Genotypic and Phenotypic Detection of Polyhydroxyalkanoate Production in Bacterial Isolates from Food
Abstract
:1. Introduction
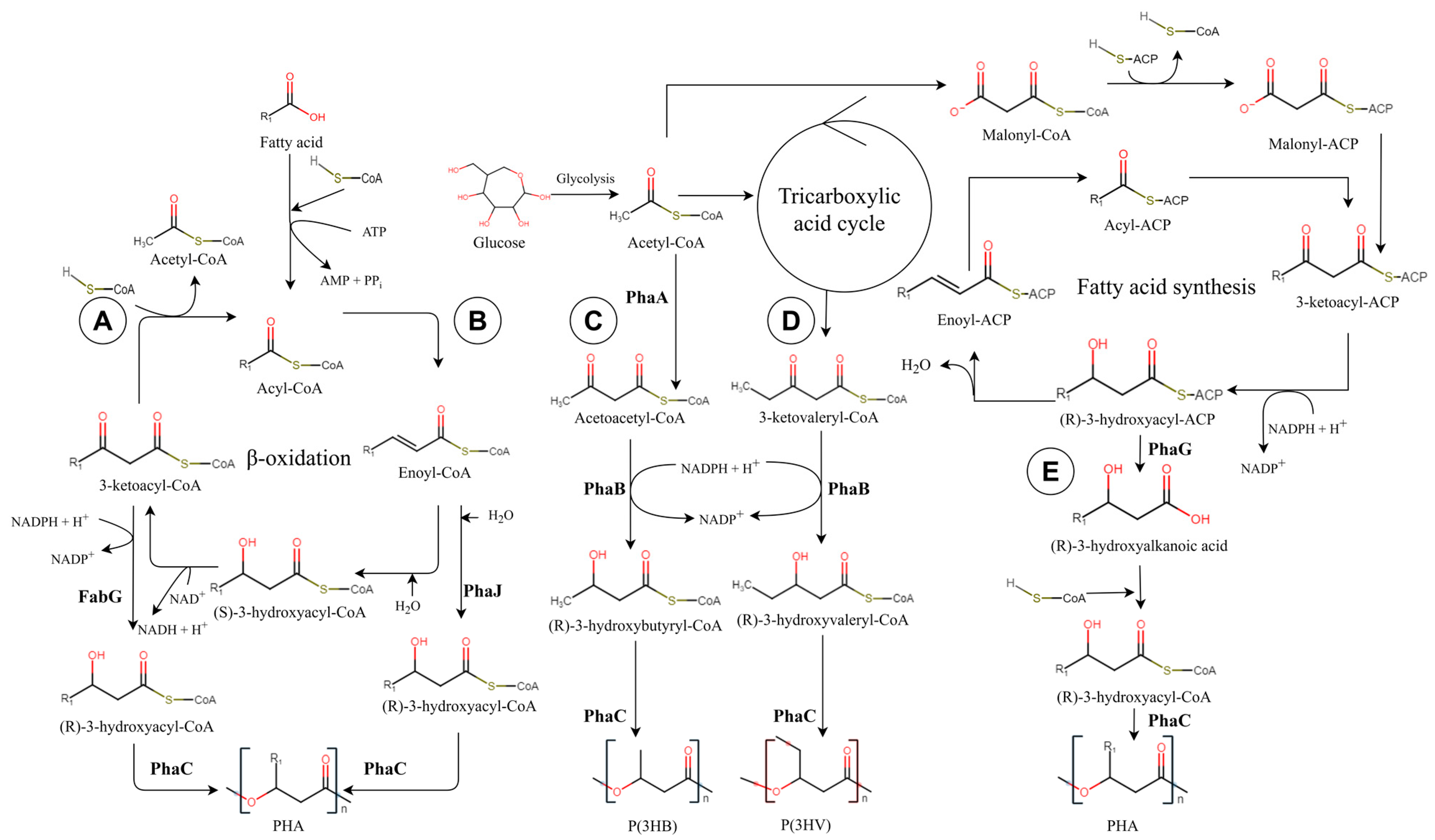
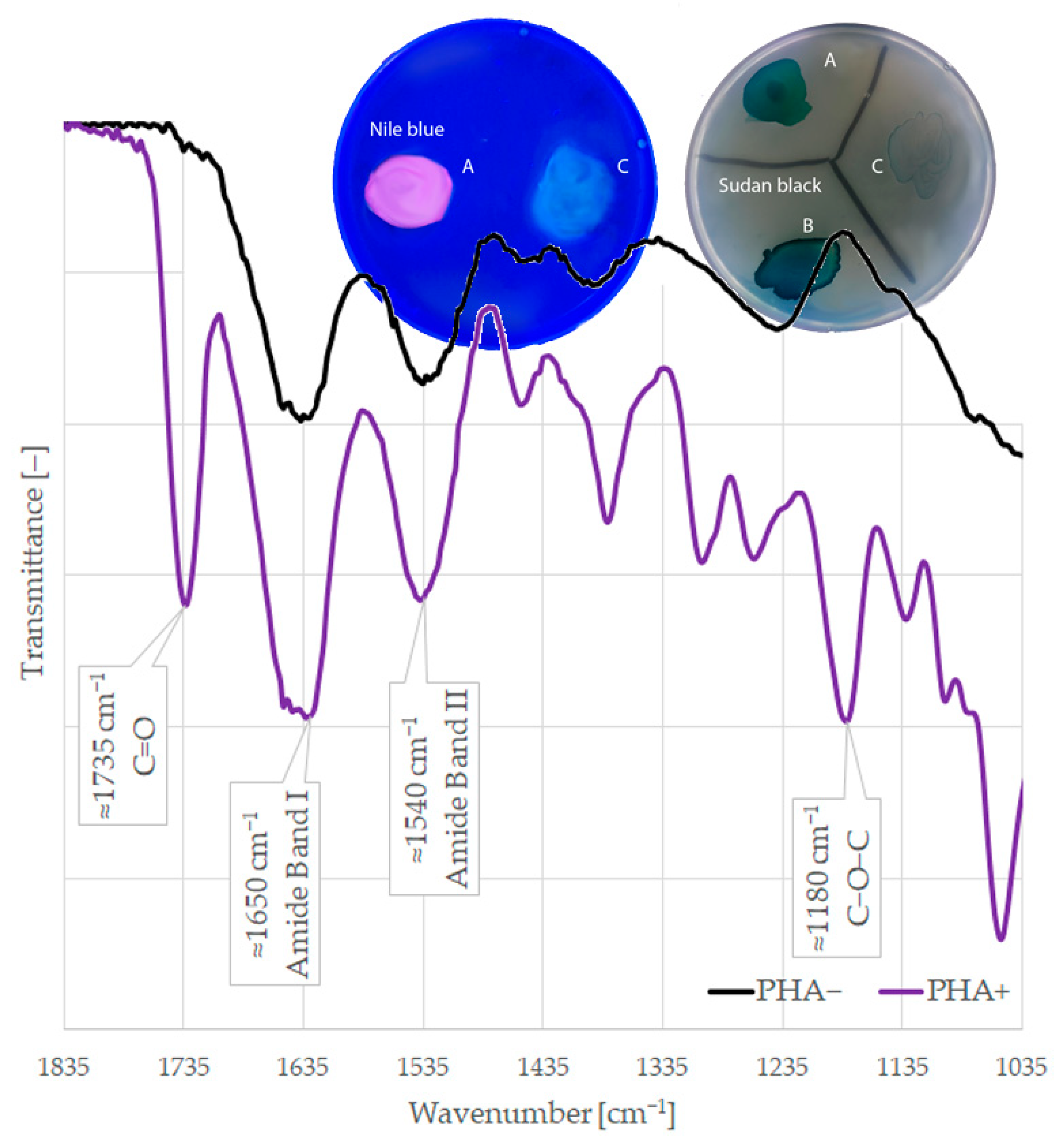
2. Results
3. Discussion
4. Materials and Methods
4.1. Materials and Chemicals
4.2. Molecular Detection
4.3. Phenotypic Detection
4.3.1. Nile Blue Staining
4.3.2. Sudan Black Staining
4.3.3. Fourier Transform Infrared Spectroscopy
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Khanna, S.; Srivastava, A.K. Recent Advances in Microbial Polyhydroxyalkanoates. Process Biochem. 2005, 40, 607–619. [Google Scholar] [CrossRef]
- Chen, G.-Q. Biofunctionalization of Polymers and Their Applications. In Biofunctionalization of Polymers and Their Applications; Springer: Berlin/Heidelberg, Germany, 2011; pp. 29–45. ISBN 978-3-642-21948-1. [Google Scholar]
- Gómez-Gast, N.; Cuellar, M.D.R.L.; Vergara-Porras, B.; Vieyra, H. Biopackaging Potential Alternatives: Bioplastic Composites of Polyhydroxyalkanoates and Vegetal Fibers. Polymers 2022, 14, 1114. [Google Scholar] [CrossRef]
- Kosior, E.; Messias, R.; Fowler, P. Lightweight Compostable Packaging: Literature Review; The Waste; Resources Action Programme: Banbury, UK, 2006. [Google Scholar]
- Sharma, S.K.; Mudhoo, A. Handbook of Applied Biopolymer Technology; Royal Society of Chemistry: Cambridge, UK, 2011; ISBN 978-1-84973-151-5. [Google Scholar]
- Kumar, A.; Srivastava, J.K.; Mallick, N.; Singh, A.K. Commercialization of Bacterial Cell Factories for the Sustainable Production of Polyhydroxyalkanoate Thermoplastics: Progress and Prospects. Recent Pat. Biotechnol. 2015, 9, 4–21. [Google Scholar] [CrossRef] [PubMed]
- Pulingam, T.; Appaturi, J.N.; Parumasivam, T.; Ahmad, A.; Sudesh, K. Biomedical Applications of Polyhydroxyalkanoate in Tissue Engineering. Polymers 2022, 14, 2141. [Google Scholar] [CrossRef] [PubMed]
- Gadgil, B.S.T.; Killi, N.; Rathna, G.V.N. Polyhydroxyalkanoates as Biomaterials. MedChemComm 2017, 8, 1774–1787. [Google Scholar] [CrossRef]
- Wong, P.A.L.; Cheung, M.K.; Lo, W.-H.; Chua, H.; Yu, P.H.F. Investigation of the Effects of the Types of Food Waste Utilized as Carbon Source on the Molecular Weight Distributions and Thermal Properties of Polyhydroxybutyrate Produced by Two Strains of Microorganisms. E-Polymers 2004, 4, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Tan, G.-Y.; Chen, C.-L.; Li, L.; Ge, L.; Wang, L.; Razaad, I.; Li, Y.; Zhao, L.; Mo, Y.; Wang, J.-Y. Start a Research on Biopolymer Polyhydroxyalkanoate (PHA): A Review. Polymers 2014, 6, 706. [Google Scholar] [CrossRef] [Green Version]
- Lu, J.; Tappel, R.C.; Nomura, C.T. Mini-Review: Biosynthesis of Poly(hydroxyalkanoates). Polym. Rev. 2009, 49, 226–248. [Google Scholar] [CrossRef]
- Steinbüchel, A.; Hein, S. Biochemical and Molecular Basis of Microbial Synthesis of Polyhydroxyalkanoates in Microorganisms. In Biopolyesters; Springer: Berlin/Heidelberg, Germany, 2001; pp. 81–123. ISBN 978-3-540-41141-3. [Google Scholar]
- Witholt, B.; Kessler, B. Perspectives of Medium Chain Length Poly(hydroxyalkanoates), a Versatile Set of Bacterial Bioplastics. Curr. Opin. Biotechnol. 1999, 10, 279–285. [Google Scholar] [CrossRef]
- Nomura, C.T.; Tanaka, T.; Eguen, T.E.; Appah, A.S.; Matsumoto, K.; Taguchi, S.; Ortiz, C.L.; Doi, Y. FabG Mediates Polyhydroxyalkanoate Production from Both Related and Nonrelated Carbon Sources in Recombinant Escherichia Coli LS5218. Biotechnol. Prog. 2008, 24, 342–351. [Google Scholar] [CrossRef]
- Tsuge, T.; Fukui, T.; Matsusaki, H.; Taguchi, S.; Kobayashi, G.; Ishizaki, A.; Doi, Y. Molecular Cloning of Two (r)-Specific Enoyl-CoA Hydratase Genes from Pseudomonas Aeruginosa and Their Use for Polyhydroxyalkanoate Synthesis. FEMS Microbiol. Lett. 2000, 184, 193–198. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lenz, R.W.; Marchessault, R.H. Bacterial Polyesters: Biosynthesis, Biodegradable Plastics and Biotechnology. Biomacromolecules 2005, 6, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Aldor, I.S.; Kim, S.-W.; Prather, K.L.J.; Keasling, J.D. Metabolic Engineering of a Novel Propionate-Independent Pathway for the Production of Poly(3-Hydroxybutyrate-Co-3-Hydroxyvalerate) in Recombinant Salmonella Enterica Serovar Typhimurium. Appl. Environ. Microbiol. 2002, 68, 3848–3854. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Satoh, Y.; Murakami, F.; Tajima, K.; Munekata, M. Enzymatic Synthesis of Poly(3-Hydroxybutyrate-Co-4-Hydroxybutyrate) with CoA Recycling Using Polyhydroxyalkanoate Synthase and Acyl-CoA Synthetase. J. Biosci. Bioeng. 2005, 99, 508–511. [Google Scholar] [CrossRef]
- Rehm, B.H.A. Polyester Synthases: Natural Catalysts for Plastics. Biochem. J. 2003, 376, 15–33. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rehm, B.H.A. Biogenesis of Microbial Polyhydroxyalkanoate Granules: A Platform Technology for the Production of Tailor-Made Bioparticles. Curr. Issues Mol. Biol. 2007, 9, 41–62. [Google Scholar] [CrossRef] [Green Version]
- Chek, M.F.; Kim, S.-Y.; Mori, T.; Arsad, H.; Samian, M.R.; Sudesh, K.; Hakoshima, T. Structure of Polyhydroxyalkanoate (PHA) Synthase PhaC from Chromobacterium Sp. USM2, Producing Biodegradable Plastics. Sci. Rep. 2017, 7, 5312. [Google Scholar] [CrossRef] [Green Version]
- Montenegro, E.; Delabary, G.; Silva, M.; Andreote, F.; Lima, A. Molecular Diagnostic for Prospecting Polyhydroxyalkanoate-Producing Bacteria. Bioengineering 2017, 4, 52. [Google Scholar] [CrossRef] [Green Version]
- Pohlmann, A.; Fricke, W.F.; Reinecke, F.; Kusian, B.; Liesegang, H.; Cramm, R.; Eitinger, T.; Ewering, C.; Pötter, M.; Schwartz, E.; et al. Genome Sequence of the Bioplastic-Producing “Knallgas” Bacterium Ralstonia eutropha H16. Nat. Biotechnol. 2006, 24, 1257–1262. [Google Scholar] [CrossRef] [Green Version]
- Mezzolla, V.; D’Urso, O.; Poltronieri, P. Role of PhaC Type i and Type II Enzymes during PHA Biosynthesis. Polymers 2018, 10, 910. [Google Scholar] [CrossRef]
- Ren, Q.; Sierro, N.; Witholt, B.; Kessler, B. FabG, an NADPH-Dependent 3-Ketoacyl Reductase of Pseudomonas aeruginosa, Provides Precursors for Medium-Chain-Length Poly-3-Hydroxyalkanoate Biosynthesis in Escherichia coli. J. Bacteriol. 2000, 182, 2978–2981. [Google Scholar] [CrossRef] [Green Version]
- Tsuge, T.; Taguchi, K.; Seiichi; Taguchi; Doi, Y. Molecular Characterization and Properties of (r)-Specific Enoyl-CoA Hydratases from Pseudomonas aeruginosa: Metabolic Tools for Synthesis of Polyhydroxyalkanoates via Fatty Acid ß-Oxidation. Int. J. Biol. Macromol. 2003, 31, 195–205. [Google Scholar] [CrossRef] [PubMed]
- Lucas, S.; Copeland, A.; Lapidus, A.; Glavina del Rio, T.; Dalin, E.; Tice, H.; Pitluck, S.; Chain, P.; Malfatti, S.; Shin, M.; et al. Complete sequence of Stenotrophomonas maltophilia R551-3. Available online:https://www.ncbi.nlm.nih.gov/nuccore/NC_011071.1?report=genbank&from=2718240&to=2719304&strand=true (accessed on 29 November 2022).
- Tsuge, T.; Hyakutake, M.; Mizuno, K. Class IV Polyhydroxyalkanoate (PHA) Synthases and PHA-Producing Bacillus. Appl. Microbiol. Biotechnol. 2015, 99, 6231–6240. [Google Scholar] [CrossRef]
- Ohji, S.; Yamazoe, A.; Hosoyama, A.; Tsuchikane, K.; Ezaki, T.; Fujita, N. The Complete Genome Sequence of Pseudomonas putida NBRC 14164 t Confirms High Intraspecies Variation. Genome Announc. 2014, 2, e00029-14. [Google Scholar] [CrossRef] [Green Version]
- Idris, S.; Rahim, R.A.; Amirul, A.-A.A. Bioprospecting and Molecular Identification of Used Transformer Oil-Degrading Bacteria for Bioplastics Production. Microorganisms 2022, 10, 583. [Google Scholar] [CrossRef]
- Lee, S.-E.; Li, Q.X.; Yu, J. Diverse Protein Regulations on PHA Formation in Ralstonia eutropha on Short Chain Organic Acids. Int. J. Biol. Sci. 2009, 5, 215–225. [Google Scholar] [CrossRef] [Green Version]
- McCool, G.J.; Cannon, M.C. Polyhydroxyalkanoate Inclusion Body-Associated Proteins and Coding Region in Bacillus megaterium. J. Bacteriol. 1999, 181, 585–592. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, M.; Kumar, P.; Patel, S.K.S.; Kalia, V.C. Production of Polyhydroxyalkanoate Co-Polymer by Bacillus thuringiensis. Indian J. Microbiol. 2013, 53, 77–83. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mohandas, S.P.; Balan, L.; Jayanath, G.; Anoop, B.S.; Philip, R.; Cubelio, S.S.; Singh, I.S.B. Biosynthesis and Characterization of Polyhydroxyalkanoate from Marine Bacillus cereus MCCB 281 Utilizing Glycerol as Carbon Source. Int. J. Biol. Macromol. 2018, 119, 380–392. [Google Scholar] [CrossRef] [PubMed]
- Ray, S.; Kalia, V.C. Polyhydroxyalkanoate Production and Degradation Patterns in Bacillus Species. Indian J. Microbiol. 2017, 57, 387–392. [Google Scholar] [CrossRef]
- Kacanski, M.; Pucher, L.; Peral, C.; Dietrich, T.; Neureiter, M. Cell Retention as a Viable Strategy for PHA Production from Diluted VFAs with Bacillus megaterium. Bioengineering 2022, 9, 122. [Google Scholar] [CrossRef]
- Biedendieck, R.; Knuuti, T.; Moore, S.J.; Jahn, D. The “Beauty in the Beast”—The Multiple Uses of Priestia megaterium in Biotechnology. Appl. Microbiol. Biotechnol. 2021, 105, 5719–5737. [Google Scholar] [CrossRef]
- Edilane, M.F.; de Lima Procópio Aldo, R.; Raimundo, C.P.J.; Sandra, P.Z.; de Lima Procópio Rudi, E. Polyhydroxyalkanoate (PHA) Production by Lysinibacillus sp. Strain UEA-20.171. Afr. J. Biotechnol. 2016, 15, 1827–1834. [Google Scholar] [CrossRef] [Green Version]
- Sagong, H.-Y.; Son, H.F.; Choi, S.Y.; Lee, S.Y.; Kim, K.-J. Structural Insights into Polyhydroxyalkanoates Biosynthesis. Trends Biochem. Sci. 2018, 43, 790–805. [Google Scholar] [CrossRef]
- Vu, D.H.; Mahboubi, A.; Root, A.; Heinmaa, I.; Taherzadeh, M.J.; Åkesson, D. Thorough Investigation of the Effects of Cultivation Factors on Polyhydroalkanoates (PHAs) Production by Cupriavidus necator from Food Waste-Derived Volatile Fatty Acids. Fermentation 2022, 8, 605. [Google Scholar] [CrossRef]
- Ekere, I.; Johnston, B.; Tchuenbou-Magaia, F.; Townrow, D.; Wojciechowski, S.; Marek, A.; Zawadiak, J.; Duale, K.; Zieba, M.; Sikorska, W.; et al. Bioconversion Process of Polyethylene from Waste Tetra Pak® Packaging to Polyhydroxyalkanoates. Polymers 2022, 14, 2840. [Google Scholar] [CrossRef] [PubMed]
- Favaro, L.; Basaglia, M.; Casella, S. Improving Polyhydroxyalkanoate Production from Inexpensive Carbon Sources by Genetic Approaches: A Review. Biofuels Bioprod. Biorefin. 2019, 13, 208–227. [Google Scholar] [CrossRef] [Green Version]
- Srirangan, K.; Liu, X.; Tran, T.T.; Charles, T.C.; Moo-Young, M.; Chou, C.P. Engineering of Escherichia coli for Direct and Modulated Biosynthesis of Poly(3-Hydroxybutyrate-Co-3-Hydroxyvalerate) Copolymer Using Unrelated Carbon Sources. Sci. Rep. 2016, 6, 36470. [Google Scholar] [CrossRef] [Green Version]
- Lopes, M.S.G.; Rocha, R.C.S.; Zanotto, S.P.; Gomez, J.G.C.; da Silva, L.F. Screening of Bacteria to Produce Polyhydroxyalkanoates from Xylose. World J. Microbiol. Biotechnol. 2009, 25, 1751–1756. [Google Scholar] [CrossRef]
- Ferreira, A.M.; Queirós, D.; Gagliano, M.C.; Serafim, L.S.; Rossetti, S. Polyhydroxyalkanoates-Accumulating Bacteria Isolated from Activated Sludge Acclimatized to Hardwood Sulphite Spent Liquor. Ann. Microbiol. 2016, 66, 833–842. [Google Scholar] [CrossRef]
- Gasser, I.; Müller, H.; Berg, G. Ecology and Characterization of Polyhydroxyalkanoate-Producing Microorganisms on and in Plants. FEMS Microbiol. Ecol. 2009, 70, 142–150. [Google Scholar] [CrossRef]
- Kumar, V.; Thakur, V.; Ambika; Kumar, S.; Singh, D. Bioplastic Reservoir of Diverse Bacterial Communities Revealed along Altitude Gradient of Pangi-Chamba Trans-Himalayan Region. FEMS Microbiol. Lett. 2018, 365, fny144. [Google Scholar] [CrossRef] [Green Version]
- Elsayed, N.S.; Aboshanab, K.M.; Aboulwafa, M.M.; Hassouna, N.A. Cost-Effective Production of the Bio-Plastic Poly-Beta-Hydroxybutyrate Using Acinetobacter baumannii Isolate P39. J. Microbiol. Biotechnol. Food Sci. 2016, 05, 552–556. [Google Scholar] [CrossRef] [Green Version]
- Pereira, J.R.; Araújo, D.; Marques, A.C.; Neves, L.A.; Grandfils, C.; Sevrin, C.; Alves, V.D.; Fortunato, E.; Reis, M.A.M.; Freitas, F. Demonstration of the Adhesive Properties of the Medium-Chain-Length Polyhydroxyalkanoate Produced by Pseudomonas chlororaphis Subsp. Aurantiaca from Glycerol. Int. J. Biol. Macromol. 2019, 122, 1144–1151. [Google Scholar] [CrossRef]
- Mozejko-Ciesielska, J.; Szacherska, K.; Marciniak, P. Pseudomonas Species as Producers of Eco-Friendly Polyhydroxyalkanoates. J. Polym. Environ. 2019, 27, 1151–1166. [Google Scholar] [CrossRef] [Green Version]
- Rai, R.; Yunos, D.M.; Boccaccini, A.R.; Knowles, J.C.; Barker, I.A.; Howdle, S.M.; Tredwell, G.D.; Keshavarz, T.; Roy, I. Poly-3-Hydroxyoctanoate p(3HO), a Medium Chain Length Polyhydroxyalkanoate Homopolymer from Pseudomonas mendocina. Biomacromolecules 2011, 12, 2126–2136. [Google Scholar] [CrossRef]
- Fernández, D.; Rodríguez, E.; Bassas, M.; Viñas, M.; Solanas, A.M.; Llorens, J.; Marqués, A.M.; Manresa, A. Agro-Industrial Oily Wastes as Substrates for PHA Production by the New Strain Pseudomonas aeruginosa NCIB 40045: Effect of Culture Conditions. Biochem. Eng. J. 2005, 26, 159–167. [Google Scholar] [CrossRef]
- Kanavaki, I.; Drakonaki, A.; Geladas, E.D.; Spyros, A.; Xie, H.; Tsiotis, G. Polyhydroxyalkanoate (PHA) Production in Pseudomonas sp. phDV1 Strain Grown on Phenol as Carbon Sources. Microorganisms 2021, 9, 1636. [Google Scholar] [CrossRef] [PubMed]
- Iqbal, B.; Khan, N.; Jamil, N. Polyhydroxybutyrate Production by Stenotrophomonas and Exiguobacterium Using Renewable Carbon Source. Annu. Res. Rev. Biol. 2016, 9, 1–9. [Google Scholar] [CrossRef]
- Kačániová, M.; Klúga, A.; Kántor, A.; Medo, J.; Žiarovská, J.; Puchalski, C.; Terentjeva, M. Comparison of MALDI-TOF MS Biotyper and 16S rDNA sequencing for the identification of Pseudomonas species isolated from fish. Microb. Pathog. 2019, 132, 313–318. [Google Scholar] [CrossRef] [PubMed]
- Kitamura, S.; Doi, Y. Staining Method of Poly(3-Hydroxyalkanoic Acids) Producing Bacteria by Nile Blue. Biotechnol. Tech. 1994, 8, 345–350. [Google Scholar] [CrossRef]
- Godbole, S. Methods for Identification, Quantification and Characterization of Polyhydroxyalkanoates. Int. J. Bioassays 2016, 5, 4977–4983. [Google Scholar] [CrossRef]
- Schlegel, H.G.; Lafferty, R.; Krauss, I. The Isolation of Mutants Not Accumulating Poly-β-Hydroxybutyric Acid. Arch. Für Mikrobiol. 1970, 71, 283–294. [Google Scholar] [CrossRef]
- Liu, M.; González, J.E.; Willis, L.B.; Walker, G.C. A Novel Screening Method for Isolating Exopolysaccharide-Deficient Mutants. Appl. Environ. Microbiol. 1998, 64, 4600–4602. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arcos-Hernandez, M.V.; Gurieff, N.; Pratt, S.; Magnusson, P.; Werker, A.; Vargas, A.; Lant, P. Rapid Quantification of Intracellular PHA Using Infrared Spectroscopy: An Application in Mixed Cultures. J. Biotechnol. 2010, 150, 372–379. [Google Scholar] [CrossRef] [PubMed]
| Strain | Genes | Related References |
|---|---|---|
| Cupriavidus necator ATCC 17699 | phaA, phaB, phaC (class I) | [23] |
| Pseudomonas mendocina ATCC 25411 | phaC (class II) | [24] |
| Pseudomonas aeruginosa ATCC 27853 | phaJ, fabG | [25,26] |
| Stenotrophomonas maltophilia | phaE (class III) | [27] |
| Priestia megaterium | phaR (class IV) | [28] |
| Pseudomonas putida | phaG | [29] |
| % * | n | phaSyn1 | phaSyn2 | phaSyn3 | phaSyn4 | phaA | phaB | phaG | fabG | phaJ |
|---|---|---|---|---|---|---|---|---|---|---|
| Total | 64 | 42.2 | 4.7 | 10.9 | 15.6 | 45.3 | 18.8 | 4.7 | 12.5 | 10.9 |
| Enterobacteriaceae | 27 | 51.9 | 0.0 | 0.0 | 18.5 | 74.1 | 11.1 | 3.7 | 3.7 | 3.7 |
| Bacillaceae | 14 | 14.3 | 0.0 | 7.1 | 28.6 | 21.4 | 14.3 | 0.0 | 7.1 | 0.0 |
| Pseudomonadaceae | 11 | 45.5 | 27.3 | 27.3 | 0.0 | 36.4 | 27.3 | 18.2 | 36.4 | 27.3 |
| Moraxellaceae | 6 | 33.3 | 0.0 | 16.7 | 0.0 | 16.7 | 33.3 | 0.0 | 16.7 | 16.7 |
| Xanthomonadaceae | 3 | 33.3 | 0.0 | 66.7 | 33.3 | 0.0 | 33.3 | 0.0 | 33.3 | 33.3 |
| Staphylococcaceae | 2 | 100.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 50.0 |
| Burkholderiaceae | 1 | 100.0 | 0.0 | 0.0 | 0.0 | 100.0 | 100.0 | 0.0 | 0.0 | 0.0 |
| % * | n | Feedstock | |||
|---|---|---|---|---|---|
| Fructose | Glucose | Sunflower Oil | Propionic Acid | ||
| Total | 64 | 29.7 | 28.1 | 9.4 | 26.6 |
| Enterobacteriaceae | 27 | 3.7 | 3.7 | 0.0 | 22.2 |
| Bacillaceae | 14 | 42.9 | 35.7 | 0.0 | 28.6 |
| Pseudomonadaceae | 11 | 72.7 | 90.9 | 36.4 | 36.4 |
| Moraxellaceae | 6 | 16.7 | 0.0 | 16.7 | 16.7 |
| Xanthomonadaceae | 3 | 33.3 | 0.0 | 0.0 | 0.0 |
| Staphylococcaceae | 2 | 50.0 | 50.0 | 50.0 | 50.0 |
| Burkholderiaceae | 1 | 100.0 | 100.0 | 0.0 | 100.0 |
| Primer | Gene | Primer Sequence (5′–3′) | Product Size (bp) | Ref. |
|---|---|---|---|---|
| phaAF | phaA | CCATGACCATCAACAAGGTG | 262 | This study |
| phaAR | TATTCCTTGGCCACGTTCTC | |||
| phaBF | phaB | AATGTGGCTGACTGGGACTC | 164 | This study |
| phaBR | GAGGTCAGGTTGGTGTCGAT | |||
| phaSyn1F | phaC (Class I) | TGCGCAACATGATGGAAGAC | 204 | This study |
| phaSyn1R | AGTACTTGTTGATGCACGGC | |||
| phaSyn2F | phaC (Class II) | TACATCGAGGCGCTCAAGGA | 594 | This study |
| phaSyn2R | GCCGAACAGATGAGCCGATT | |||
| phaSyn3F | phaE (Class III) | CAGTGGATGCTGCAGGGC | 463 | This study |
| phaSyn3R | GCAGGCCGATACGTTCACTC | |||
| phaSyn4F | phaR (Class IV) | AAATGAAGTAACGGGGCGCT | 326 | This study |
| phaSyn4R | CCTGAAGCTGCTCACCTTGA | |||
| phaJF | phaJ | CGAGTACACCAGCAGCATCG | 287 | This study |
| phaJR | GGTTCTTCGGCAGCTTCTCG | |||
| fabGF | fabG | TGGCTCGAGAGAGAGAAAGGAGA | 750 | [25] |
| fabGR | TTCCGCAACGAATTCTAGAC | |||
| phaGF | phaG | AGAAGGCAGTGGTGAGTTCG | 425 | This study |
| phaGR | ACAGGCGGTCTTGTTTTCCA |
| Step Name | Temperature (°C) | Time (min) | Number of Cycles |
|---|---|---|---|
| Initial denaturation | 95 | 5 | 1 |
| Denaturation | 95 | 0.5 | 40 |
| Annealing | 65 | 0.5 | |
| Elongation | 72 | 1 | |
| Final elongation | 72 | 10 | 1 |
| Cooling | 4 | ∞ | 1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Máčalová, D.; Janalíková, M.; Sedlaříková, J.; Rektoříková, I.; Koutný, M.; Pleva, P. Genotypic and Phenotypic Detection of Polyhydroxyalkanoate Production in Bacterial Isolates from Food. Int. J. Mol. Sci. 2023, 24, 1250. https://doi.org/10.3390/ijms24021250
Máčalová D, Janalíková M, Sedlaříková J, Rektoříková I, Koutný M, Pleva P. Genotypic and Phenotypic Detection of Polyhydroxyalkanoate Production in Bacterial Isolates from Food. International Journal of Molecular Sciences. 2023; 24(2):1250. https://doi.org/10.3390/ijms24021250
Chicago/Turabian StyleMáčalová, Daniela, Magda Janalíková, Jana Sedlaříková, Iveta Rektoříková, Marek Koutný, and Pavel Pleva. 2023. "Genotypic and Phenotypic Detection of Polyhydroxyalkanoate Production in Bacterial Isolates from Food" International Journal of Molecular Sciences 24, no. 2: 1250. https://doi.org/10.3390/ijms24021250






