The LIV-1 Subfamily of Zinc Transporters: From Origins to Present Day Discoveries
Abstract
:1. Introduction
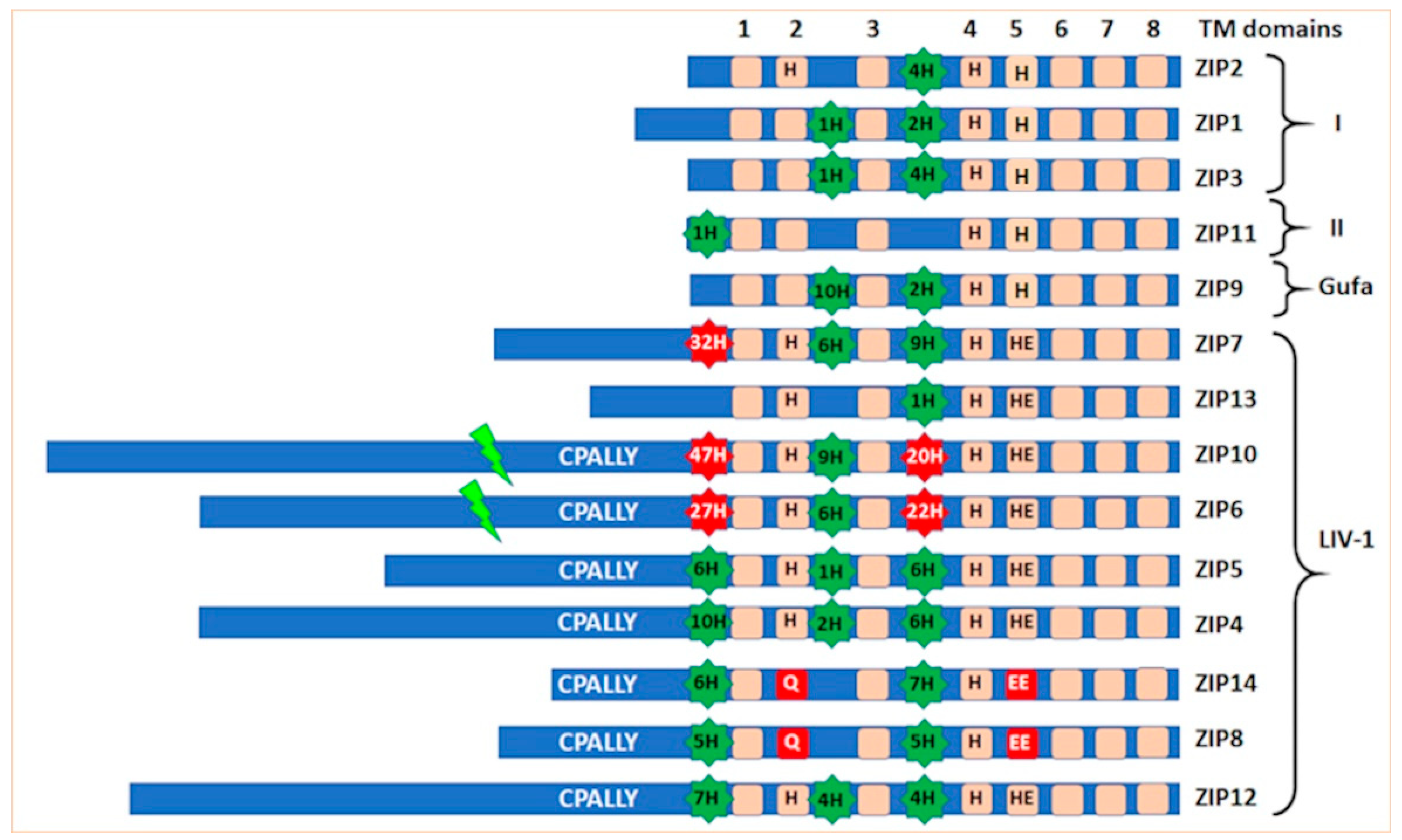
2. The Origin of the LIV-1 Family of Zinc Transporters
Discovery of the LIV-1 Subfamily
3. Special Features of the LIV-1 Subfamily of Zinc Transporters
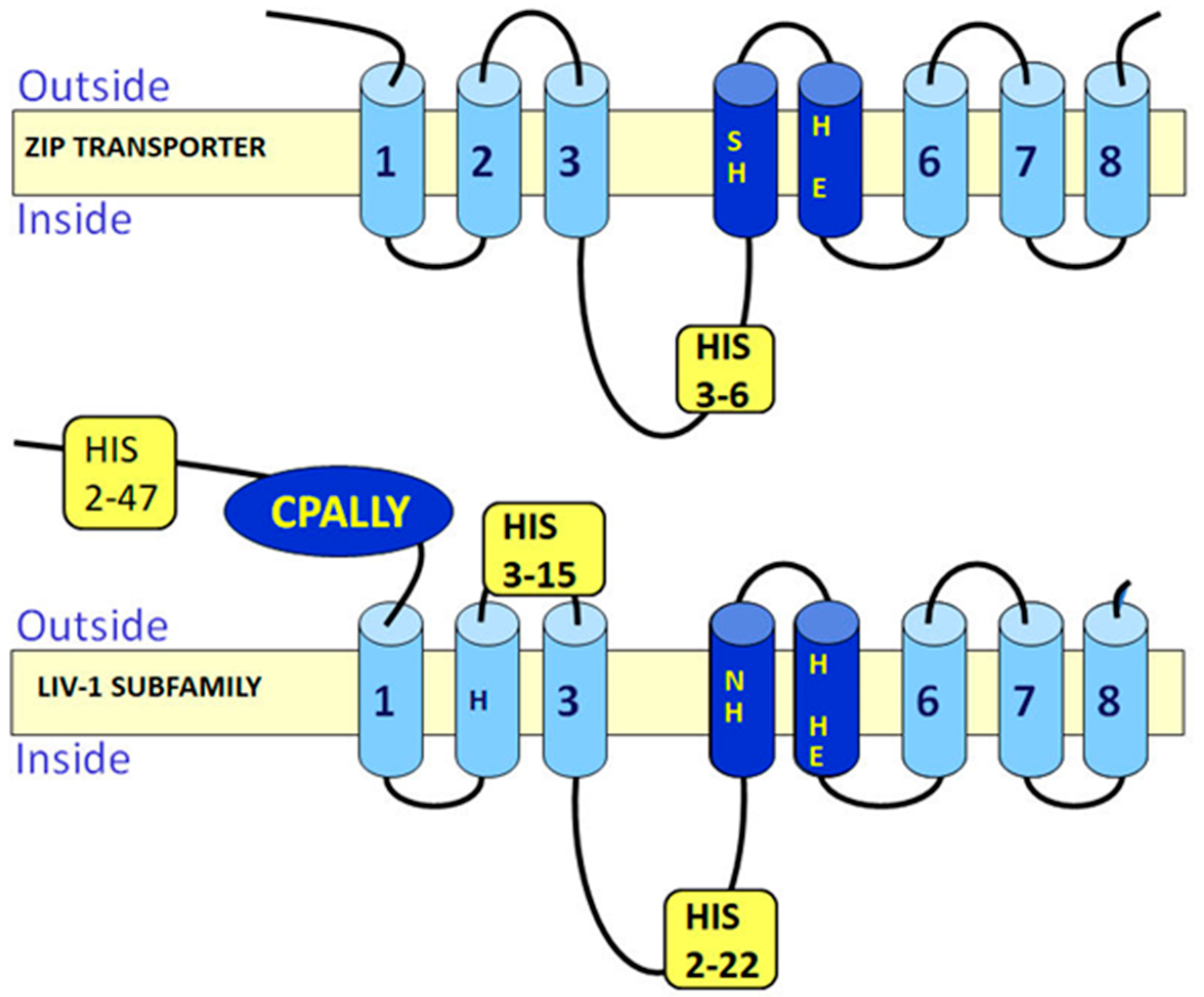
3.1. HEXPHE Motif
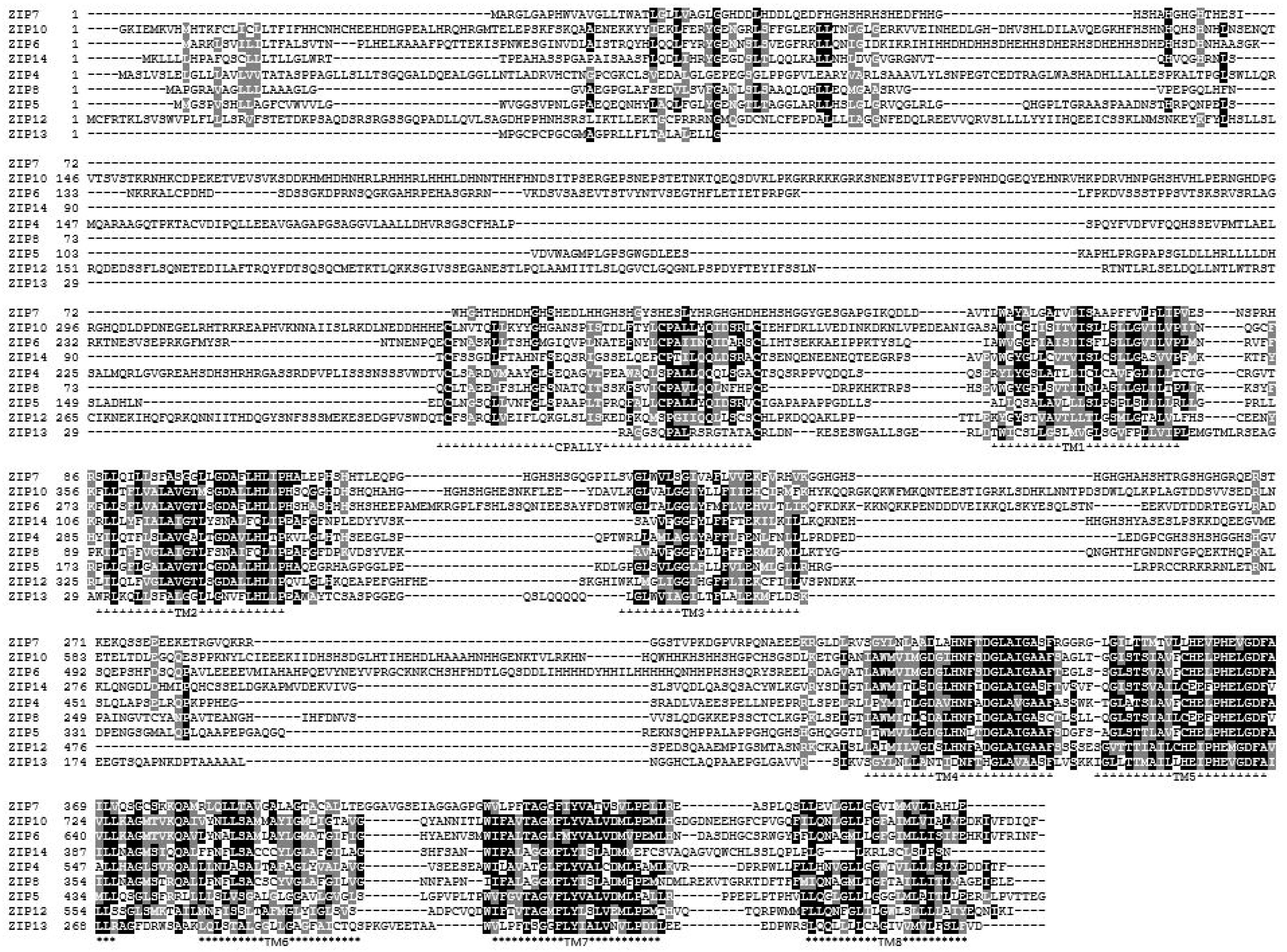
3.2. Conserved Proline Residues within TM Domains
3.3. Histidine Residues
3.4. CPALLY Motif and Link to Prions
4. The Role of Post-Translational Modification in LIV-1 Family Function
4.1. Proteolytic Cleavage
4.2. Phosphorylation
5. The Role of ZIP Transporters in Cancer
5.1. ZIP7 Drives Anti-Hormone-Resistant Breast Cancer
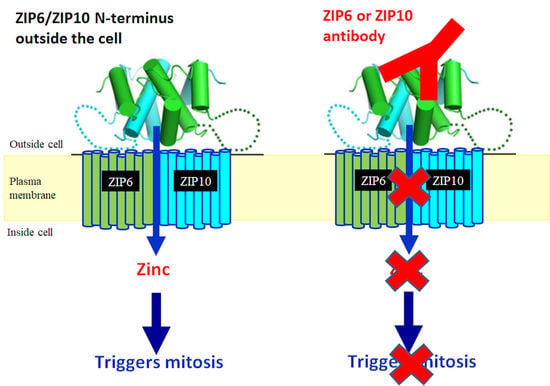
5.2. ZIP6 and ZIP10 Are Essential for Cell Mitosis
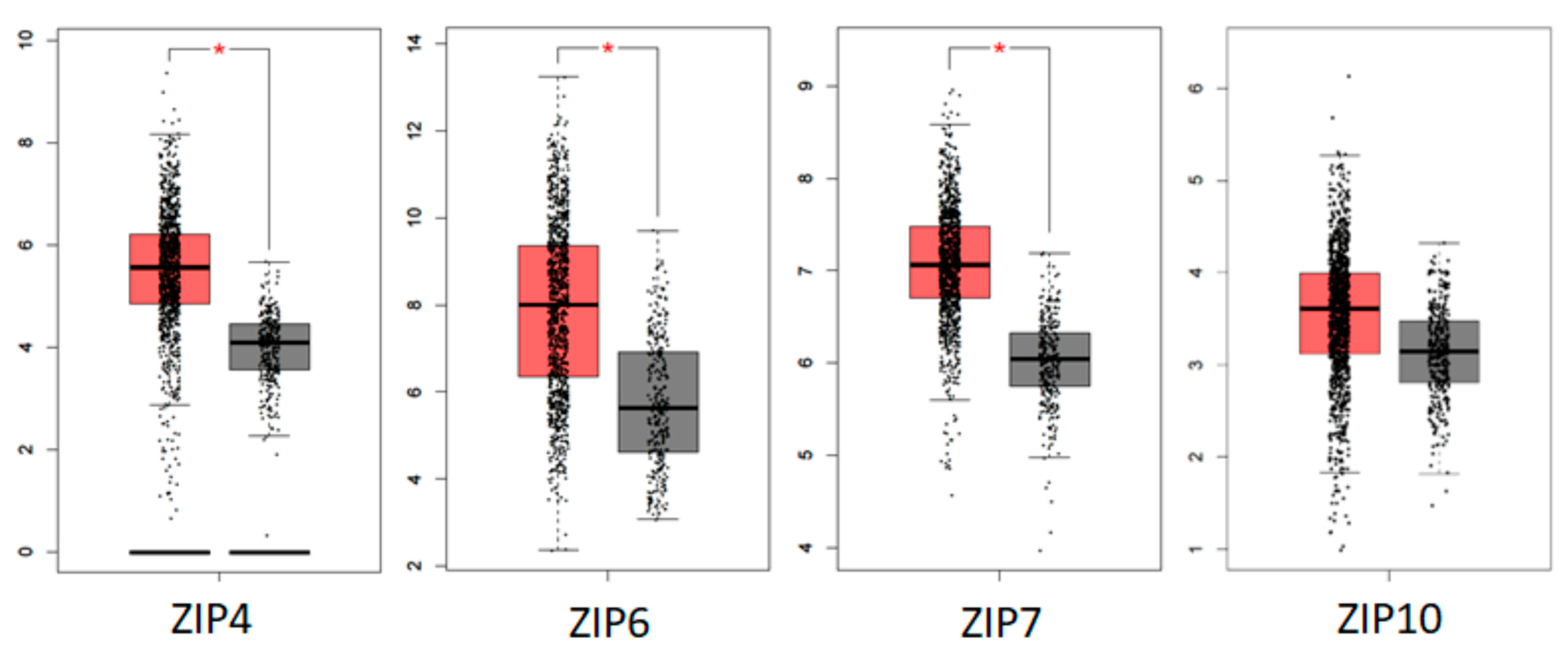
5.3. Human Tumour Database Evidence of the Role of ZIP Transporters in Breast Cancer
6. Additional Areas of Investigation
6.1. ZIP7 in Immunity and B Cell Activation
6.2. The Effect of ZIP Knockdown on Other ZIP Transporters
6.3. Zinc and Calcium
7. Conclusions
8. Patents
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Manning, D.L.; McClelland, R.A.; Knowlden, J.M.; Bryant, S.; Gee, J.M.; Green, C.D.; Robertson, J.F.; Blamey, R.W.; Sutherland, R.L.; Ormandy, C.J.; et al. Differential Expression of Oestrogen Regulated Genes in Breast Cancer. Acta Oncol. 1995, 34, 641–646. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manning, D.; Robertson, J.; Ellis, I.; Elston, C.; McClelland, R.; Gee, J.; Jones, R.; Green, C.; Cannon, P.; Blamey, R.; et al. Oestrogen-regulated genes in breast cancer: Association of pLIV1 with lymph node involvement. Eur. J. Cancer 1994, 30, 675–678. [Google Scholar] [CrossRef]
- McClelland, R.; Manning, D.; Gee, J.; Willsher, P.; Robertson, J.; Ellis, I.; Blamey, R.; Nicholson, R. Oestrogen-regulated genes in breast cancer: Association of pLIV1 with response to endocrine therapy. Br. J. Cancer 1998, 77, 1653–1656. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taylor, K.M. LIV-1 Breast Cancer Protein Belongs to New Family of Histidine-Rich Membrane Proteins with Potential to Control Intracellular Zn2+ Homeostasis. IUBMB Life 2000, 49, 249–253. [Google Scholar] [CrossRef] [PubMed]
- Hooper, N.M. Families of zinc metalloproteases. FEBS Lett. 1994, 354, 1–6. [Google Scholar] [CrossRef] [Green Version]
- Dardel, F.; Ragusa, S.; Lazennec, C.; Blanquet, S.; Meinnel, T. Solution structure of nickel-peptide deformylase. J. Mol. Biol. 1998, 280, 501–513. [Google Scholar] [CrossRef]
- Jiang, W.; Bond, J.S. Families of metalloendopeptidases and their relationships. FEBS Lett. 1992, 312, 110–114. [Google Scholar] [CrossRef] [Green Version]
- Taylor, K.M.; Nicholson, R.I. The LZT proteins; the LIV-1 subfamily of zinc transporters. Biochim. Biophys. Acta (BBA)—Biomembr. 2003, 1611, 16–30. [Google Scholar] [CrossRef] [Green Version]
- Eide, D.J. The SLC39 family of metal ion transporters. Pflugers Arch. 2004, 447, 796–800. [Google Scholar] [CrossRef]
- Kambe, T.; Taylor, K.M.; Fu, D. Zinc transporters and their functional integration in mammalian cells. J. Biol. Chem. 2021, 296, 100320. [Google Scholar] [CrossRef]
- Zhang, T.; Liu, J.; Fellner, M.; Zhang, C.; Sui, D.; Hu, J. Crystal structures of a ZIP zinc transporter reveal a binuclear metal center in the transport pathway. Sci. Adv. 2017, 3, e1700344. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef] [PubMed]
- Krężel, A.; Maret, W. The biological inorganic chemistry of zinc ions. Arch. Biochem. Biophys. 2016, 611, 3–19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gomis-Rüth, F.X. Structural Aspects of the Metzincin Clan of Metalloendopeptidases. Mol. Biotechnol. 2003, 24, 157–202. [Google Scholar] [CrossRef]
- Aydemir, T.B.; Cousins, R.J. The Multiple Faces of the Metal Transporter ZIP14 (SLC39A14). J. Nutr. 2018, 148, 174–184. [Google Scholar] [CrossRef] [Green Version]
- Fujishiro, H.; Kambe, T. Manganese transport in mammals by zinc transporter family proteins, ZNT and ZIP. J. Pharmacol. Sci. 2022, 148, 125–133. [Google Scholar] [CrossRef]
- Winslow, J.W.; Limesand, K.H.; Zhao, N. The Functions of ZIP8, ZIP14, and ZnT10 in the Regulation of Systemic Manganese Homeostasis. Int. J. Mol. Sci. 2020, 21, 3304. [Google Scholar] [CrossRef]
- Widhalm, R.; Ellinger, I.; Granitzer, S.; Forsthuber, M.; Bajtela, R.; Gelles, K.; Hartig, P.Y.; Hengstschlager, M.; Zeisler, H.; Salzer, H.; et al. Human placental cell line HTR-8/SVneo accumulates cadmium by divalent metal transporters DMT1 and ZIP14. Metallomics 2020, 12, 1822–1833. [Google Scholar] [CrossRef]
- Kosman, D.J. A holistic view of mammalian (vertebrate) cellular iron uptake. Metallomics 2020, 12, 1323–1334. [Google Scholar] [CrossRef]
- Knutson, M.D. Non-transferrin-bound iron transporters. Free Radic. Biol. Med. 2019, 133, 101–111. [Google Scholar] [CrossRef]
- Fujishiro, H.; Miyamoto, S.; Sumi, D.; Kambe, T.; Himeno, S. Effects of individual amino acid mutations of zinc transporter ZIP8 on manganese- and cadmium-transporting activity. Biochem. Biophys. Res. Commun. 2022, 616, 26–32. [Google Scholar] [CrossRef] [PubMed]
- Wiuf, A.; Steffen, J.H.; Becares, E.R.; Grønberg, C.; Mahato, D.R.; Rasmussen, S.G.F.; Andersson, M.; Croll, T.; Gotfryd, K.; Gourdon, P. The two-domain elevator-type mechanism of zinc-transporting ZIP proteins. Sci. Adv. 2022, 8, eabn4331. [Google Scholar] [CrossRef] [PubMed]
- Hegedűs, T.; Geisler, M.; Lukács, G.L.; Farkas, B. Ins and outs of AlphaFold2 transmembrane protein structure predictions. Cell. Mol. Life Sci. 2022, 79, 73. [Google Scholar] [CrossRef] [PubMed]
- Nilsson, I.; Sääf, A.; Whitley, P.; Gafvelin, G.; Waller, C.; von Heijne, G. Proline-induced disruption of a transmembrane α-helix in its natural environment. J. Mol. Biol. 1998, 284, 1165–1175. [Google Scholar] [CrossRef] [Green Version]
- Woolfson, D.N.; Williams, D.H. The influence of proline residues on α-helical structure. FEBS Lett. 1990, 277, 185–188. [Google Scholar] [CrossRef] [Green Version]
- Glatzova, D.; Mavila, H.; Saija, M.C.; Chum, T.; Cwiklik, L.; Brdicka, T.; Cebecauer, M. The role of prolines and glycine in the transmembrane domain of LAT. FEBS J. 2021, 288, 4039–4052. [Google Scholar] [CrossRef]
- Tieleman, D.P.; Shrivastava, I.H.; Ulmschneider, M.R.; Sansom, M.S. Proline-induced hinges in transmembrane helices: Possible roles in ion channel gating. Proteins Struct. Funct. Bioinform. 2001, 44, 63–72. [Google Scholar] [CrossRef]
- Palmieri, F.; Pierri, C.L. Structure and function of mitochondrial carriers—Role of the transmembrane helix P and G residues in the gating and transport mechanism. FEBS Lett. 2010, 584, 1931–1939. [Google Scholar] [CrossRef] [Green Version]
- Olenic, S.; Buchanan, F.; VanPortfliet, J.; Parrell, D.; Kroos, L. Conserved Proline Residues of Bacillus subtilis Intramembrane Metalloprotease SpoIVFB Are Important for Substrate Interaction and Cleavage. J. Bacteriol. 2022, 204, e0038621. [Google Scholar] [CrossRef]
- Partridge, A.W.; Therien, A.G.; Deber, C.M. Missense mutations in transmembrane domains of proteins: Phenotypic propensity of polar residues for human disease. Proteins Struct. Funct. Bioinform. 2004, 54, 648–656. [Google Scholar] [CrossRef]
- Waldron, K.J.; Rutherford, J.C.; Ford, D.; Robinson, N.J. Metalloproteins and metal sensing. Nature 2009, 460, 823–830. [Google Scholar] [CrossRef] [PubMed]
- Vallee, B.L.; Auld, D.S. Zinc coordination, function, and structure of zinc enzymes and other proteins. Biochemistry 1990, 29, 5647–5659. [Google Scholar] [CrossRef] [PubMed]
- Schmitt-Ulms, G.; Ehsani, S.; Watts, J.C.; Westaway, D.; Wille, H. Evolutionary Descent of Prion Genes from the ZIP Family of Metal Ion Transporters. PLoS ONE 2009, 4, e7208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perera, W.S.; Hooper, N.M. Ablation of the metal ion-induced endocytosis of the prion protein by disease-associated mutation of the octarepeat region. Curr. Biol. 2001, 11, 519–523. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ehsani, S.; Mehrabian, M.; Pocanschi, C.L.; Schmitt-Ulms, G. The ZIP-prion connection. Prion 2012, 6, 317–321. [Google Scholar] [CrossRef] [Green Version]
- Brethour, D.; Mehrabian, M.; Williams, D.; Wang, X.; Ghodrati, F.; Ehsani, S.; Rubie, E.A.; Woodgett, J.R.; Sevalle, J.; Xi, Z.; et al. A ZIP6-ZIP10 heteromer controls NCAM1 phosphorylation and integration into focal adhesion complexes during epithelial-to-mesenchymal transition. Sci. Rep. 2017, 7, 40313. [Google Scholar] [CrossRef] [Green Version]
- Ma, Z.; Li, Z.; Wang, S.; Zhou, Q.; Ma, Z.; Liu, C.; Huang, B.; Zheng, Z.; Yang, L.; Zou, Y.; et al. SLC39A10 Upregulation Predicts Poor Prognosis, Promotes Proliferation and Migration, and Correlates with Immune Infiltration in Hepatocellular Carcinoma. J. Hepatocell. Carcinoma 2021, 8, 899–912. [Google Scholar] [CrossRef]
- Wan, Z.; Wang, X. Role of SLC39A6 in the development and progression of liver cancer. Oncol. Lett. 2022, 23, 77. [Google Scholar] [CrossRef]
- Li, H.; Shen, X.; Ma, M.; Liu, W.; Yang, W.; Wang, P.; Cai, Z.; Mi, R.; Lu, Y.; Zhuang, J.; et al. ZIP10 drives osteosarcoma proliferation and chemoresistance through ITGA10-mediated activation of the PI3K/AKT pathway. J. Exp. Clin. Cancer Res. 2021, 40, 340. [Google Scholar] [CrossRef]
- Hogstrand, C.; Kille, P.; Ackland, M.L.; Hiscox, S.; Taylor, K.M. A mechanism for epithelial–mesenchymal transition and anoikis resistance in breast cancer triggered by zinc channel ZIP6 and STAT3 (signal transducer and activator of transcription 3). Biochem. J. 2013, 455, 229–237. [Google Scholar] [CrossRef]
- Kambe, T.; Andrews, G.K. Novel Proteolytic Processing of the Ectodomain of the Zinc Transporter ZIP4 (SLC39A4) during Zinc Deficiency Is Inhibited by Acrodermatitis Enteropathica Mutations. Mol. Cell. Biol. 2009, 29, 129–139. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ehsani, S.; Salehzadeh, A.; Huo, H.; Reginold, W.; Pocanschi, C.L.; Ren, H.; Wang, H.; So, K.; Sato, C.; Mehrabian, M.; et al. LIV-1 ZIP Ectodomain Shedding in Prion-Infected Mice Resembles Cellular Response to Transition Metal Starvation. J. Mol. Biol. 2012, 422, 556–574. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rogers, S.; Wells, R.; Rechsteiner, M. Amino Acid Sequences Common to Rapidly Degraded Proteins: The PEST Hypothesis. Science 1986, 234, 364–368. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Zhang, X. New insights into S2P signaling cascades: Regulation, variation, and conservation. Protein Sci. 2010, 19, 2015–2030. [Google Scholar] [CrossRef] [Green Version]
- Maret, W. Zinc in Cellular Regulation: The Nature and Significance of “Zinc Signals”. Int. J. Mol. Sci. 2017, 18, 2285. [Google Scholar] [CrossRef] [Green Version]
- Nimmanon, T.; Ziliotto, S.; Morris, S.; Flanagan, L.; Taylor, K.M. Phosphorylation of zinc channel ZIP7 drives MAPK, PI3K and mTOR growth and proliferation signalling. Metallomics 2017, 9, 471–481. [Google Scholar] [CrossRef] [Green Version]
- Taylor, K.M.; Kille, P.; Hogstrand, C. Protein kinase CK2 opens the gate for zinc signaling. Cell Cycle 2012, 11, 1863–1864. [Google Scholar] [CrossRef] [Green Version]
- Ziliotto, S.; Gee, J.M.W.; O Ellis, I.; Green, A.R.; Finlay, P.; Gobbato, A.; Taylor, K.M. Activated zinc transporter ZIP7 as an indicator of anti-hormone resistance in breast cancer. Metallomics 2019, 11, 1579–1592. [Google Scholar] [CrossRef] [Green Version]
- Hornbeck, P.V.; Zhang, B.; Murray, B.; Kornhauser, J.M.; Latham, V.; Skrzypek, E. PhosphoSitePlus, 2014: Mutations, PTMs and recalibrations. Nucleic Acids Res. 2015, 43, D512–D520. [Google Scholar] [CrossRef] [Green Version]
- Blom, N.; Sicheritz-Pontén, T.; Gupta, R.; Gammeltoft, S.; Brunak, S. Prediction of post-translational glycosylation and phosphorylation of proteins from the amino acid sequence. Proteomics 2004, 4, 1633–1649. [Google Scholar] [CrossRef]
- Fuhs, S.R.; Hunter, T. pHisphorylation: The emergence of histidine phosphorylation as a reversible regulatory modification. Curr. Opin. Cell Biol. 2017, 45, 8–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manning, B.D.; Cantley, L.C. AKT/PKB Signaling: Navigating Downstream. Cell 2007, 129, 1261–1274. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fruman, D.A.; Chiu, H.; Hopkins, B.D.; Bagrodia, S.; Cantley, L.C.; Abraham, R.T. The PI3K Pathway in Human Disease. Cell 2017, 170, 605–635. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, Y.J.; Pan, W.W.; Liu, S.B.; Shen, Z.F.; Xu, Y.; Hu, L.L. ERK/MAPK signalling pathway and tumorigenesis. Exp. Ther. Med. 2020, 19, 1997–2007. [Google Scholar] [CrossRef] [Green Version]
- Sabatini, D.M. Twenty-five years of mTOR: Uncovering the link from nutrients to growth. Proc. Natl. Acad. Sci. USA 2017, 114, 11818–11825. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Racaud-Sultan, C.; Vergnolle, N. GSK3β, a Master Kinase in the Regulation of Adult Stem Cell Behavior. Cells 2021, 10, 225. [Google Scholar] [CrossRef]
- Bellomo, E.; Massarotti, A.; Hogstrand, C.; Maret, W. Zinc ions modulate protein tyrosine phosphatase 1B activity. Metallomics 2014, 6, 1229–1239. [Google Scholar] [CrossRef] [Green Version]
- Hutcheson, I.R.; Knowlden, J.M.; Madden, T.-A.; Barrow, D.; Gee, J.M.; Wakeling, A.E.; Nicholson, R.I. Oestrogen Receptor-Mediated Modulation of the EGFR/MAPK Pathway in Tamoxifen-Resistant MCF-7 Cells. Breast Cancer Res. Treat. 2003, 81, 81–93. [Google Scholar] [CrossRef]
- Taylor, K.M.; Hiscox, S.; Nicholson, R.I.; Hogstrand, C.; Kille, P. Protein Kinase CK2 Triggers Cytosolic Zinc Signaling Pathways by Phosphorylation of Zinc Channel ZIP7. Sci. Signal. 2012, 5, ra11. [Google Scholar] [CrossRef] [Green Version]
- Nimmanon, T.; Ziliotto, S.; Ogle, O.; Burt, A.; Gee, J.M.W.; Andrews, G.K.; Kille, P.; Hogstrand, C.; Maret, W.; Taylor, K.M. The ZIP6/ZIP10 heteromer is essential for the zinc-mediated trigger of mitosis. Cell. Mol. Life Sci. 2021, 78, 1781–1798. [Google Scholar] [CrossRef]
- Ng, D.; Lin, B.H.; Lim, C.P.; Huang, G.; Zhang, T.; Poli, V.; Cao, X. Stat3 regulates microtubules by antagonizing the depolymerization activity of stathmin. J. Cell Biol. 2006, 172, 245–257. [Google Scholar] [CrossRef] [PubMed]
- Silva, V.C.; Cassimeris, L. Stathmin and microtubules regulate mitotic entry in HeLa cells by controlling activation of both Aurora kinase A and Plk1. Mol. Biol. Cell 2013, 24, 3819–3831. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, Z.; Li, C.; Kang, B.; Gao, G.; Li, C.; Zhang, Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017, 45, W98–W102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- The Cancer Genome Atlas Research Network; Weinstein, J.N.; Collisson, E.A.; Mills, G.B.; Shaw, K.R.M.; Ozenberger, B.A.; Ellrott, K.; Shmulevich, I.; Sander, C.; Stuart, J.M. The Cancer Genome Atlas Pan-Cancer analysis project. Nat. Genet. 2013, 45, 1113–1120. [Google Scholar] [CrossRef] [PubMed]
- Consortium, G.T. The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 2013, 45, 580–585. [Google Scholar]
- Wu, X.; Wu, H.; Liu, L.; Qiang, G.; Zhu, J. Serum zinc level and tissue ZIP4 expression are related to the prognosis of patients with stages I–III colon cancer. Transl. Cancer Res. 2020, 9, 5585–5594. [Google Scholar] [CrossRef] [PubMed]
- Ollig, J.; Kloubert, V.; Taylor, K.M.; Rink, L. B cell activation and proliferation increase intracellular zinc levels. J. Nutr. Biochem. 2019, 64, 72–79. [Google Scholar] [CrossRef]
- Cildir, G.; Low, K.C.; Tergaonkar, V. Noncanonical NF-kappaB Signaling in Health and Disease. Trends Mol. Med. 2016, 22, 414–429. [Google Scholar] [CrossRef]
- Matsuzawa, A.; Tseng, P.-H.; Vallabhapurapu, S.; Luo, J.-L.; Zhang, W.; Wang, H.; Vignali, D.A.A.; Gallagher, E.; Karin, M. Essential Cytoplasmic Translocation of a Cytokine Receptor–Assembled Signaling Complex. Science 2008, 321, 663–668. [Google Scholar] [CrossRef] [Green Version]
- Ewing, R.M.; Chu, P.; Elisma, F.; Li, H.; Taylor, P.; Climie, S.; McBroom-Cerajewski, L.; Robinson, M.; O’Connor, L.; Li, M.; et al. Large-scale mapping of human protein–protein interactions by mass spectrometry. Mol. Syst. Biol. 2007, 3, 89. [Google Scholar] [CrossRef]
- Kumar, M.; Michael, S.; Alvarado-Valverde, J.; Mészáros, B.; Sámano-Sánchez, H.; Zeke, A.; Dobson, L.; Lazar, T.; Örd, M.; Nagpal, A.; et al. The Eukaryotic Linear Motif resource: 2022 release. Nucleic Acids Res. 2021, 50, D497–D508. [Google Scholar] [CrossRef] [PubMed]
- Anzilotti, C.; Swan, D.J.; Boisson, B.; Deobagkar-Lele, M.; Oliveira, C.; Chabosseau, P.; Engelhardt, K.R.; Xu, X.; Chen, R.; Alvarez, L.; et al. An essential role for the Zn2+ transporter ZIP7 in B cell development. Nat. Immunol. 2019, 20, 350–361. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Inoue, K.; O’Bryant, Z.; Xiong, Z.G. Zinc-Permeable Ion Channels: Effects on Intracellular Zinc Dynamics and Potential Physiological/Pathophysiological Significance. Curr. Med. Chem. 2015, 22, 1248–1257. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abiria, S.A.; Krapivinsky, G.; Sah, R.; Santa-Cruz, A.G.; Chaudhuri, D.; Zhang, J.; Adstamongkonkul, P.; DeCaen, P.G.; Clapham, D.E. TRPM7 senses oxidative stress to release Zn2+ from unique intracellular vesicles. Proc. Natl. Acad. Sci. USA 2017, 114, E6079–E6088. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamasaki, S.; Sakata-Sogawa, K.; Hasegawa, A.; Suzuki, T.; Kabu, K.; Sato, E.; Kurosaki, T.; Yamashita, S.; Tokunaga, M.; Nishida, K.; et al. Zinc is a novel intracellular second messenger. J. Cell Biol. 2007, 177, 637–645. [Google Scholar] [CrossRef] [Green Version]
- Woodier, J.; Rainbow, R.; Stewart, A.J.; Pitt, S.J. Intracellular Zinc Modulates Cardiac Ryanodine Receptor-mediated Calcium Release. J. Biol. Chem. 2015, 290, 17599–17610. [Google Scholar] [CrossRef] [Green Version]
- Pitt, S.J.; Stewart, A.J. Examining a new role for zinc in regulating calcium release in cardiac muscle. Biochem. Soc. Trans. 2015, 43, 359–363. [Google Scholar] [CrossRef]
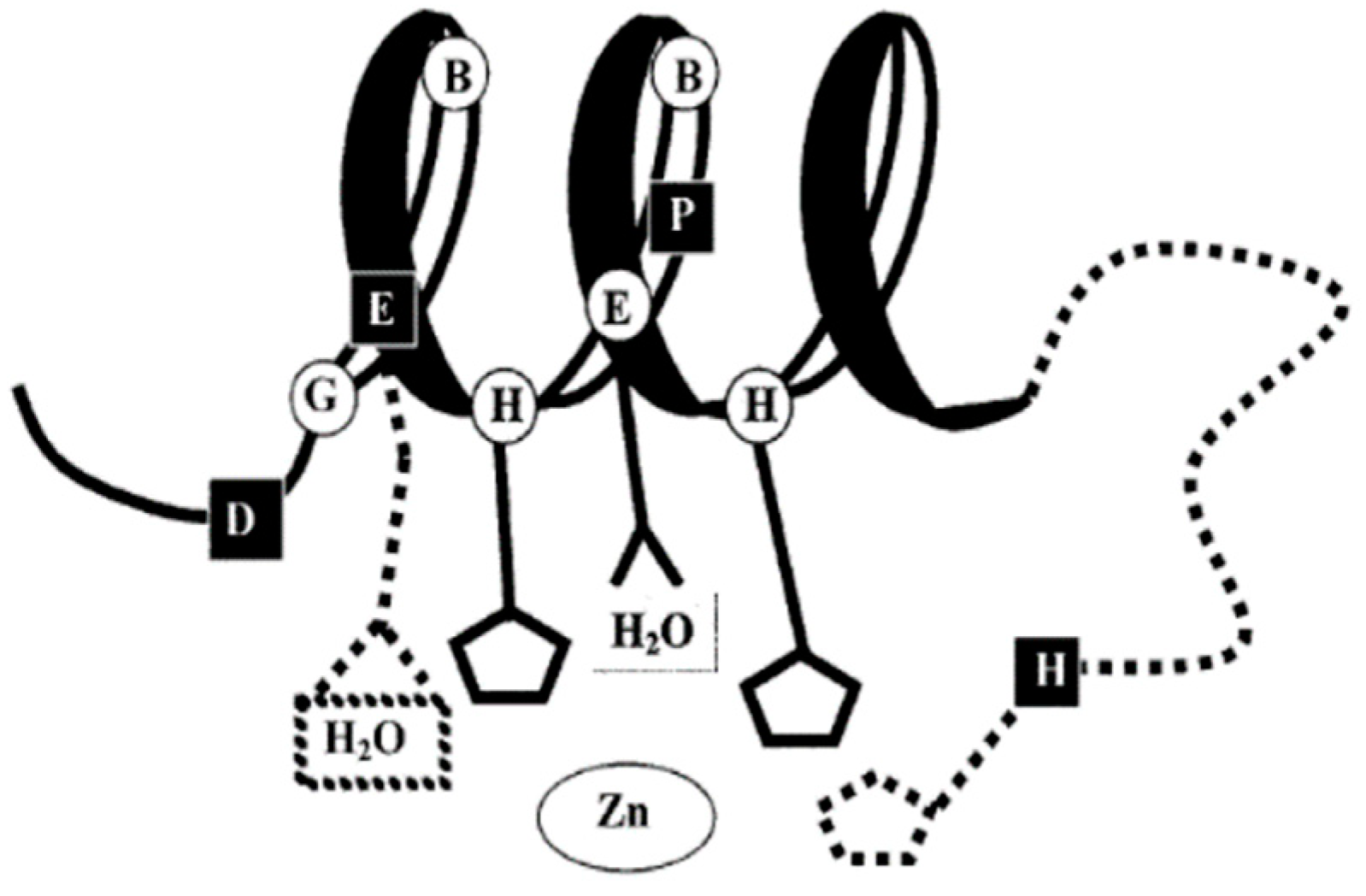

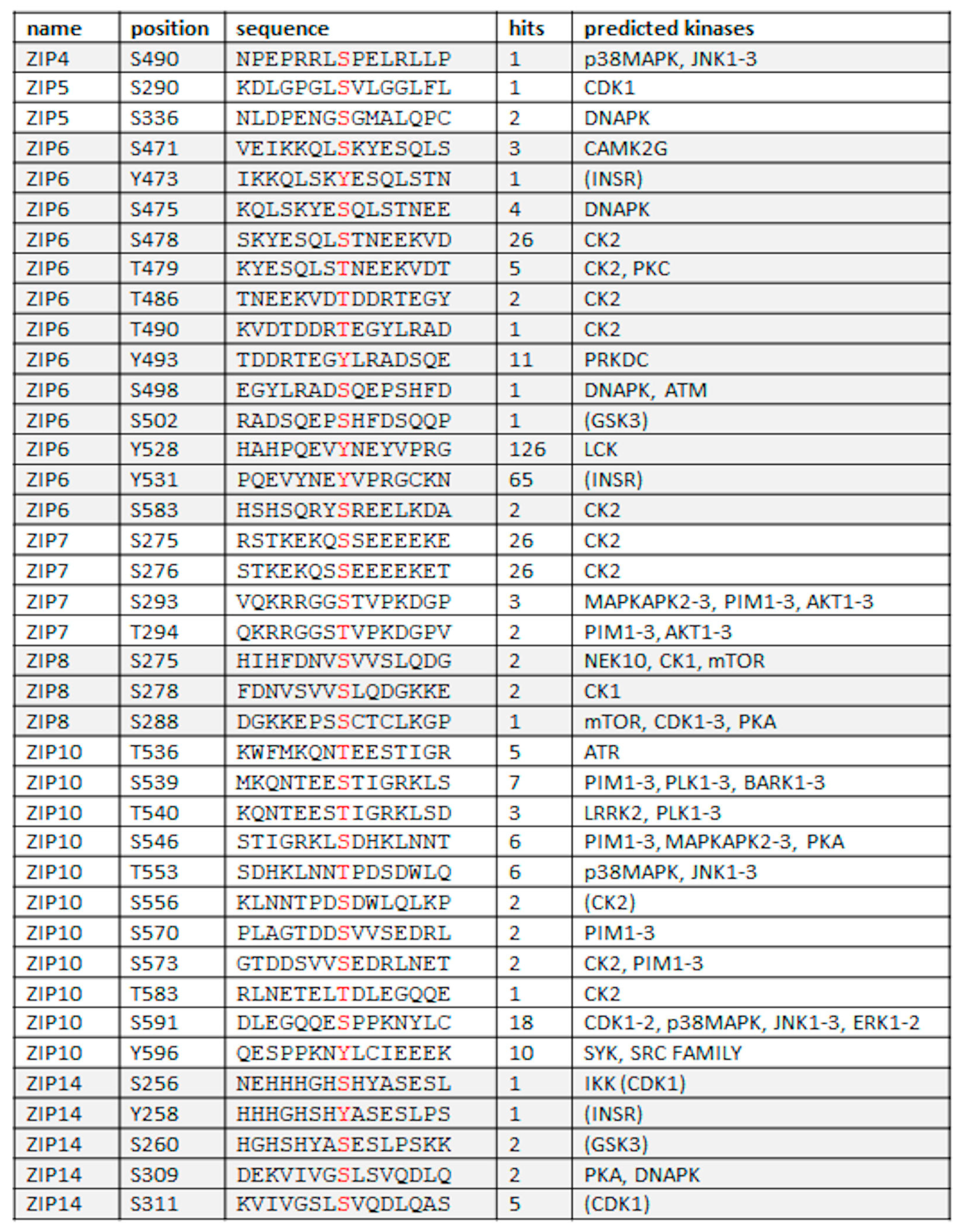
| Protease Family | TM Motif | TM Motif | TM Motif | TM Motif | C-Terminal Motif |
|---|---|---|---|---|---|
| Presenilin | YD  | LGLGDFI 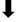 | LPALPI | ||
| SPP | YD 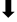 | LGLGD  | QPALLYXXP | ||
| S2P animals | HEIGH 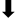 | LDG 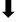 | |||
| S2P bacteria | HEIGH  | LDG  | |||
| spoIVFB | HEXXH  | LDGG  | NLLP  |
| Location + Direction | TM2  | TM4  | TM5  | TM5  | TM7  | TM8  | N-TERMINUS |
|---|---|---|---|---|---|---|---|
| ZIP4 | HLTP | DGLA | HEXPH | LGDFA | HLTP | GLLXG | SPALLQQ |
| ZIP5 | HLLP | DGLA | HEXPH | LGDFA | HLLP | GLLXG | CPALLYQ |
| ZIP6 | HLLP | DGLA | HEXPH | LGDFA | HLLP | GMLXG | CPAIINQ |
| ZIP7 | HLIP | DGLA | HEXPH | VDGFA | HLIP | GLLXG | |
| ZIP8 | QLIP | DGLA | EEXPH | LDGFA | QLIP | GMLXG | CPAVLQQ |
| ZIP10 | HLLP | DGLA | HEXPH | LDGFA | HLLP | GLLXG | CPALLYQ |
| ZIP12 | HLIP | DGLA | HEXPH | MGDFA | HLIP | GLIXG | SPGIIQQ |
| ZIP13 | HLLP | HGLA | HEXPH | VGDFA | HLLP | LLCXG | |
| ZIP14 | QLIP | DGLA | EEXPH | LDGFA | QLIP | GLLXG | CPTILQQ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Taylor, K.M. The LIV-1 Subfamily of Zinc Transporters: From Origins to Present Day Discoveries. Int. J. Mol. Sci. 2023, 24, 1255. https://doi.org/10.3390/ijms24021255
Taylor KM. The LIV-1 Subfamily of Zinc Transporters: From Origins to Present Day Discoveries. International Journal of Molecular Sciences. 2023; 24(2):1255. https://doi.org/10.3390/ijms24021255
Chicago/Turabian StyleTaylor, Kathryn M. 2023. "The LIV-1 Subfamily of Zinc Transporters: From Origins to Present Day Discoveries" International Journal of Molecular Sciences 24, no. 2: 1255. https://doi.org/10.3390/ijms24021255
APA StyleTaylor, K. M. (2023). The LIV-1 Subfamily of Zinc Transporters: From Origins to Present Day Discoveries. International Journal of Molecular Sciences, 24(2), 1255. https://doi.org/10.3390/ijms24021255