Phosphatidylethanolamines Are Associated with Nonalcoholic Fatty Liver Disease (NAFLD) in Obese Adults and Induce Liver Cell Metabolic Perturbations and Hepatic Stellate Cell Activation
Abstract
1. Introduction
2. Results
2.1. Demographic, Histological, and Biochemical Features of the Study Subjects
2.2. Elevation of Serum PLs in SS, B-NASH, and NASH
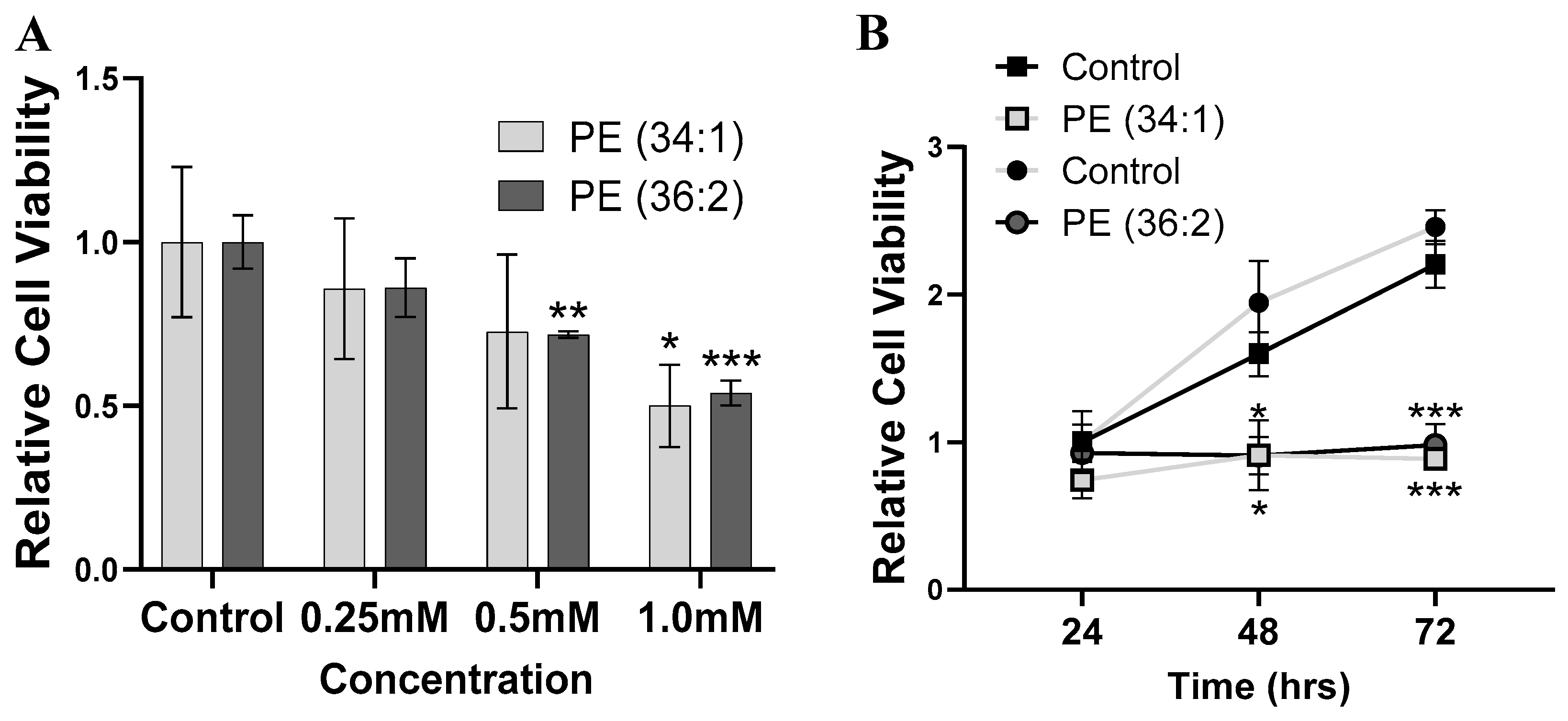
2.3. PEs Inhibited Cell Viability in HepG2 Cells
2.4. PEs Increased Lipid Droplet Accumulation in HepG2 Cells
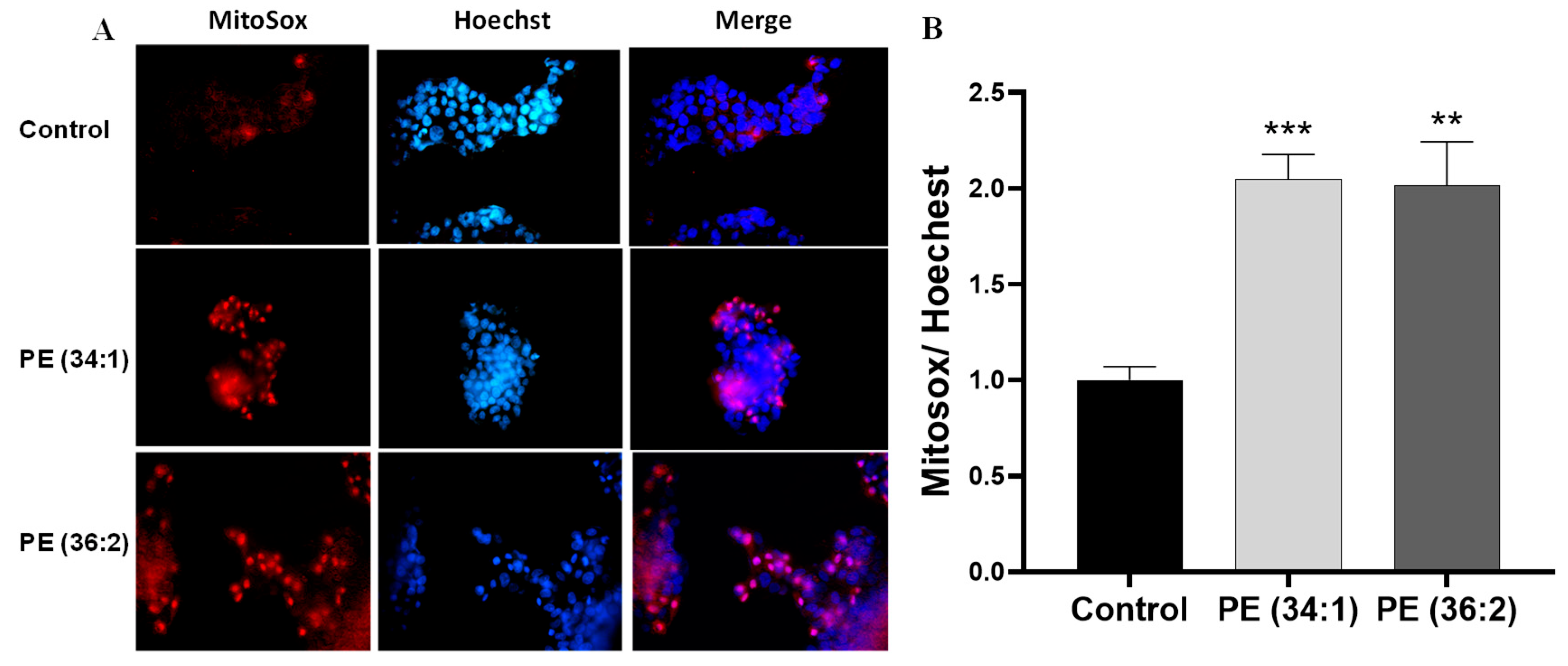
2.5. PEs Increased Mitochondrial Dysfunction in HepG2 Cells
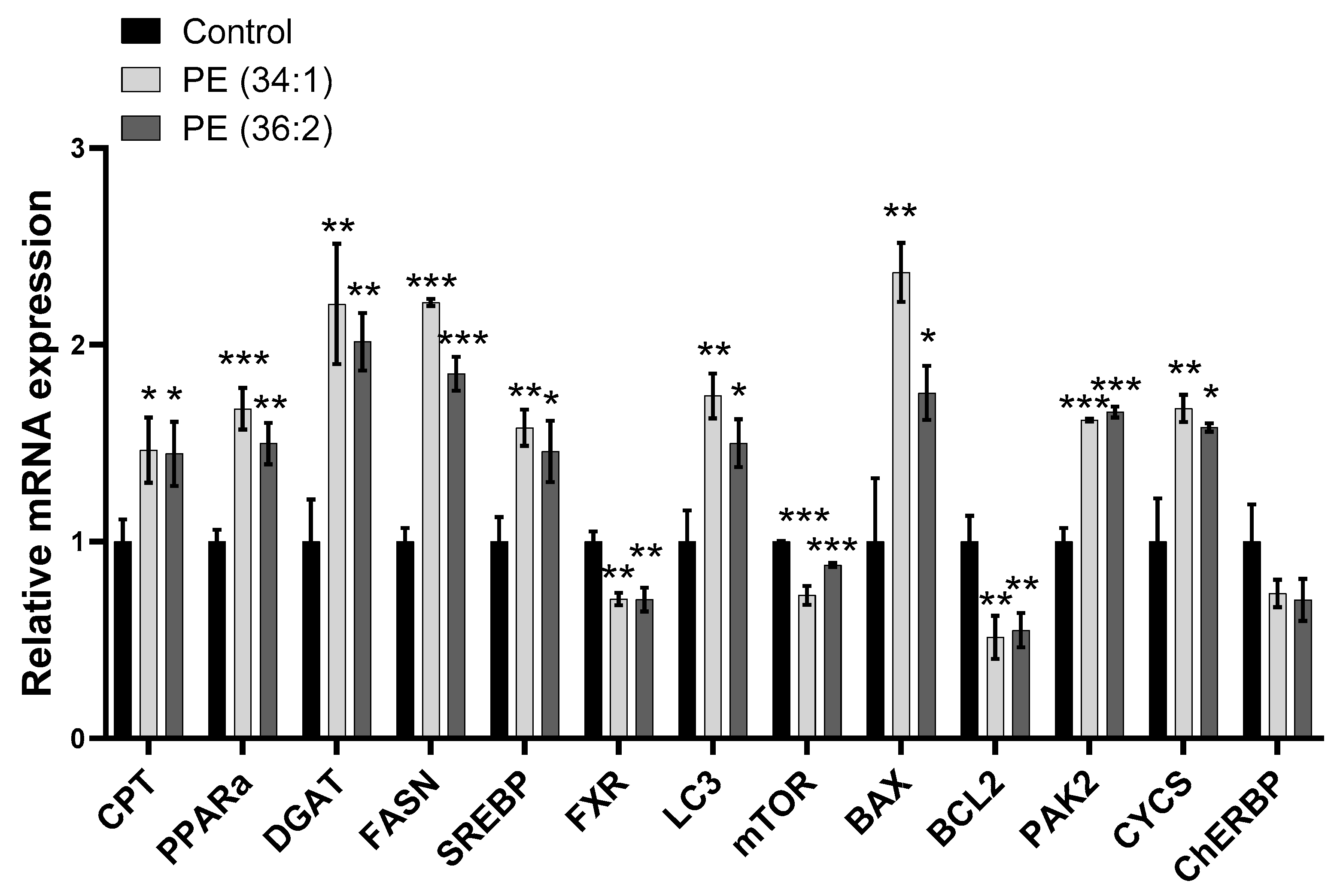
2.6. PE Altered Lipid Homeostasis Gene Expression in HepG2 Cells
2.7. PEs Altered the Expression of Mitophagy and Apoptosis/Cell Proliferation-Related Genes in HepG2 Cells
2.8. PEs Induced LX2 Cell Migration
2.9. PE Treatments Increased mRNA Expression of Inflammation and Fibrosis-Related Genes in LX2 Cells
3. Discussion
4. Materials and Methods
4.1. Materials
Subjects
4.2. Methods
4.2.1. Lipidomic Analysis
4.2.2. Cell Culture
4.2.3. Phospholipid Treatment
4.2.4. Cell Viability Assay
4.2.5. Lipid Droplet/Nucleus Staining with BODIPY/Hoechst
4.2.6. Mitochondrial ROS Production/Nucleus Staining by MitoSox™/Hoechst
4.2.7. Mitochondrial Membrane Potential (ΔΨm) and Mitochondrial Mass/Nucleus Staining
4.2.8. Cell Migration Assay
4.2.9. RNA Extraction and Reverse Transcription Polymerase Chain Reaction (RT-PCR)
4.2.10. Statistics
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Maurice, J.A.; Manousou, P. Non-alcoholic fatty liver disease. J. Clin. Med. 2018, 8, 245–250. [Google Scholar] [CrossRef] [PubMed]
- Reddy, J.K.; Rao, M.S. Lipid Metabolism and Liver Inflammation. II. Fatty liver disease and fatty acid oxidation. Am. J. Physiol. Gastrointest. Liver Physiol. 2006, 290, G852–G858. [Google Scholar] [CrossRef]
- Angulo, P. Non-alcoholic fatty liver disease. N. Engl. J. Med. 2002, 346, 1221–1231. [Google Scholar] [CrossRef]
- Anstee, Q.M.; Daly, A.K.; Day, C.P. Genetics of alcoholic and nonalcoholic fatty liver disease. Semin. Liver Dis. 2011, 31, 128–146. [Google Scholar] [CrossRef]
- Eslam, M.; Sanyal, A.J.; George, J.; International Consensus Panel. MAFLD: A Consensus-Driven Proposed Nomenclature for Metabolic Associated Fatty Liver Disease. Gastroenterology 2020, 158, 1999–2014.e1. [Google Scholar] [CrossRef]
- Eslam, M.; Newsome, P.N.; Sarin, S.K.; Anstee, Q.M.; Targher, G.; Romero-Gomez, M.; Zelber-Sagi, S.; Wong, V.W.-S.; Dufour, J.-F.; Schattenberg, J.M.; et al. A new definition for metabolic dysfunction-associated fatty liver disease: An international expert consensus statement. J. Hepatol. 2020, 73, 202–209. [Google Scholar] [CrossRef]
- Hanshaw, R.G.; Smith, B.D. New reagents for phosphatidylserine recognition and detection of apoptosis. Bioorg. Med. Chem. 2005, 13, 5035–5042. [Google Scholar] [CrossRef]
- Chaurio, R.A.; Janko, C.; Muñoz, L.E.; Frey, B.; Herrmann, M.; Gaipl, U.S. Phospholipids: Key players in apoptosis and immune regulation. Molecules 2009, 14, 4892–4914. [Google Scholar] [CrossRef]
- Vance, J.E.; Vance, D.E. The assembly of lipids into lipoproteins during secretion. Experientia 1990, 46, 560–569. [Google Scholar] [CrossRef]
- Reshetnyak, I.V. Physiological and molecular biochemical mechanisms of bile formation. World J. Gastroenterol. 2013, 19, 7341–7360. [Google Scholar] [CrossRef]
- Dabak, T.K.; Sertkaya, O.; Acar, N.; Donmez, B.O.; Ustunel, I. The Effect of Phospholipids (Surfactant) on Adhesion and Biomechanical Properties of Tendon: A Rat Achilles Tendon Repair Model. Biomed. Res. Int. 2015, 2015, 689314. [Google Scholar] [CrossRef]
- Giordano, F. Non-vesicular lipid trafficking at the endoplasmic reticulum–mitochondria interface. Biochem. Soc. Trans. 2018, 46, 437–452. [Google Scholar] [CrossRef] [PubMed]
- Xie, T.; Zhou, X.; Wang, S.; Lu, Y.; Zhu, H.; Kang, A.; Deng, H.; Xu, J.; Shen, C.; Di, L.; et al. Development and application of a comprehensive lipidomic analysis to investigate Tripterygium wilfordii-induced liver injury. Anal. Bioanal. Chem. 2016, 408, 4341–4355. [Google Scholar] [CrossRef]
- Ming, Y.-N.; Zhang, J.-Y.; Wang, X.-L.; Li, C.-M.; Ma, S.-C.; Wang, Z.-Y.; Liu, X.-L.; Li, X.-B.; Mao, Y.-M. Liquid chromatography mass spectrometry-based profiling of phosphatidylcholine and phosphatidylethanolamine in the plasma and liver of acetaminophen-induced liver injured mice. Lipids Health Dis. 2017, 16, 153. [Google Scholar] [CrossRef]
- Tiwari-Heckler, S.; Gan-Schreier, H.; Stremmel, W.; Chamulitrat, W.; Pathil, A. Circulating phospholipid patterns in NAFLD patients associated with a combination of metabolic risk factors. Nutrients 2018, 10, 649. [Google Scholar] [CrossRef] [PubMed]
- Ikon, N.; Ryan, R.O. Cardiolipin and mitochondrial cristae organization. Biochim. Biophys. Acta Biomembr. 2017, 1859, 1156–1163. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.Y.; Cheng, X.L.; Lin, R. Chapter One—Lipidomics applications for discovering biomarkers of diseases in clinical chemistry. Int. Rev. Cell Mol. Biol. 2014, 313, 1–26. [Google Scholar]
- Li, Z.; Agellon, L.B.; Allen, T.M.; Umeda, M.; Jewell, L.; Mason, A.; Vance, D.E. The ratio of phosphatidylcholine to phosphatidylethanolamine influences membrane integrity and steatohepatitis. Cell Metab. 2006, 3, 321–331. [Google Scholar] [CrossRef]
- Arendt, B.M.; Ma, D.W.; Simons, B.; Noureldin, S.A.; Therapondos, G.; Guindi, M.; Sherman, M.; Allard, J.P. Nonalcoholic fatty liver disease is associated with lower hepatic and erythrocyte ratios of phosphatidylcholine to phosphatidylethanolamine. Appl. Physiol. Nutr. Metab. 2013, 38, 334–340. [Google Scholar] [CrossRef]
- Ma, D.W.; Arendt, B.M.; Hillyer, L.M.; Fung, S.K.; McGilvray, I.; Guindi, M.; Allard, J.P. Plasma phospholipids and fatty acid composition differ between liver biopsy-proven nonalcoholic fatty liver disease and healthy subjects. Nutr. Diabetes 2016, 6, e220. [Google Scholar] [CrossRef]
- Nakatsuka, A.; Matsuyama, M.; Yamaguchi, S.; Katayama, A.; Eguchi, J.; Murakami, K.; Teshigawara, S.; Ogawa, D.; Wada, N.; Yasunaka, T.; et al. Insufficiency of phosphatidylethanolamine N-methyltransferase is risk for lean non-alcoholic steatohepatitis. Sci. Rep. 2016, 6, 21721. [Google Scholar] [CrossRef]
- Bale, G.; Vishnubhotla, R.V.; Mitnala, S.; Sharma, M.; Padaki, R.N.; Pawar, S.C.; Duvvur, R.N. Whole-Exome Sequencing Identifies a Variant in Phosphatidylethanolamine N-Methyltransferase Gene to be Associated with Lean-Nonalcoholic Fatty Liver Disease. J. Clin. Exp. Hepatol. 2019, 9, 561–568. [Google Scholar] [CrossRef]
- Tan, H.L.; Mohamed, R.; Mohamed, Z.; Zain, S.M. Phosphatidylethanolamine N-methyltransferase gene rs7946 polymorphism plays a role in risk of nonalcoholic fatty liver disease: Evidence from meta-analysis. Pharm. Genom. 2016, 26, 88–95. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; da Costa, K.A.; Fischer, L.M.; Kohlmeier, M.; Kwock, L.; Wang, S.; Zeisel, S.H. Polymorphism of the PEMT gene and susceptibility to nonalcoholic fatty liver disease (NAFLD). FASEB J. 2005, 19, 1266–1271. [Google Scholar] [CrossRef]
- Draijer, L.G.; Froon-Torenstra, D.; van Weeghel, M.; Vaz, F.M.; Bohte, A.E.; Holleboom, A.G.; Benninga, M.A.; Koot, B.G.P. Lipidomics in Nonalcoholic Fatty Liver Disease: Exploring Serum Lipids as Biomarkers for Pediatric Nonalcoholic Fatty Liver Disease. J. Pediatr. Gastroenterol. Nutr. 2020, 71, 433–439. [Google Scholar] [CrossRef]
- Kleiner, D.E.; Brunt, E.M.; Van Natta, M.; Behling, C.; Contos, M.J.; Cummings, O.W.; Ferrell, L.D.; Liu, Y.-C.; Torbenson, M.S.; Unalp-Arida, A.; et al. Nonalcoholic Steatohepatitis Clinical Research Network. Design and validation of a histological scoring system for nonalcoholic fatty liver disease. Hepatology 2005, 41, 1313–1321. [Google Scholar] [CrossRef] [PubMed]
- Wattacheril, J.; Seeley, E.H.; Angel, P.; Chen, H.; Bowen, B.P.; Lanciault, C.; Caprioli, R.M.; Abumrad, N.; Flynn, C.R. Differential Intrahepatic Phospholipid Zonation in Simple Steatosis and Nonalcoholic Steatohepatitis. PLoS ONE 2013, 8, e57165. [Google Scholar] [CrossRef] [PubMed]
- Tan, C.Y.; Virtue, S.; Murfitt, S.; Roberts, L.D.; Phua, Y.H.; Dale, M.; Griffin, J.L.; Tinahones, F.; Scherer, P.E.; Vidal-Puig, A. Adipose tissue fatty acid chain length and mono-unsaturation increases with obesity and insulin resistance. Sci. Rep. 2015, 5, 18366. [Google Scholar] [CrossRef]
- Puri, P.; Baillie, R.A.; Wiest, M.M.; Mirshahi, F.; Choudhury, J.; Cheung, O.; Sargeant, C.; Contos, M.J.; Sanyal, A.J. A lipidomic analysis of nonalcoholic fatty liver disease. Hepatology 2007, 46, 1081–1090. [Google Scholar] [CrossRef]
- Kim, M.K.; Reaven, G.M.; Kim, S.H. Dissecting the relationship between obesity and hyperinsulinemia: Role of insulin secretion and insulin clearance. Obesity 2017, 25, 378–383. [Google Scholar] [CrossRef] [PubMed]
- Yammine, S.G.; Naja, F.; Tamim, H.; Nasrallah, M.; Biessy, C.; Aglago, E.K.; Matta, M.; Romieu, I.; Gunter, M.J.; Nasreddine, L.; et al. Association between Serum Phospholipid Fatty Acid Levels and Adiposity among Lebanese Adults: A Cross-Sectional Study. Nutrients 2018, 10, 1371. [Google Scholar] [CrossRef] [PubMed]
- Law, S.H.; Chan, M.L.; Marathe, G.K.; Parveen, F.; Chen, C.H.; Ke, L.-Y. An Updated Review of Lysophosphatidylcholine Metabolism in Human Diseases. Int. J. Mol. Sci. 2019, 20, 1149. [Google Scholar] [CrossRef]
- Nass, K.J.; van den Berg, E.H.; Gruppen, E.G.; Dullaart, R.P.F. Plasma lecithin: Cholesterol acyltransferase and phospholipid transfer protein activity independently associate with nonalcoholic fatty liver disease. Eur. J. Clin. Investig. 2018, 48, e12988. [Google Scholar] [CrossRef]
- Kakisaka, K.; Suzuki, Y.; Fujiwara, Y.; Suzuki, A.; Kanazawa, J.; Takikawa, Y. Caspase-independent hepatocyte death: A result of the decrease of lysophosphatidylcholine acyltransferase 3 in non-alcoholic steatohepatitis. J. Gastroenterol. Hepatol. 2019, 34, 1256–1262. [Google Scholar] [CrossRef] [PubMed]
- Wan, S.; van der Veen, J.N.; Bakala N’Goma, J.C.; Nelson, R.C.; Vance, D.E.; Jacobs, R.L. Hepatic PEMT activity mediates liver health, weight gain, and insulin resistance. FASEB J. 2019, 33, 10986–10995. [Google Scholar] [CrossRef] [PubMed]
- Müller, F.A.; Sturla, S.J. Human in vitro models of nonalcoholic fatty liver disease. Curr. Opin. Toxicol. 2019, 16, 9–16. [Google Scholar] [CrossRef]
- Kanuri, G.; Bergheim, L. In Vitro and in Vivo Models of Non-Alcoholic Fatty Liver Disease (NAFLD). Int. J. Mol. Sci. 2013, 14, 11963–11980. [Google Scholar] [CrossRef]
- Van der Veen, J.N.; Kennelly, J.P.; Wan, S.; Vance, J.E.; Vance, D.E.; Jacobs, R.L. The critical role of phosphatidylcholine and phosphatidylethanolamine metabolism in health and disease. Biochim. Biophys. Acta (BBA)—Biomembr. 2017, 1859, 1558–1572. [Google Scholar] [CrossRef]
- Grishko, V.; Rachek, L.; Musiyenko, S.; Ledoux, S.P.; Wilson, G.L. Involvement of mtDNA damage in free fatty acid-induced apoptosis. Free Radic. Biol. Med. 2005, 38, 755–762. [Google Scholar] [CrossRef] [PubMed]
- Chiu, H.-C.; Kovacs, A.; Ford, D.A.; Hsu, F.-F.; Garcia, R.; Herrero, P.; Saffitz, J.E.; Schaffer, J.E. A novel mouse model of lipotoxic cardiomyopathy. J. Clin. Investig. 2001, 107, 813–822. [Google Scholar] [CrossRef]
- Yao, Y.; Huang, C.; Li, Z.F.; Wang, A.Y.; Liu, L.Y. Exogenous phosphatidylethanolamine induces apoptosis of human hepatoma HepG2 cells via the bcl-2/bax pathway. World J. Gastroenterol. 2009, 15, 1751–1758. [Google Scholar] [CrossRef]
- Xue, L.; Li, M.; Chen, T.; Sun, H.; Zhu, J.; Li, X.; Wu, F.; Wang, B.; Li, J.; Chen, Y. PE-induced apoptosis in SMMC-7721 cells: Involvement of Erk and Stat signaling pathways. Int. J. Mol. Med. 2014, 34, 119–129. [Google Scholar] [CrossRef] [PubMed]
- Arendt, B.M.; Comelli, E.M.; Ma, D.W.; Lou, W.; Teterina, A.; Kim, T.; Fung, S.K.; Wong, D.K.; McGilvray, I.; Fischer, S.E.; et al. Altered hepatic gene expression in nonalcoholic fatty liver disease is associated with lower hepatic n-3 and n-6 polyunsaturated fatty acids. Hepatology 2015, 61, 1565–1577. [Google Scholar] [CrossRef] [PubMed]
- Takahara, Y.; Takahashi, M.; Wagatsuma, H.; Yokoya, F.; Zhang, Q.W.; Yamaguchi, M.; Aburatani, H.; Kawada, N. Gene expression profiles of hepatic cell-type specific marker genes in progression of liver fibrosis. World J. Gastroenterol. 2006, 12, 6473–6499. [Google Scholar] [CrossRef]
- Gawrieh, S.; Marion, M.C.; Komorowski, R.; Wallace, J.; Charlton, M.; Kissebah, A.; Langefeld, C.D.; Olivier, M. Genetic variation in the peroxisome proliferator activated receptor-gamma gene is associated with histologically advanced NAFLD. Dig. Dis. Sci. 2012, 57, 952–957. [Google Scholar] [CrossRef]
- Wang, L.; Athinarayanan, S.; Jiang, G.; Chalasani, N.; Zhang, M.; Liu, W. Fatty acid desaturase 1 gene polymorphisms control human hepatic lipid composition. Hepatology 2015, 61, 119–128. [Google Scholar] [CrossRef] [PubMed]
- Devaiah, S.P.; Roth, M.R.; Baughman, E.; Li, M.; Tamura, P.; Jeannotte, R.; Welti, R.; Wang, X. Quantitative profiling of polar glycerolipid species from organs of wild-type Arabidopsis and a phospholipase Dalpha1 knockout mutant. Phytochemistry 2006, 67, 1907–1924. [Google Scholar] [CrossRef]
- Folch, J.; Lees, M.; Sloane Stanley, G.H. A simple method for the isolation and purification of total lipids from animal tissues. J. Biol. Chem. 1957, 226, 497–509. [Google Scholar] [CrossRef]
- Baumann, J.M.; Kokabee, L.; Wang, X.; Sun, Y.; Wong, J.; Conklin, D.S. Metabolic Assays for Detection of Neutral Fat Stores. Bio-Protocol 2015, 5, e1511. [Google Scholar] [CrossRef]
- Justus, C.R.; Leffler, N.; Ruiz-Echevarria, M.; Yang, L.V. In vitro cell migration and invasion assays. J. Vis. Exp. 2014, 88, e51046. [Google Scholar]






| Control (n = 80) | Simple Steatosis (n = 93) | Borderline NASH (n = 16) | NASH (n = 14) | All (n = 203) | p * | |
|---|---|---|---|---|---|---|
| Age (years)–Median (range) | 42 (20, 66) | 46 (20, 66) | 43.5 (32, 58) | 49.5 (26, 58) | 44 (20, 66) | 0.348 |
| Gender–no. (%) | 0.091 | |||||
| Female | 73 (91) | 86 (92) | 14 (88) | 10 (71) | 183 (90) | |
| Male | 7 (9) | 7 (8) | 2 (12) | 4 (29) | 20 (10) | |
| Race–no. (%) | 0.545 | |||||
| EA | 69 (86) | 77 (83) | 12 (75) | 13 (93) | 171 (84) | |
| Non-EA | 11 (14) | 16 (17) | 4 (25) | 1 (7) | 32 (16) | |
| Height (feet)–Median (range) | 5.5 (4.92, 6.17) | 5.42 (4.92, 6.25) | 5.42 (5, 5.67) | 5.54 (4.75, 6.25) | 5.42 (4.75, 6.25) | 0.248 |
| Weight (lbs)–Median (range) | 278.5 (194, 481) | 291 (194, 381) | 288 (207, 502) | 310.5 (217, 416) | 281.5 (194, 502) | 0.768 |
| Diabetes Mellitus–no. (%) ‡ | <0.001 | |||||
| No | 68 (85) | 61 (66) | 8 (50) | 5 (36) | 142 (70) | |
| Yes | 12 (15) | 32 (34) ‡ | 8 (50) ‡ | 9 (64) ‡ | 61 (30) | |
| Hypertension–no. (%) | 0.148 | |||||
| No | 43 (54) | 45 (48) | 9 (56) | 3 (21) | 100 (49) | |
| Yes | 37 (46) | 48 (52) | 7 (44) | 11 (79) | 103 (51) | |
| Dyslipidemia–no. (%) | 0.587 | |||||
| No | 59 (74) | 68 (73) | 10 (62) | 12 (86) | 149 (73) | |
| Yes | 21 (26) | 25 (27) | 6 (38) | 2 (14) | 54 (27) | |
| Depression–no. (%) | 0.673 | |||||
| No | 45 (56) | 59 (63) | 10 (62) | 7 (50) | 121 (60) | |
| Yes | 35 (44) | 34 (37) | 6 (38) | 7 (50) | 82 (40) |
| Control (n = 80) | Simple Steatosis (n = 93) | Borderline NASH (n = 16) | NASH (n = 14) | All (n = 203) | p * | |
|---|---|---|---|---|---|---|
| Steatosis grade–no. (%) | <0.001 | |||||
| <5% | 80 (100) | 0 (0) | 0 (0) | 1 (7) | 81 (40) | |
| 5–33% | 0 (0) | 58 (62) | 9 (56) | 4 (29) | 71 (35) | |
| 33–66% | 0 (0) | 24 (26) | 6 (38) | 6 (43) | 36 (18) | |
| >66% | 0 (0) | 11 (12) | 1 (6) | 3 (21) | 15 (7) | |
| Steatosis distribution–no. (%) | <0.001 | |||||
| Zone 3 | 76 (95) | 32 (34) | 3 (19) | 6 (43) | 117 (58) | |
| Zone 1 | 0 (0) | 2 (2) | 1 (6) | 1 (7) | 4 (2) | |
| Azonal | 2 (2) | 39 (42) | 10 (62) | 3 (21) | 54 (27) | |
| Panacinar | 0 (0) | 20 (22) | 2 (12) | 4 (29) | 26 (13) | |
| Missing | 2 (2) | 0 (0) | 0 (0) | 0 (0) | 2 (1) | |
| Microvesicular steatosis–no. (%) | <0.001 | |||||
| Not present | 78 (98) | 22 (24) | 1 (6) | 1 (7) | 102 (50) | |
| Present | 2 (2) | 70 (75) | 15 (94) | 13 (93) | 100 (49) | |
| Missing | 0 (0) | 1 (1) | 0 (0) | 0 (0) | 1 (0) | |
| Ballooning–no. (%) | <0.001 | |||||
| None | 80 (100) | 93 (100) | 10 (62) | 5 (36) | 188 (93) | |
| Few balloon cells | 0 (0) | 0 (0) | 6 (38) | 8 (57) | 14 (7) | |
| Many cells/prominent ballooning | 0 (0) | 0 (0) | 0 (0) | 1 (7) | 1 (0) | |
| Lobular inflammation–no. (%) | <0.001 | |||||
| No foci | 80 (100) | 93 (100) | 13 (81) | 9 (64) | 195 (96) | |
| 2 foci per 200 field | 0 (0) | 0 (0) | 3 (19) | 5 (36) | 8 (4) | |
| Portal inflammation–no. (%) | <0.001 | |||||
| None | 74 (92) | 68 (73) | 6 (38) | 2 (14) | 150 (74) | |
| Mild | 6 (8) | 24 (26) | 9 (56) | 10 (71) | 49 (24) | |
| Moderate | 0 (0) | 1 (1) | 1 (6) | 2 (14) | 4 (2) | |
| Fibrosis–no. (%) | <0.001 | |||||
| None | 80 (100) | 91 (98) | 8 (50) | 5 (36) | 184 (91) | |
| Perisinusoidal or periportal | 0 (0) | 2 (2) | 4 (25) | 6 (43) | 12 (6) | |
| Perisinusoidal and portal/periportal | 0 (0) | 0 (0) | 1 (6) | 0 (0) | 1 (0) | |
| Cirrhosis | 0 (0) | 0 (0) | 3 (19) | 3 (21) | 6 (3) | |
| Mallory bodies–no. (%) | <0.001 | |||||
| None to rare | 80 (100) | 93 (100) | 15 (94) | 11 (79) | 199 (98) | |
| Many | 0 (0) | 0 (0) | 1 (6) | 3 (21) | 4 (2) | |
| NAS score–median (range) ‡ | 0 (0, 0) | 1 (1, 3) | 2 (1, 3) | 3 (0, 5) | 1 (0, 5) | <0.001 |
| Median (Range) | p * | |||||
|---|---|---|---|---|---|---|
| Control (n = 80) | Simple Steatosis (n = 93) | Borderline NASH (n = 16) | NASH (n = 14) | All (n = 203) | ||
| T Chol | 169.5 (104, 278) | 178.5 (112, 268) | 174 (107, 295) | 171 (117, 266) | 173 (104, 295) | 0.702 |
| Missing | 0 | 3 | 0 | 1 | 4 | |
| TG | 107 (41, 600) | 147.5 (49, 693) | 144.5 (63, 968) | 218 (66, 823) | 126 (41, 968) | <0.001 |
| Missing | 0 | 3 | 0 | 1 | 4 | |
| HDL | 45.5 (30, 81) | 41 (24, 69) | 38.5 (26, 64) | 40 (29, 61) | 43 (24, 81) | 0.001 |
| Missing | 0 | 3 | 0 | 1 | 4 | |
| LDL | 99.5 (38, 163) | 103 (46, 188) | 108 (49, 138) | 91.5 (43, 154) | 103 (38, 188) | 0.895 |
| Missing | 2 | 6 | 2 | 4 | 14 | |
| Albumin | 4.2 (3.8, 64) | 4.2 (0.1, 73) | 4.2 (3.5, 5.1) | 4.25 (3.8, 4.5) | 4.2 (0.1, 73) | 0.968 |
| Missing | 1 | 2 | 0 | 0 | 3 | |
| HbA1c | 6.3 (5.7, 10.1) | 6.5 (3.1, 13.9) | 6.8 (5.5, 11.4) | 6.5 (5.3, 9.7) | 6.5 (3.1, 13.9) | 0.612 |
| Missing | 70 | 62 | 6 | 7 | 145 | |
| Insulin level | 20.43 (7.04, 44.58) | 25.56 (7.14, 112) | 24.13 (11.76, 178.9) | 32.88 (11.47, 52.69) | 24.09 (7.04, 178.9) | 0.058 |
| Missing | 44 | 39 | 7 | 5 | 95 | |
| ALT | 17 (8, 74) | 22 (9, 65) | 40.5 (10, 99) | 30 (7, 85) | 20 (7, 99) | <0.001 |
| Missing | 1 | 2 | 0 | 0 | 3 | |
| AST | 16 (7, 55) | 19 (11, 76) | 35.5 (15, 127) | 29.5 (15, 55) | 19 (7, 127) | <0.001 |
| Missing | 1 | 2 | 0 | 0 | 3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shama, S.; Jang, H.; Wang, X.; Zhang, Y.; Shahin, N.N.; Motawi, T.K.; Kim, S.; Gawrieh, S.; Liu, W. Phosphatidylethanolamines Are Associated with Nonalcoholic Fatty Liver Disease (NAFLD) in Obese Adults and Induce Liver Cell Metabolic Perturbations and Hepatic Stellate Cell Activation. Int. J. Mol. Sci. 2023, 24, 1034. https://doi.org/10.3390/ijms24021034
Shama S, Jang H, Wang X, Zhang Y, Shahin NN, Motawi TK, Kim S, Gawrieh S, Liu W. Phosphatidylethanolamines Are Associated with Nonalcoholic Fatty Liver Disease (NAFLD) in Obese Adults and Induce Liver Cell Metabolic Perturbations and Hepatic Stellate Cell Activation. International Journal of Molecular Sciences. 2023; 24(2):1034. https://doi.org/10.3390/ijms24021034
Chicago/Turabian StyleShama, Samaa, Hyejeong Jang, Xiaokun Wang, Yang Zhang, Nancy Nabil Shahin, Tarek Kamal Motawi, Seongho Kim, Samer Gawrieh, and Wanqing Liu. 2023. "Phosphatidylethanolamines Are Associated with Nonalcoholic Fatty Liver Disease (NAFLD) in Obese Adults and Induce Liver Cell Metabolic Perturbations and Hepatic Stellate Cell Activation" International Journal of Molecular Sciences 24, no. 2: 1034. https://doi.org/10.3390/ijms24021034
APA StyleShama, S., Jang, H., Wang, X., Zhang, Y., Shahin, N. N., Motawi, T. K., Kim, S., Gawrieh, S., & Liu, W. (2023). Phosphatidylethanolamines Are Associated with Nonalcoholic Fatty Liver Disease (NAFLD) in Obese Adults and Induce Liver Cell Metabolic Perturbations and Hepatic Stellate Cell Activation. International Journal of Molecular Sciences, 24(2), 1034. https://doi.org/10.3390/ijms24021034








