ame-miR-34 Modulates the Larval Body Weight and Immune Response of Apis mellifera Workers to Ascosphara apis Invasion
Abstract
:1. Introduction
2. Results
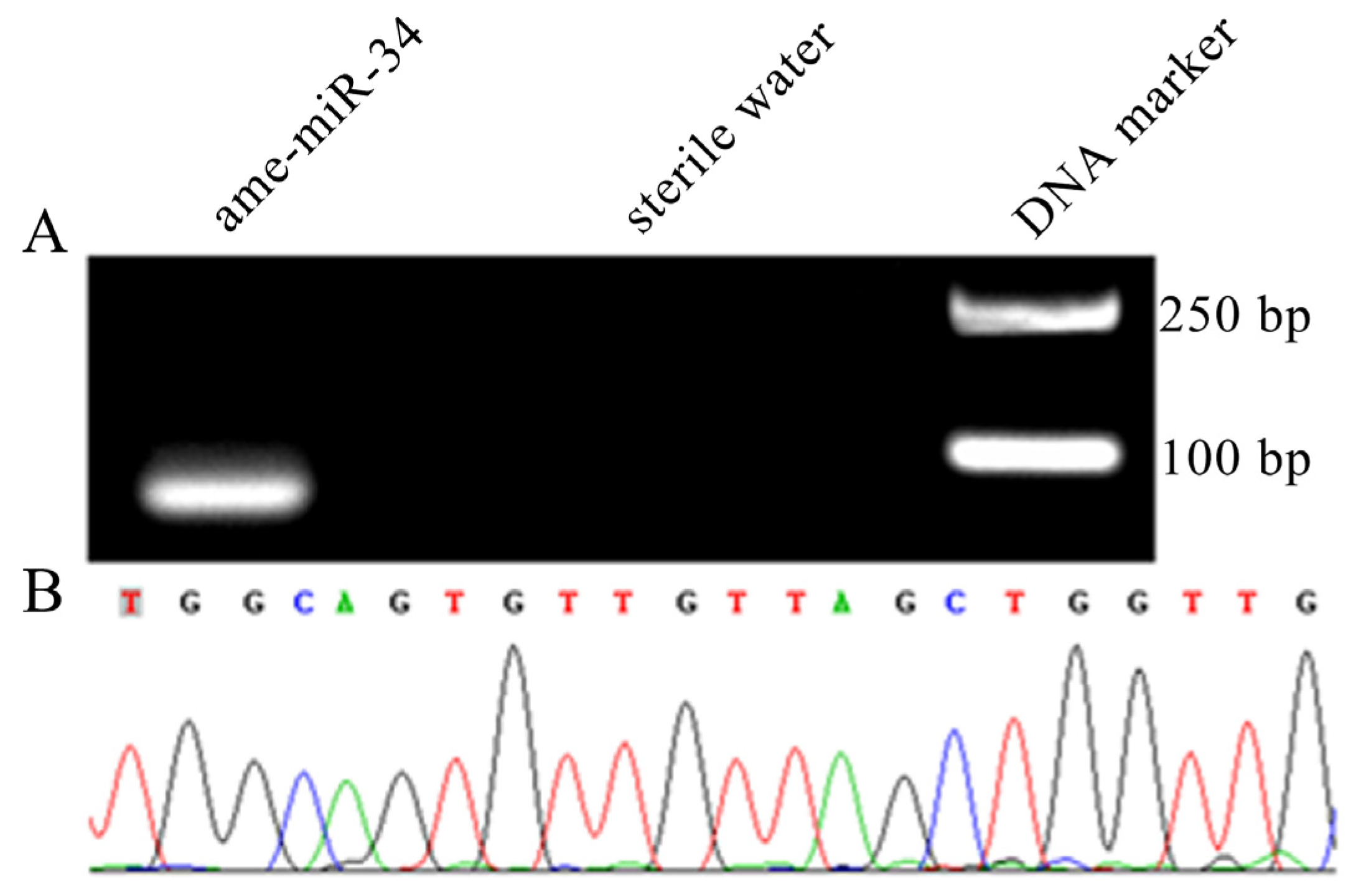
2.1. Molecular Verification of ame-miR-34 Expression and Sequence
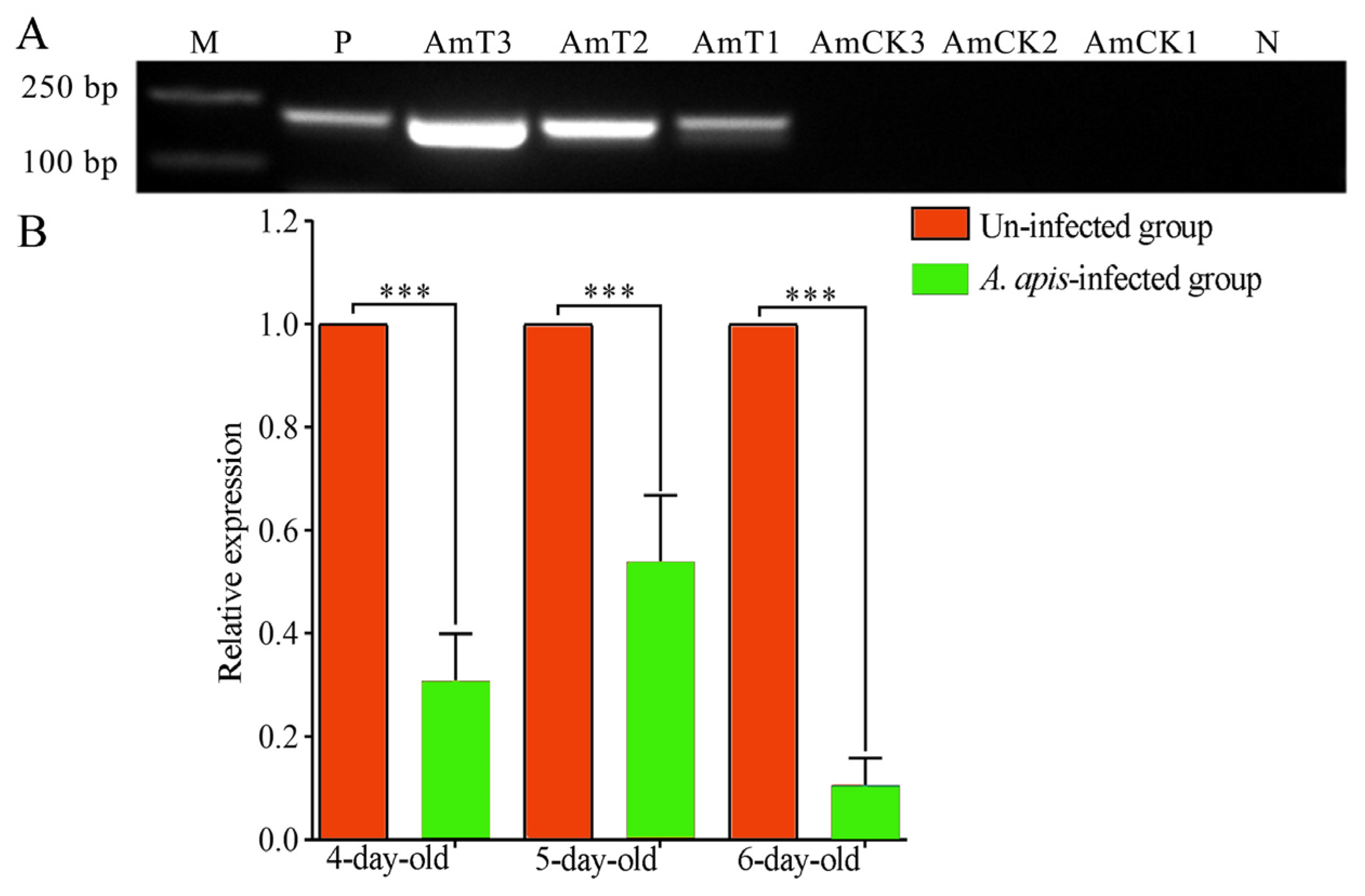
2.2. Expression Level of ame-miR-34 in the Larval Guts was Altered Due to A. apis Infection
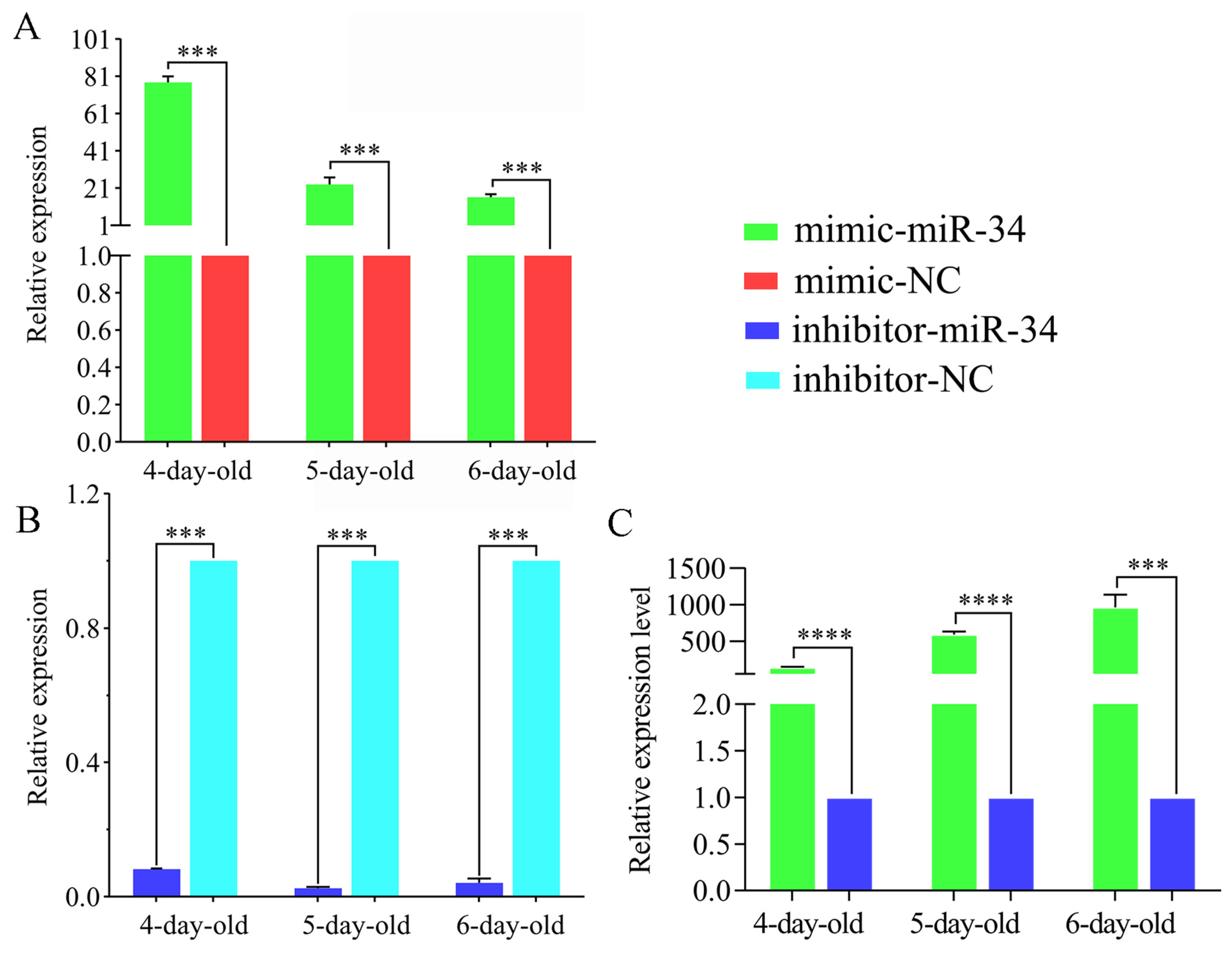
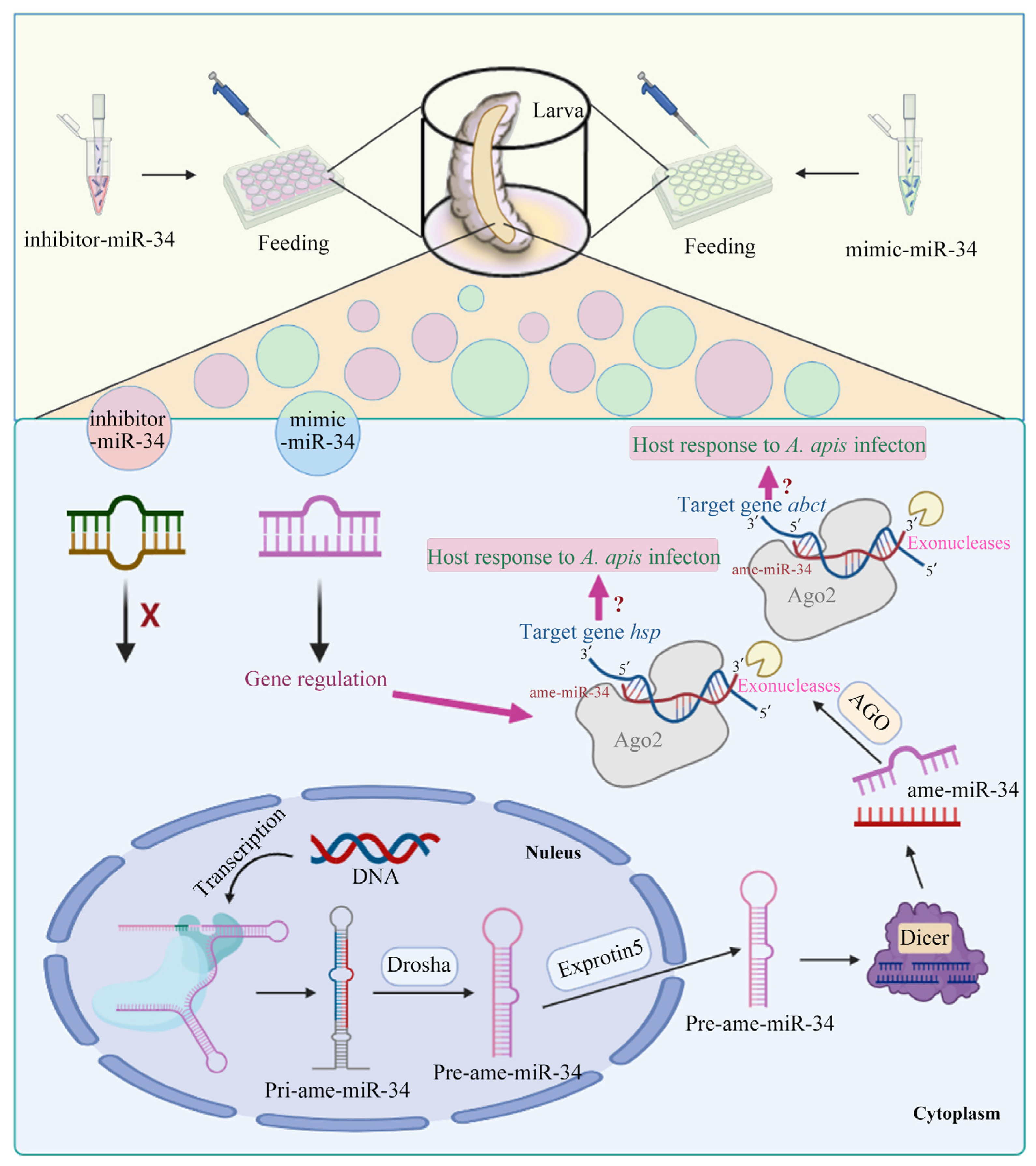
2.3. Overexpression and Knockdown of ame-miR-34 in Uninfected and A. apis-Infected Larval Guts
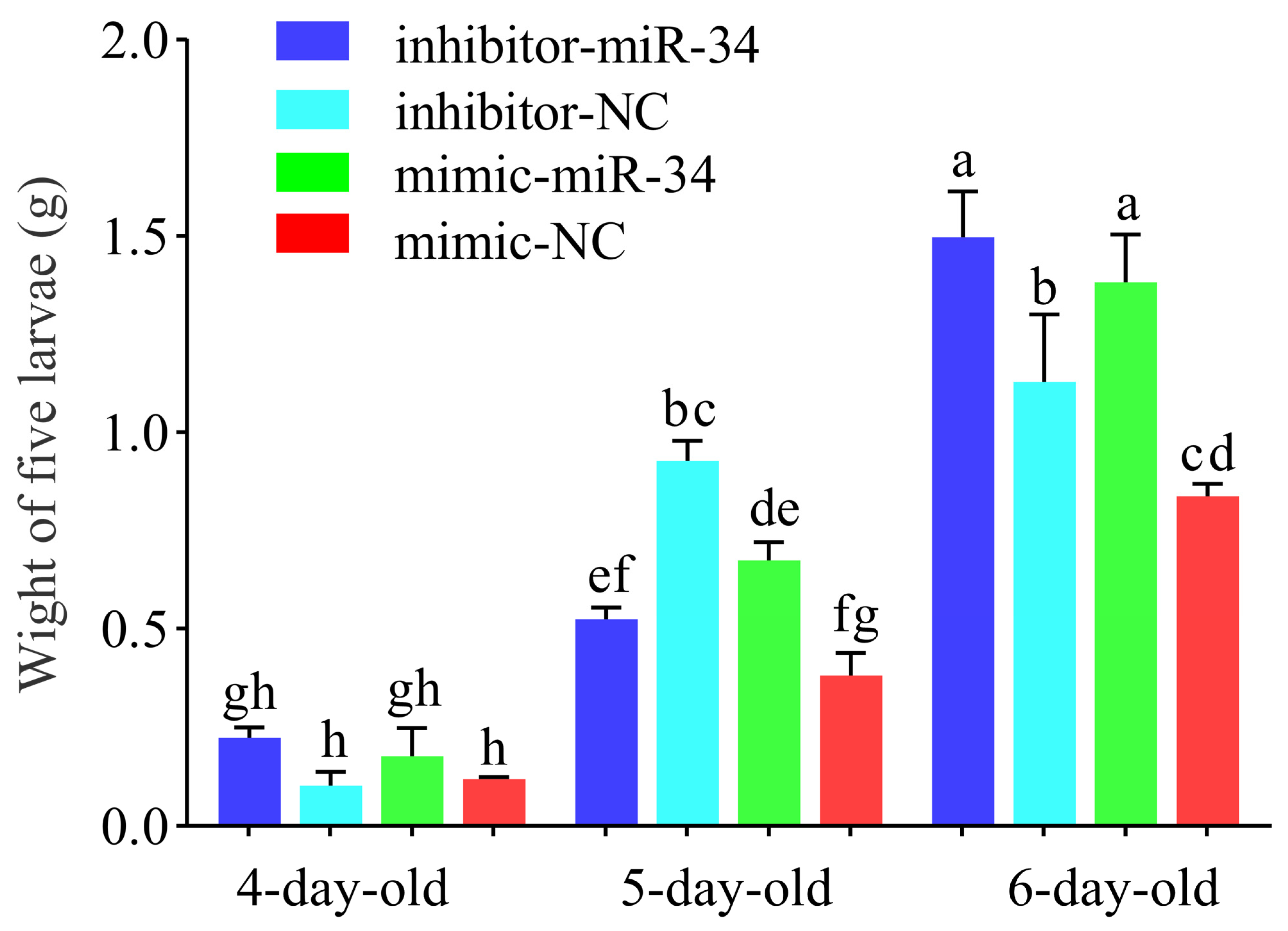
2.4. Effect of ame-miR-34 Overexpression and Knockdown on Body Weights of A. mellifera Larvae
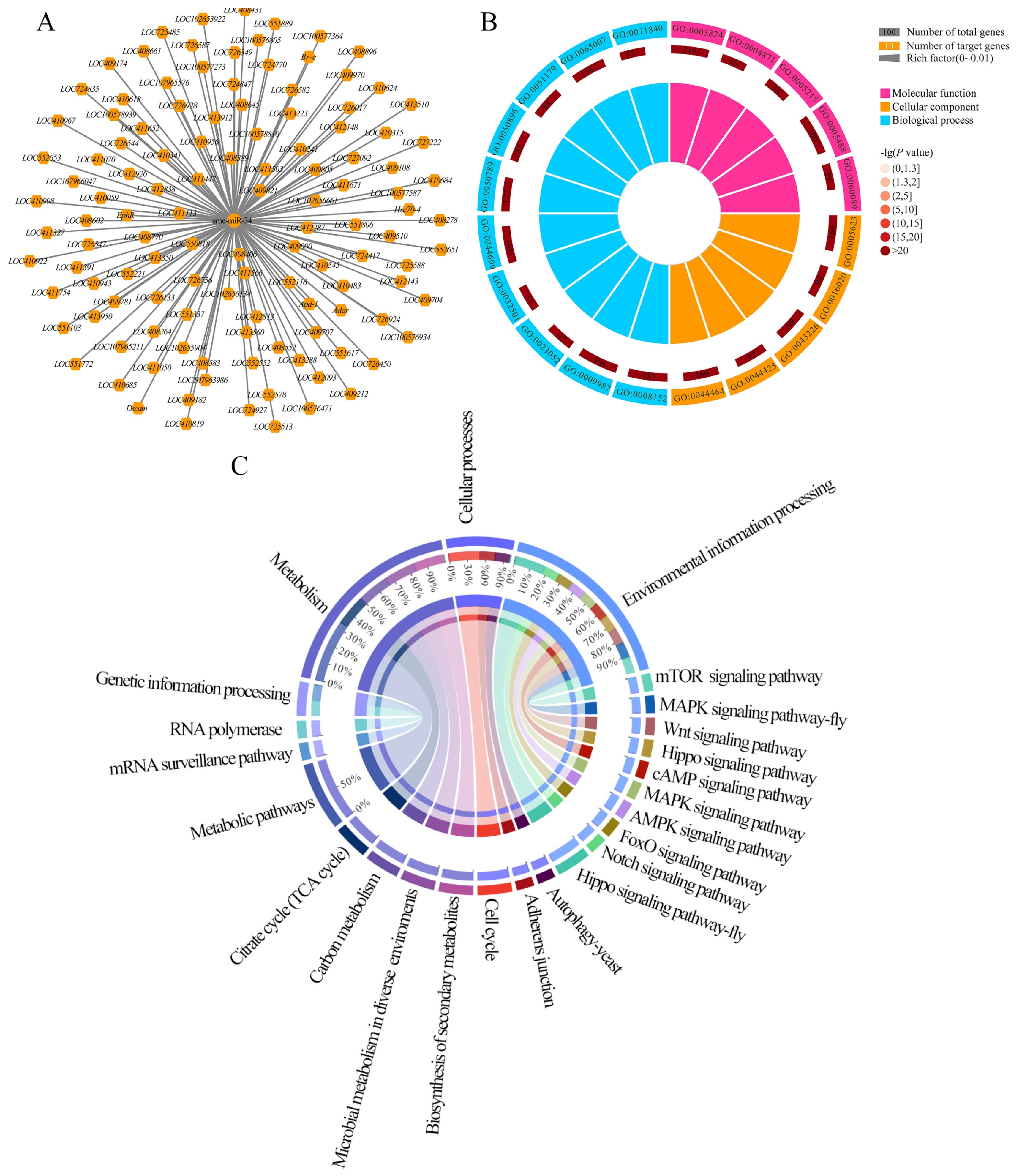
2.5. Analysis of ame-miR-34-Targeted Genes and the Corresponding Regulatory Network
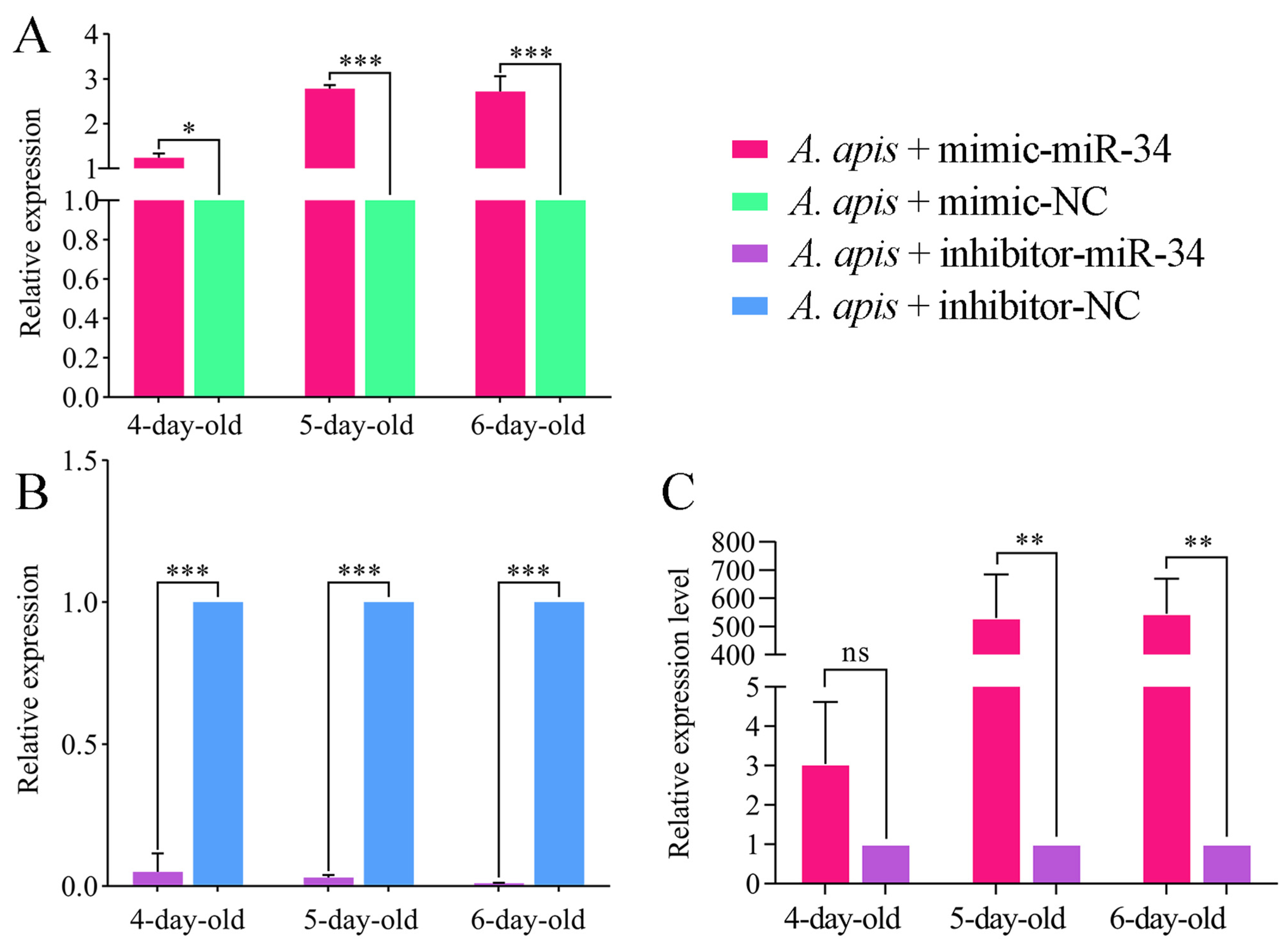
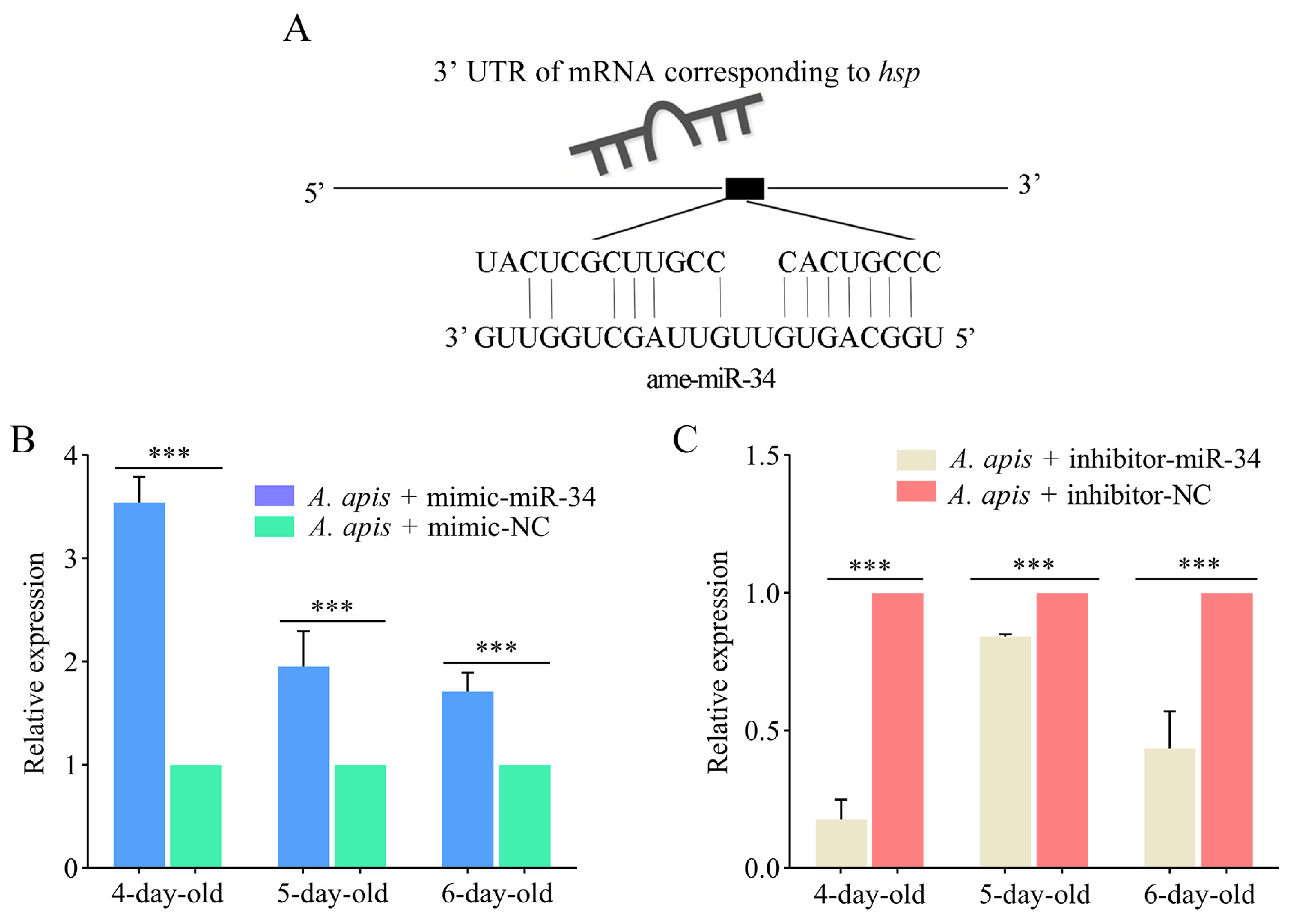
2.6. Effect of ame-miR-34 Overexpression and Knockdown on the Expression of hsp and abct in A. apis-Infected Larval Guts
3. Discussion
4. Materials and Methods
4.1. Honey Bee and Microsporidian Spore
4.2. Stem-Loop RT-PCR and Sanger Sequencing of ame-miR-34
4.3. Experimental Inoculation and Gut Sample Preparation
4.4. RT-qPCR Detection of ame-miR-34
4.5. Overexpression and Knockdown of ame-miR-34 in Normal Larval Guts
4.6. Measurement of Body Weight
4.7. RNA Isolation, cDNA Synthesis, and RT-qPCR Detection
4.8. Overexpression and Knockdown of ame-miR-34 in A. apis-Inoculated Larval Guts
4.9. Target Prediction and Analysis of ame-miR-34
4.10. RT-qPCR Determination of ame-miR-34-Targeted Genes
4.11. Statistical Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Morfin, N.; Anguiano-Baez, R.; Guzman-Novoa, E. Honey Bee (Apis mellifera) Immunity. Vet. Clin. North Am. Food Anim. Pract. 2021, 37, 521–533. [Google Scholar] [CrossRef]
- Zeng, Z.J. Apiculture, 3rd ed.; China Agriculture Press: Beijing, China, 2017; pp. 10–11. (In Chinese) [Google Scholar]
- Aronstein, K.A.; Murray, K.D. Chalkbrood disease in honey bees. J. Invertebr. Pathol. 2010, 103, S20–S29. [Google Scholar] [CrossRef] [PubMed]
- Griffiths-Jones, S.; Grocock, R.J.; van Dongen, S.; Bateman, A.; Enright, A.J. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006, 34, D140–D144. [Google Scholar] [CrossRef]
- Klinge, C.M. miRNAs regulated by estrogens, tamoxifen, and endocrine disruptors and their downstream gene targets. Mol. Cell. Endocrinol. 2015, 418 Pt 3, 273–297. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, C.; Freter, C. Lipid metabolism, apoptosis and cancer therapy. Int. J. Mol. Sci. 2015, 16, 924–949. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.; Heikkinen, L.; Wang, C.L.; Yang, Y.; Sun, H.Y.; Wong, G. Trends in the development of miRNA bioinformatics tools. Brief. Bioinform. 2019, 20, 1836–1852. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kishore, A.; Petrek, M. Roles of macrophage polarization and macrophage-derived miRNAs in pulmonary fibrosis. Front. Immunol. 2021, 12, 678457. [Google Scholar] [CrossRef]
- Shi, T.F.; Zhu, Y.J.; Liu, P.; Ye, L.; Jiang, X.C.; Cao, H.Q.; Yu, L.S. Age and behavior-dependent differential miRNAs expression in the hypopharyngeal glands of Honeybees (Apis mellifera L.). Insects 2021, 12, 764. [Google Scholar] [CrossRef]
- Vieira, J.; Freitas, F.; Cristino, A.S.; Moda, L.; Martins, J.R.; Bitondi, M.; Simões, Z.; Barchuk, A.R. miRNA-34 and miRNA-210 target hexamerin genes enhancing their differential expression during early brain development of honeybee (Apis mellifera) castes. Insect Mol. Biol. 2021, 30, 594–604. [Google Scholar] [CrossRef]
- Chen, X.; Fu, J. The microRNA miR-14 regulates egg-laying by targeting EcR in Honeybees (Apis mellifera). Insects 2021, 12, 351. [Google Scholar] [CrossRef]
- Li, L.; Liu, F.; Li, W.F.; Li, Z.G.; Pan, J.; Yan, L.M.; Zhang, S.W.; Huang, Z.Y.; Su, S.K. Differences in microRNAs and their expressions between foraging and dancing honey bees, Apis mellifera L. J. Insect Physiol. 2012, 58, 1438–1443. [Google Scholar] [CrossRef]
- Lourenço, A.P.; Guidugli-Lazzarini, K.R.; de Freitas, N.; Message, D.; Bitondi, M.; Simões, Z.; Teixeira, É.W. Immunity and physiological changes in adult honey bees (Apis mellifera) infected with Nosema ceranae: The natural colony environment. J. Insect Physiol. 2021, 131, 104237. [Google Scholar] [CrossRef] [PubMed]
- Cristino, A.S.; Barchuk, A.R.; Freitas, F.C.; Narayanan, R.K.; Biergans, S.D.; Zhao, Z.; Simoes, Z.L.; Reinhard, J.; Claudianos, C. Neuroligin-associated microRNA-932 targets actin and regulates memory in the honeybee. Nat. Commun. 2014, 5, 5529. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Freitas, F.C.; Pires, C.V.; Claudianos, C.; Cristino, A.S.; Simões, Z.L. MicroRNA-34 directly targets pair-rule genes and cytoskeleton component in the honey bee. Sci. Rep. 2017, 7, 40884. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Michely, J.; Kraft, S.; Müller, U. miR-12 and miR-124 contribute to defined early phases of long-lasting and transient memory. Sci. Rep. 2017, 7, 7910. [Google Scholar] [CrossRef] [Green Version]
- Liu, F.; Shi, T.F.; Yin, W.; Su, X.; Qi, L.; Huang, Z.Y.; Zhang, S.W.; Yu, L.S. The microRNA ame-miR-279a regulates sucrose responsiveness of forager honey bees (Apis mellifera). Insect Biochem. Mol. Biol. 2017, 90, 34–42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seong, K.M.; Coates, B.S.; Pittendrigh, B.R. Post-transcriptional modulation of cytochrome P450s, Cyp6g1 and Cyp6g2, by miR-310s cluster is associated with DDT-resistant Drosophila melanogaster strain 91-R. Sci. Rep. 2020, 10, 14394. [Google Scholar] [CrossRef]
- Gerasymchuk, M.; Cherkasova, V.; Kovalchuk, O.; Kovalchuk, I. The role of microRNAs in organismal and skin aging. Int. J. Mol. Sci. 2020, 21, 5281. [Google Scholar] [CrossRef]
- Zhang, G.B.; Liu, Z.G.; Wang, J.; Fan, W. MiR-34 promotes apoptosis of lens epithelial cells in cataract rats via the TGF-β/Smads signaling pathway. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 3485–3491. [Google Scholar]
- Ye, X.H.; Xu, L.; Li, X.; He, K.; Hua, H.X.; Cao, Z.H.; Xu, J.D.; Ye, W.Y.; Zhang, J.; Yuan, Z.T.; et al. miR-34 modulates wing polyphenism in planthopper. PLoS Genet. 2019, 15, e1008235. [Google Scholar] [CrossRef] [Green Version]
- Xiong, X.P.; Kurthkoti, K.; Chang, K.Y.; Li, J.L.; Ren, X.; Ni, J.Q.; Rana, T.M.; Zhou, R. miR-34 modulates innate immunity and ecdysone signaling in Drosophila. PLoS Pathog. 2016, 12, e1006034. [Google Scholar] [CrossRef]
- Lai, Y.W.; Chu, S.Y.; Wei, J.Y.; Cheng, C.Y.; Li, J.C.; Chen, P.L.; Chen, C.H.; Yu, H.H. Drosophila microRNA-34 impairs axon pruning of mushroom body γ neurons by downregulating the expression of ecdysone receptor. Sci. Rep. 2016, 6, 39141. [Google Scholar] [CrossRef]
- Zhu, K.G.; Liu, M.H.; Fu, Z.; Zhou, Z.; Kong, Y.; Liang, H.W.; Lin, Z.G.; Luo, J.; Zheng, H.Q.; Wan, P.; et al. Plant microRNAs in larval food regulate honeybee caste development. PLoS Genet. 2017, 13, e1006946. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, Z.W.; Wang, J.; Long, Q.; Xu, Y.J.; Feng, R.R.; Liu, J.M.; Zhao, H.D.; Zhu, L.R.; Hou, H.Q.; Chen, D.F.; et al. Impact of overexpression and knockdown of ame-miR-13b on the expression of genes in larval gut of Apis mellifera ligustica. Acta Entomol. Sin. 2022, 65, 460–468. (In Chinese) [Google Scholar]
- Zhang, K.Y.; Liu, J.M.; Zhang, W.D.; Hu, Y.; Kang, Y.X.; Wang, Z.X.; Wang, S.Y.; Qian, J.J.; Zhao, X.; Zhang, J.X.; et al. ame-miR-79 negatively regulates the expression of target genes CYP450 and FG in the larval guts of Apis mellifera ligustica workers. Acta Entomol. Sin. 2022, 65, 1256–1265. (In Chinese) [Google Scholar]
- Hu, Y.; Zhang, W.D.; Wang, S.Y.; Zhang, K.Y.; Ren, Z.M.; Ji, T.; Lin, Z.G.; Zhang, H.X.; Chen, D.F.; Guo, R. Regulation of the expression of target genes USP and P300 by ame-miR-bantam in the larval gut of Apis mellifera ligustica workers. Acta Entomol. Sin. 2022, 65, 1401–1410. (In Chinese) [Google Scholar]
- Zhang, Q.; Dou, W.; Taning, C.N.T.; Yu, S.S.; Yuan, G.R.; Shang, F.; Smagghe, G.; Wang, J.J. miR-309a is a regulator of ovarian development in the oriental fruit fly Bactrocera dorsalis. PLoS Genet. 2022, 18, e1010411. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Guo, X.; Li, Y.; Zhang, J.; Fu, Y. miR-34-5p, encoded by Spodoptera frugiperda, participates in anti-baculovirus by regulating innate immunity in the insect host. Int. J. Biol. Macromol. 2022, 222, 2190–2199. [Google Scholar] [CrossRef] [PubMed]
- Fang, H.; Wang, X.; Liu, X.; Michaud, J.P.; Wu, Y.; Zhang, H.; Li, Y.; Li, Z. Molecular characterization of insulin receptor (IR) in oriental fruit moth, Grapholita molesta (Lepidoptera: Tortricidae), and elucidation of its regulatory roles in glucolipid homeostasis and metamorphosis through interaction with miR-982490. Insect Mol. Biol. 2022, 31, 659–670. [Google Scholar] [CrossRef] [PubMed]
- Sato, A.; Takamatsu, M.; Kobayashi, S.; Ogawa, M.; Shiwa, Y.; Watanabe, S.; Chibazakura, T.; Yoshikawa, H. Novel heat shock response mechanism mediated by the initiation nucleotide of transcription. J. Gen Appl. Microbiol. 2022, 68, 95–108. [Google Scholar] [CrossRef]
- Musa, M.; Dionisio, P.A.; Casqueiro, R.; Milosevic, I.; Raimundo, N.; Krisko, A. Lack of peroxisomal catalase affects heat shock response in Caenorhabditis elegans. Life Sci. Alliance 2022, 6, e202201737. [Google Scholar] [CrossRef]
- Al-Ghzawi, A.A.A.; Al-Zghoul, M.B.; Zaitoun, S.; Al-Omary, I.M.; Alahmad, N.A. Dynamics of heat shock proteins and heat shock factor expression during heat stress in daughter workers in pre-heat-treated (rapid heat hardening) Apis mellifera mother queens. J. Therm. Biol. 2022, 104, 103194. [Google Scholar] [CrossRef] [PubMed]
- Bi, H.; Miao, J.; He, J.; Chen, Q.; Qian, J.; Li, H.; Xu, Y.; Ma, D.; Zhao, Y.; Tian, X.; et al. Characterization of the wheat heat shock factor TaHsfA2e-5D conferring heat and drought tolerance in Arabidopsis. Int. J. Mol. Sci. 2022, 23, 2784. [Google Scholar] [CrossRef] [PubMed]
- Dores-Silva, P.R.; Cauvi, D.M.; Coto, A.; Silva, N.; Borges, J.C.; De Maio, A. Human heat shock cognate protein (HSC70/HSPA8) interacts with negatively charged phospholipids by a different mechanism than other HSP70s and brings HSP90 into membranes. Cell Stress Chaperones. 2021, 26, 671–684. [Google Scholar] [CrossRef]
- Tian, F.; Hu, X.L.; Yao, T.; Yang, X.; Chen, J.G.; Lu, M.Z.; Zhang, J. Recent advances in the roles of HSFs and HSPs in heat stress response in woody plants. Front. Plant Sci. 2021, 12, 704905. [Google Scholar] [CrossRef] [PubMed]
- Yuan, F.F.; Cai, J.Z.; Wu, J.F.; Tang, Y.T.; Zhao, K.; Liang, F.; Li, F.L.; Yang, X.Y.; He, Z.H.; Billiar, T.R.; et al. Z-DNA binding protein 1 promotes heatstroke-induced cell death. Science 2022, 376, 609–615. [Google Scholar] [CrossRef]
- Dokladny, K.; Myers, O.B.; Moseley, P.L. Heat shock response and autophagy--cooperation and control. Autophagy 2015, 11, 200–213. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dean, M.; Rzhetsky, A.; Allikmets, R. The human ATP-binding cassette (ABC) transporter superfamily. Genome Res. 2001, 11, 1156–1166. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Cao, H.; Kimari, M.; Maronitis, G.; Williams, M.J.; Schiöth, H.B. Multidrug resistance like protein 1 activity in malpighian tubules regulates lipid homeostasis in Drosophila. Membranes 2021, 11, 432. [Google Scholar] [CrossRef]
- Wang, L.X.; Tao, S.; Zhang, Y.C.; Pei, X.G.; Gao, Y.; Song, X.Y.; Yu, Z.T.; Gao, C.F. Overexpression of ATP-binding cassette transporter Mdr49-like confers resistance to imidacloprid in the field populations of brown planthopper, Nilaparvata lugens. Pest Manag. Sci. 2022, 78, 579–590. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Wang, Y.; Sinakevitch, I.; Lei, H.; Smith, B.H. Comparison of RNAi knockdown effect of tyramine receptor 1 induced by dsRNA and siRNA in brains of the honey bee, Apis mellifera. J. Insect Physiol. 2018, 111, 47–52. [Google Scholar] [CrossRef]
- Rodríguez-García, C.; Heerman, M.C.; Cook, S.C.; Evans, J.D.; DeGrandi-Hoffman, G.; Banmeke, O.; Zhang, Y.; Huang, S.; Hamilton, M.; Chen, Y.P. Transferrin-mediated iron sequestration suggests a novel therapeutic strategy for controlling Nosema disease in the honey bee, Apis mellifera. PLoS Pathog. 2021, 17, e1009270. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.L.; Yu, K.J.; Zhao, X.; Qian, J.J.; Zhao, H.D.; Zhang, J.; Zhang, Y.; Zhao, H.X.; Xu, X.J.; Luo, Q.; et al. Bioinformatic analysis and functional study of nkd gene in larvae of Apis mellifera ligustica workers. Acta Microbiol. Sin. 2022, 62, 5005–5017. (In Chinese) [Google Scholar]
- Leonard, S.P.; Powell, J.E.; Perutka, J.; Geng, P.; Heckmann, L.C.; Horak, R.D.; Davies, B.W.; Ellington, A.D.; Barrick, J.E.; Moran, N.A. Engineered symbionts activate honey bee immunity and limit pathogens. Science 2020, 367, 573–576. [Google Scholar] [CrossRef] [PubMed]
- Guo, R.; Chen, D.F.; Chen, H.Z.; Fu, Z.M.; Xiong, C.L.; Hou, C.S.; Zheng, Y.Z.; Guo, Y.L.; Wang, H.P.; Du, Y.; et al. Systematic investigation of circular RNAs in Ascosphaera apis, a fungal pathogen of honeybee larvae. Gene 2018, 678, 17–22. [Google Scholar] [CrossRef]
- Guo, R.; Chen, D.F.; Xiong, C.L.; Hou, C.S.; Zheng, Y.Z.; Fu, Z.M.; Diao, Q.Y.; Zhang, L.; Wang, H.Q.; Hou, Z.X.; et al. Identification of long non-coding RNAs in the chalkbrood disease pathogen Ascospheara apis. J. Invertebr. Pathol. 2018, 156, 1–5. [Google Scholar] [CrossRef]
- Zhu, Z.W.; Fu, Z.M.; Long, Q.; Du, Y.; Zhang, W.D.; Hu, Y.; Zhao, X.; Shi, X.Y.; Xu, X.J.; Chen, D.F.; et al. Expression profiles and potential function of three miRNAs during the pupal development process of Apis mellifera ligustica worker. Acta Entomol. Sin. 2018, 65, 53–62. (In Chinese) [Google Scholar]
- Chen, D.F.; Guo, R.; Xu, X.J.; Xiong, C.L.; Liang, Q.; Zheng, Y.Z.; Luo, Q.; Zhang, Z.N.; Huang, Z.J.; Kumar, D.; et al. Uncovering the immune responses of Apis mellifera ligustica larval gut to Ascosphaera apis infection utilizing transcriptome sequencing. Gene 2017, 621, 40–50. [Google Scholar] [CrossRef]
- Jensen, A.B.; Aronstein, K.; Flores, J.M.; Vojvodic, S.; Palacio, M.A.; Spivak, M. Standard methods for fungal brood disease research. J. Apic. Res. 2013, 52. [Google Scholar] [CrossRef] [Green Version]
- Brødsgaard, C.F.; Ritter, W.; Hansen, H. Response of in vitro reared honey bee larvae to various doses of Paenibacillus larvae spores. Apidologie 1998, 29, 569–578. [Google Scholar] [CrossRef] [Green Version]
- Guo, R.; Chen, D.; Diao, Q.; Xiong, C.; Zheng, Y.; Hou, C. Transcriptomic investigation of immune responses of the Apis cerana cerana larval gut infected by Ascosphaera apis. J. Invertebr. Pathol. 2019, 166, 107210. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Wang, X.R.; Li, X.M.; Pu, Q.; Luo, C.Y.; Xu, L.L.; Peng, X.Y.; Liu, S.P. Genetic manipulation of micrornas in the silk gland of Silkworm, Bombyx Mori. Biol. Proced. Online 2019, 21, 16. [Google Scholar] [CrossRef] [PubMed]
- Peng, C.Y.; Mussen, E.; Fong, A.; Montague, M.A.; Tyler, T. Effect of chlortetracycline of honeybee worker larvae reared in vitro. J. Invertebr. Pathol. 1992, 60, 127–133. [Google Scholar] [CrossRef]
- Borsuk, G.; Ptaszyńska, A.A.; Olszewski, K.; Domaciuk, M.; Krutmuang, P.; Paleolog, J. A new method for quick and easy hemolymph collection from apidae adults. PLoS ONE 2017, 12, e0170487. [Google Scholar] [CrossRef]
- Xiong, C.L.; Du, Y.; Feng, R.R.; Jiang, H.B.; Shi, X.Y.; Wang, H.P.; Fan, X.X.; Wang, J.; Zhu, Z.W.; Fan, Y.C.; et al. Differential expression pattern and regulation network of microRNAs in Ascosphaera apis invading Apis cerana cerana 6-day-old larvae. Acta Microbiol. Sin. 2020, 60, 992–1009. (In Chinese) [Google Scholar]
- Jagla, T.; Dubińska-Magiera, M.; Poovathumkadavil, P.; Daczewska, M.; Jagla, K. Developmental expression and functions of the small heat shock proteins in Drosophila. Int. J. Mol. Sci. 2018, 19, 3441. [Google Scholar] [CrossRef] [PubMed]

| Name | Sequence (5′–3′) | Purpose |
|---|---|---|
| ame-miR-34-F | TGGCAGTGTTGTTA | Reverse transcription of ame-miR-34 |
| ame-miR-34-Loop | CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGCAACCAGC | |
| ame-miR-34-R | CTCAACTGGTGTCGTGGA | |
| AmU6-F | GTTAGGCTTTGACGATTTCG | Internal reference for qPCR of ame-miR-34 |
| AmU6-R | GGCATTTCTCCACCAGGTA | |
| actin-F | ATGCCAACACTGTCCTTTCTGG | Internal reference for qPCR of target genes |
| actin-R | GACCCACCAATCCATACGGA | |
| hsp-F | TCCTGTGTTGGTGTATTCCAGCATG | qPCR detection |
| hsp-R | GCAACTTGGTTCTTGGCAGCATC | |
| abct-F | ACGACGACTATACCTGGCAGTGG | |
| abct-R | CAGTTGAGACGAGACAGCATCCG |
| Name | Sequence (5′–3′) | Purpose |
|---|---|---|
| mimic-miR-34-sense | UGGCAGUGUUGUUAGCUGGUUG | ame-miR-34 overexpression |
| mimic-miR-34-antisense | ACCAGCUAACAACACUGCCAUU | |
| inhibitor-miR-34 | CAACCAGCUAACAACACUGCCA | ame-miR-34 knockdown |
| mimic-NC-sense | UUCUCCGAACGUGUCACGUTT | Negative control for ame-miR-34 overexpression |
| mimic-NC-antisense | ACGUGACACGUUCGGAGAATT | |
| inhibitor-NC | CAGUACUUUUGUGUAGUACAA | Negative control for ame-miR-34 knockdown |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, Y.; Guo, Y.; Fan, X.; Zhao, H.; Zhang, Y.; Guo, S.; Jing, X.; Liu, Z.; Feng, P.; Liu, X.; et al. ame-miR-34 Modulates the Larval Body Weight and Immune Response of Apis mellifera Workers to Ascosphara apis Invasion. Int. J. Mol. Sci. 2023, 24, 1214. https://doi.org/10.3390/ijms24021214
Wu Y, Guo Y, Fan X, Zhao H, Zhang Y, Guo S, Jing X, Liu Z, Feng P, Liu X, et al. ame-miR-34 Modulates the Larval Body Weight and Immune Response of Apis mellifera Workers to Ascosphara apis Invasion. International Journal of Molecular Sciences. 2023; 24(2):1214. https://doi.org/10.3390/ijms24021214
Chicago/Turabian StyleWu, Ying, Yilong Guo, Xiaoxue Fan, Haodong Zhao, Yiqiong Zhang, Sijia Guo, Xin Jing, Zhitan Liu, Peilin Feng, Xiaoyu Liu, and et al. 2023. "ame-miR-34 Modulates the Larval Body Weight and Immune Response of Apis mellifera Workers to Ascosphara apis Invasion" International Journal of Molecular Sciences 24, no. 2: 1214. https://doi.org/10.3390/ijms24021214
APA StyleWu, Y., Guo, Y., Fan, X., Zhao, H., Zhang, Y., Guo, S., Jing, X., Liu, Z., Feng, P., Liu, X., Zou, P., Li, Q., Na, Z., Zhang, K., Chen, D., & Guo, R. (2023). ame-miR-34 Modulates the Larval Body Weight and Immune Response of Apis mellifera Workers to Ascosphara apis Invasion. International Journal of Molecular Sciences, 24(2), 1214. https://doi.org/10.3390/ijms24021214







