H.; Zhang, H.; Li, G.; Liu, Z.; Zhang, Y.; Zhang, H.; Guo, X. Expression of maize heat shock transcription factor gene ZmHsf06 enhances the thermotolerance and drought-stress tolerance of transgenic Arabidopsis. Funct. Plant Biol. 2015, 42, 1080–1091. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, Z.; Li, M.; Zhang, H.; Zhao, B.; Liu, Z.; Duan, S.; Meng, X.; Li, G.; Guo, X. Alternative Splicing of TaHsfA2-7 Is Involved in the Improvement of Thermotolerance in Wheat. Int. J. Mol. Sci. 2023, 24, 1014. https://doi.org/10.3390/ijms24021014
Ma Z, Li M, Zhang H, Zhao B, Liu Z, Duan S, Meng X, Li G, Guo X. Alternative Splicing of TaHsfA2-7 Is Involved in the Improvement of Thermotolerance in Wheat. International Journal of Molecular Sciences. 2023; 24(2):1014. https://doi.org/10.3390/ijms24021014
Chicago/Turabian StyleMa, Zhenyu, Mingyue Li, Huaning Zhang, Baihui Zhao, Zihui Liu, Shuonan Duan, Xiangzhao Meng, Guoliang Li, and Xiulin Guo. 2023. "Alternative Splicing of TaHsfA2-7 Is Involved in the Improvement of Thermotolerance in Wheat" International Journal of Molecular Sciences 24, no. 2: 1014. https://doi.org/10.3390/ijms24021014
APA StyleMa, Z., Li, M., Zhang, H., Zhao, B., Liu, Z., Duan, S., Meng, X., Li, G., & Guo, X. (2023). Alternative Splicing of TaHsfA2-7 Is Involved in the Improvement of Thermotolerance in Wheat. International Journal of Molecular Sciences, 24(2), 1014. https://doi.org/10.3390/ijms24021014





