Structural Insights Uncover the Specific Phosphoinositide Recognition by the PH1 Domain of Arap3
Abstract
1. Introduction
2. Results
2.1. The PH1 Domain of Arap3 Is Sufficient to Bind PIPs and Prefers PI(3,4,5)P3
2.2. Crystal Structures of Unliganded and diC4-PI(3,4,5)P3-Bound Arap3-PH1 Domain
2.3. NMR Characterize of Arap3-PH1 Domain Binding to PI(3,4,5)P3 and PI(4,5)P2 Head Groups in Solution
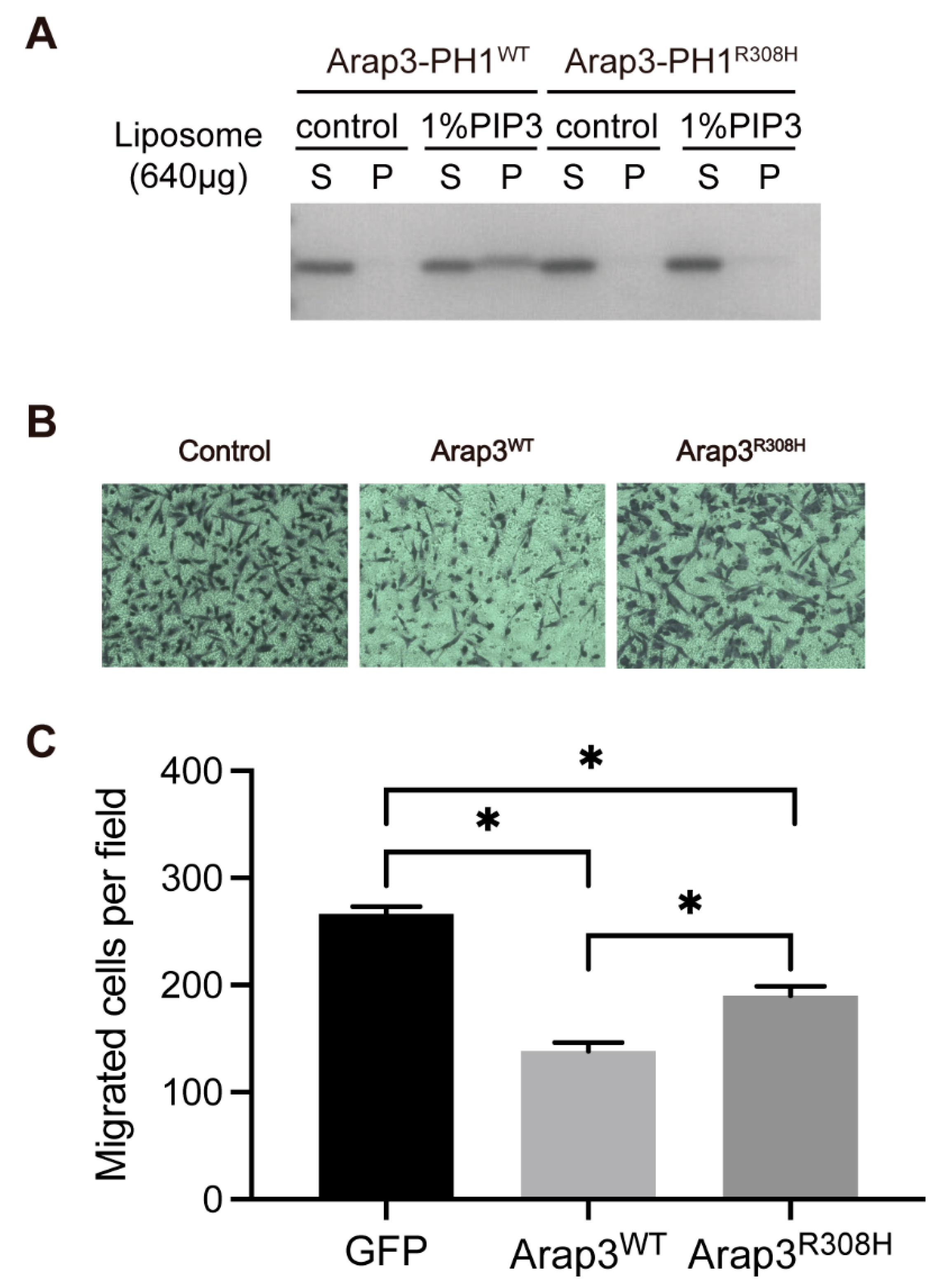
2.4. The PIPs-Binding Ability of Arap3-PH1 Domain Is Required for Arap3 to Inhibit Breast Cancer Cell Invasion In Vitro
3. Discussion
4. Materials and Methods
4.1. Plasmid Constructions
4.2. Protein Expression and Purification
4.3. Liposome Preparation and Liposome Pull-Down Assay
4.4. Surface Plasmon Resonance (SPR)
4.5. Crystallization and Structure
4.6. Data Collection and Structure Determination
4.7. Nuclear Magnetic Resonance (NMR) Experiments
4.8. Cell Cultures, Transfection
4.9. Western Blotting and Antibodies
4.10. Matrigel Invasion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wymann, M.P.; Schneiter, R. Lipid signalling in disease. Nat. Rev. Mol. Cell Biol. 2008, 9, 162–176. [Google Scholar] [CrossRef] [PubMed]
- Cho, W.H.; Stahelin, R.V. Membrane-protein interactions in cell signaling and membrane trafficking. Annu. Rev. Biophys. Biomol. 2005, 34, 119–151. [Google Scholar] [CrossRef] [PubMed]
- Balla, T. Inositol-lipid binding motifs: Signal integrators through protein-lipid and protein-protein interactions. J. Cell Sci. 2005, 118, 2093–2104. [Google Scholar] [CrossRef]
- Stahelin, R.V.; Scott, J.L.; Frick, C.T. Cellular and molecular interactions of phosphoinositides and peripheral proteins. Chem. Phys. Lipids 2014, 182, 3–18. [Google Scholar] [CrossRef] [PubMed]
- Lemmon, M.A. Membrane recognition by phospholipid-binding domains. Nat. Rev. Mol. Cell Biol. 2008, 9, 99–111. [Google Scholar] [CrossRef] [PubMed]
- Vanhaesebroeck, B.; Guillermet-Guibert, J.; Graupera, M.; Bilanges, B. The emerging mechanisms of isoform-specific PI3K signalling. Nat. Rev. Mol. Cell Biol. 2010, 11, 329–341. [Google Scholar] [CrossRef]
- Leevers, S.J.; Vanhaesebroeck, B.; Waterfield, M.D. Signalling through phosphoinositide 3-kinases: The lipids take centre stage. Curr. Opin. Cell Biol. 1999, 11, 219–225. [Google Scholar] [CrossRef] [PubMed]
- Vanhaesebroeck, B.; Leevers, S.J.; Ahmadi, K.; Timms, J.; Katso, R.; Driscoll, P.C.; Woscholski, R.; Parker, P.J.; Waterfield, M.D. Synthesis and function of 3-phosphorylated inositol lipids. Annu. Rev. Biochem. 2001, 70, 535–602. [Google Scholar] [CrossRef]
- Pirola, L.; Zvelebil, M.J.; Bulgarelli-Leva, G.; Van Obberghen, E.; Waterfield, M.D.; Wymann, M.P. Activation loop sequences confer substrate specificity to phosphoinositide 3-kinase alpha (PI3Kalpha). Functions of lipid kinase-deficient PI3Kalpha in signaling. J. Biol. Chem. 2001, 276, 21544–21554. [Google Scholar] [CrossRef]
- Gulluni, F.; De Santis, M.C.; Margaria, J.P.; Martini, M.; Hirsch, E. Class II PI3K Functions in Cell Biology and Disease. Trends Cell Biol. 2019, 29, 339–359. [Google Scholar] [CrossRef]
- Krugmann, S.; Anderson, K.E.; Ridley, S.H.; Risso, N.; McGregor, A.; Coadwell, J.; Davidson, K.; Eguinoa, A.; Ellson, C.D.; Lipp, P.; et al. Identification of ARAP3, a novel PI3K effector regulating both Arf and Rho GTPases, by selective capture on phosphoinositide affinity matrices. Mol. Cell 2002, 9, 95–108. [Google Scholar] [CrossRef] [PubMed]
- Krugmann, S.; Williams, R.; Stephens, L.; Hawkins, P.T. ARAP3 is a PI3K- and rap-regulated GAP for RhoA. Curr. Biol. 2004, 14, 1380–1384. [Google Scholar] [CrossRef] [PubMed]
- Ha, V.L.; Bharti, S.; Inoue, H.; Vass, W.C.; Campa, F.; Nie, Z.Z.; de Gramont, A.; Ward, Y.; Randazzo, P.A. ASAP3 is a focal adhesion-associated Arf GAP that functions in cell migration and invasion. J. Biol. Chem. 2008, 283, 14915–14926. [Google Scholar] [CrossRef] [PubMed]
- Krugmann, S.; Andrews, S.; Stephens, L.; Hawkins, P.T. ARAP3 is essential for formation of lamellipodia after growth factor stimulation. J. Cell Sci. 2006, 119, 425–432. [Google Scholar] [CrossRef] [PubMed]
- Gambardella, L.; Anderson, K.E.; Nussbaum, C.; Segonds-Pichon, A.; Margarido, T.; Norton, L.; Ludwig, T.; Sperandio, M.; Hawkins, P.T.; Stephens, L.; et al. The GTPase-activating protein ARAP3 regulates chemotaxis and adhesion-dependent processes in neutrophils. Blood 2011, 118, 1087–1098. [Google Scholar] [CrossRef]
- Gambardella, L.; Hemberger, M.; Hughes, B.; Zudaire, E.; Andrews, S.; Vermeren, S. PI3K signaling through the dual GTPase-activating protein ARAP3 is essential for developmental angiogenesis. Sci. Signal 2010, 3, ra76. [Google Scholar] [CrossRef]
- Kartopawiro, J.; Bower, N.I.; Karnezis, T.; Kazenwadel, J.; Betterman, K.L.; Lesieur, E.; Koltowska, K.; Astin, J.; Crosier, P.; Vermeren, S.; et al. Arap3 is dysregulated in a mouse model of hypotrichosis-lymphedema-telangiectasia and regulates lymphatic vascular development. Hum. Mol. Genet. 2014, 23, 1286–1297. [Google Scholar] [CrossRef]
- Yagi, R.; Tanaka, M.; Sasaki, K.; Kamata, R.; Nakanishi, Y.; Kanai, Y.; Sakai, R. ARAP3 inhibits peritoneal dissemination of scirrhous gastric carcinoma cells by regulating cell adhesion and invasion. Oncogene 2011, 30, 1413–1421. [Google Scholar] [CrossRef]
- Loskutov, Y.V.; Kozyulina, P.Y.; Kozyreva, V.K.; Ice, R.J.; Jones, B.C.; Roston, T.J.; Smolkin, M.B.; Ivanov, A.V.; Wysolmerski, R.B.; Pugacheva, E.N. NEDD9/Arf6-dependent endocytic trafficking of matrix metalloproteinase 14: A novel mechanism for blocking mesenchymal cell invasion and metastasis of breast cancer. Oncogene 2015, 34, 3662–3675. [Google Scholar] [CrossRef]
- Han, J.J.; Du, B.R.; Zhang, C.H. Bioinformatic analysis of prognostic value of ARAP3 in breast cancer and the associated signaling pathways. Eur. Rev. Med. Pharmacol. Sci. 2017, 21, 2405–2412. [Google Scholar]
- Krugmann, S.; Stephens, L.; Hawkins, P.T. Purification of ARAP3 and characterization of GAP activities. Method Enzymol. 2006, 406, 91–103. [Google Scholar] [CrossRef]
- Song, Y.; Jiang, J.; Vermeren, S.; Tong, W. ARAP3 functions in hematopoietic stem cells. PLoS ONE 2014, 9, e116107. [Google Scholar] [CrossRef] [PubMed]
- Craig, H.E.; Coadwell, J.; Guillou, H.; Vermeren, S. ARAP3 binding to phosphatidylinositol-(3,4,5)-trisphosphate depends on N-terminal tandem PH domains and adjacent sequences. Cell Signal 2010, 22, 257–264. [Google Scholar] [CrossRef]
- Kavran, J.M.; Klein, D.E.; Lee, A.; Falasca, M.; Isakoff, S.J.; Skolnik, E.Y.; Lemmon, M.A. Specificity and promiscuity in phosphoinositide binding by pleckstrin homology domains. J. Biol. Chem. 1998, 273, 30497–30508. [Google Scholar] [CrossRef]
- Park, W.S.; Heo, W.D.; Whalen, J.H.; O’Rourke, N.A.; Bryan, H.M.; Meyer, T.; Teruel, M.N. Comprehensive identification of PIP3-regulated PH domains from C. elegans to H. sapiens by model prediction and live imaging. Mol. Cell 2008, 30, 381–392. [Google Scholar] [CrossRef] [PubMed]
- Singh, N.; Reyes-Ordonez, A.; Compagnone, M.A.; Moreno, J.F.; Leslie, B.J.; Ha, T.; Chen, J. Redefining the specificity of phosphoinositide-binding by human PH domain-containing proteins. Nat. Commun. 2021, 12, 4339. [Google Scholar] [CrossRef]
- Moravcevic, K.; Oxley, C.L.; Lemmon, M.A. Conditional peripheral membrane proteins: Facing up to limited specificity. Structure 2012, 20, 15–27. [Google Scholar] [CrossRef] [PubMed]
- Cash, J.N.; Davis, E.M.; Tesmer, J.J.G. Structural and Biochemical Characterization of the Catalytic Core of the Metastatic Factor P-Rex1 and Its Regulation by PtdIns(3,4,5)P3. Structure 2016, 24, 730–740. [Google Scholar] [CrossRef]
- Posor, Y.; Kampyli, C.; Bilanges, B.; Ganguli, S.; Koch, P.A.; Wallroth, A.; Morelli, D.; Jenkins, M.; Alliouachene, S.; Deltcheva, E.; et al. Local synthesis of the phosphatidylinositol-3,4-bisphosphate lipid drives focal adhesion turnover. Dev. Cell 2022, 57, 1694–1711.e7. [Google Scholar] [CrossRef]
- Le Huray, K.I.P.; Wang, H.; Sobott, F.; Kalli, A.C. Systematic simulation of the interactions of pleckstrin homology domains with membranes. Sci. Adv. 2022, 8, eabn6992. [Google Scholar] [CrossRef]
- Vonkova, I.; Saliba, A.E.; Deghou, S.; Anand, K.; Ceschia, S.; Doerks, T.; Galih, A.; Kugler, K.G.; Maeda, K.; Rybin, V.; et al. Lipid Cooperativity as a General Membrane-Recruitment Principle for PH Domains. Cell Rep. 2015, 12, 1519–1530. [Google Scholar] [CrossRef] [PubMed]
- Ni, T.; Kalli, A.C.; Naughton, F.B.; Yates, L.A.; Naneh, O.; Kozorog, M.; Anderluh, G.; Sansom, M.S.; Gilbert, R.J. Structure and lipid-binding properties of the kindlin-3 pleckstrin homology domain. Biochem. J. 2017, 474, 539–556. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, E.; Domanski, J.; Naughton, F.B.; Best, R.B.; Kalli, A.C.; Stansfeld, P.J.; Sansom, M.S.P. Multiple lipid binding sites determine the affinity of PH domains for phosphoinositide-containing membranes. Sci. Adv. 2020, 6, eaay5736. [Google Scholar] [CrossRef] [PubMed]
- Aleshin, A.E.; Yao, Y.; Iftikhar, A.; Bobkov, A.A.; Yu, J.; Cadwell, G.; Klein, M.G.; Dong, C.; Bankston, L.A.; Liddington, R.C.; et al. Structural basis for the association of PLEKHA7 with membrane-embedded phosphatidylinositol lipids. Structure 2021, 29, 1029–1039.e3. [Google Scholar] [CrossRef] [PubMed]
- Soubias, O.; Pant, S.; Heinrich, F.; Zhang, Y.; Roy, N.S.; Li, J.; Jian, X.; Yohe, M.E.; Randazzo, P.A.; Losche, M.; et al. Membrane surface recognition by the ASAP1 PH domain and consequences for interactions with the small GTPase Arf1. Sci. Adv. 2020, 6, eabd1882. [Google Scholar] [CrossRef]
- Ravula, T.; Hardin, N.Z.; Ramadugu, S.K.; Cox, S.J.; Ramamoorthy, A. Formation of pH-Resistant Monodispersed Polymer-Lipid Nanodiscs. Angew. Chem. Int. Ed. Engl. 2018, 57, 1342–1345. [Google Scholar] [CrossRef]
- Barnaba, C.; Ramamoorthy, A. Picturing the Membrane-assisted Choreography of Cytochrome P450 with Lipid Nanodiscs. Chemphyschem 2018, 19, 2603–2613. [Google Scholar] [CrossRef]
- Barnaba, C.; Gentry, K.; Sumangala, N.; Ramamoorthy, A. The catalytic function of cytochrome P450 is entwined with its membrane-bound nature. F1000Res 2017, 6, 662. [Google Scholar] [CrossRef]
- Sahoo, B.R.; Genjo, T.; Bekier, M.; Cox, S.J.; Stoddard, A.K.; Ivanova, M.; Yasuhara, K.; Fierke, C.A.; Wang, Y.; Ramamoorthy, A. Alzheimer’s amyloid-beta intermediates generated using polymer-nanodiscs. Chem. Commun. 2018, 54, 12883–12886. [Google Scholar] [CrossRef]
- Wu, B.; Wang, F.S.; Zhang, J.H.; Zhang, Z.Y.; Qin, L.Y.; Peng, J.H.; Li, F.D.; Liu, J.P.; Lu, G.W.; Gong, Q.G.; et al. Identification and structural basis for a novel interaction between Vav2 and Arap3. J. Struct. Biol. 2012, 180, 84–95. [Google Scholar] [CrossRef]
- Ge, L.; Wu, B.; Zhang, Y.; Wang, J.; Zhao, H.; Wang, J. Biochemical and NMR characterization of the interactions of Vav2-SH2 domain with lipids and the EphA2 juxtamembrane region on membrane. Biochem. J. 2020, 477, 3791–3801. [Google Scholar] [CrossRef] [PubMed]
- Otwinowski, Z.; Minor, W. [20] Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997, 276, 307–326. [Google Scholar] [CrossRef] [PubMed]
- Collaborative Computational Project, Number 4. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 1994, 50, 760–763. [Google Scholar] [CrossRef]
- McCoy, A.J.; Grosse-Kunstleve, R.W.; Adams, P.D.; Winn, M.D.; Storoni, L.C.; Read, R.J. Phaser crystallographic software. J. Appl. Crystallogr. 2007, 40, 658–674. [Google Scholar] [CrossRef]
- Murshudov, G.N.; Vagin, A.A.; Dodson, E.J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 1997, 53, 240–255. [Google Scholar] [CrossRef] [PubMed]
- Adams, P.D.; Afonine, P.V.; Bunkoczi, G.; Chen, V.B.; Davis, I.W.; Echols, N.; Headd, J.J.; Hung, L.W.; Kapral, G.J.; Grosse-Kunstleve, R.W.; et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 213–221. [Google Scholar] [CrossRef]
- Emsley, P.; Cowtan, K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004, 60, 2126–2132. [Google Scholar] [CrossRef]
- Delaglio, F.; Grzesiek, S.; Vuister, G.W.; Zhu, G.; Pfeifer, J.; Bax, A. Nmrpipe—A Multidimensional Spectral Processing System Based on Unix Pipes. J. Biomol. NMR 1995, 6, 277–293. [Google Scholar] [CrossRef]
- Hashimoto, S.; Onodera, Y.; Hashimoto, A.; Tanaka, M.; Hamaguchi, M.; Yamada, A.; Sabe, H. Requirement for Arf6 in breast cancer invasive activities. Proc. Natl. Acad. Sci. USA 2004, 101, 6647–6652. [Google Scholar] [CrossRef]



| Arap3-PH1 | Arap3-PH1/diC4-PI(3,4,5)P3 Complex | |
|---|---|---|
| Data Collection | ||
| Wavelength (Å) | 0.9798 | 0.9798 |
| Space group | P6122 | P6122 |
| Cell dimension | ||
| a, b, c (Å) | 102.63, 102.63, 45.51 | 74.15, 74.15, 109.85 |
| α, β, γ (°) | 90, 90, 120 | 90, 90, 120 |
| Resolution * (Å) | 50.00–2.10 (2.14–2.10) | 50.00–3.30 (3.36–3.30) |
| Rmerge (%) | 11.3 (35.7) | 19.6 (48.7) |
| I/σI | 20.5 (7.0) | 16.0 (5.0) |
| Completeness (%) | 99.4 (100.0) | 100.0 (100.0) |
| Redundancy | 8.9 (8.4) | 9.6 (9.5) |
| Refinement | ||
| No. reflections used/free | 8150/424 | 2633/251 |
| Resolution range (Å) | 33.60–2.10 | 32.11–3.30 |
| Rwork/Rfree (%) | 18.45/23.00 | 22.44/27.6 |
| R.m.s. deviations | ||
| Bonds lengths (Å) | 0.009 | 0.004 |
| Bond angles (°) | 0.976 | 0.644 |
| B-factors (Å2) | ||
| Protein | 37.28 | 57.14 |
| Non-protein | 37.04 | 61.53 |
| No. atoms | ||
| Protein | 799 | 795 |
| Non-protein | 53 | 43 |
| Ramachandran plot | ||
| Favored/allowed/outlier (%) | 98.97/1.03/0 | 94.79/5.21/0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.; Ge, L.; Xu, L.; Liu, Y.; Wang, J.; Liu, C.; Zhao, H.; Xing, L.; Wang, J.; Wu, B. Structural Insights Uncover the Specific Phosphoinositide Recognition by the PH1 Domain of Arap3. Int. J. Mol. Sci. 2023, 24, 1125. https://doi.org/10.3390/ijms24021125
Zhang Y, Ge L, Xu L, Liu Y, Wang J, Liu C, Zhao H, Xing L, Wang J, Wu B. Structural Insights Uncover the Specific Phosphoinositide Recognition by the PH1 Domain of Arap3. International Journal of Molecular Sciences. 2023; 24(2):1125. https://doi.org/10.3390/ijms24021125
Chicago/Turabian StyleZhang, Youjia, Liang Ge, Li Xu, Yongrui Liu, Jiarong Wang, Chongxu Liu, Hongxin Zhao, Lei Xing, Junfeng Wang, and Bo Wu. 2023. "Structural Insights Uncover the Specific Phosphoinositide Recognition by the PH1 Domain of Arap3" International Journal of Molecular Sciences 24, no. 2: 1125. https://doi.org/10.3390/ijms24021125
APA StyleZhang, Y., Ge, L., Xu, L., Liu, Y., Wang, J., Liu, C., Zhao, H., Xing, L., Wang, J., & Wu, B. (2023). Structural Insights Uncover the Specific Phosphoinositide Recognition by the PH1 Domain of Arap3. International Journal of Molecular Sciences, 24(2), 1125. https://doi.org/10.3390/ijms24021125






