CXADR: From an Essential Structural Component to a Vital Signaling Mediator in Spermatogenesis
Abstract
:1. Introduction
2. Structure of the CXADR Protein and Gene
2.1. Structure of the CXADR Gene
2.2. Structure of the CXADR Protein
3. CXADR Knockout and Overexpression Mouse Models
4. Pathogeneses Associated with CXADR Dysregulation
5. Functions of CXADR
5.1. CXADR Functions as Structural Component of Tight and Adherens Junctions
5.2. CXADR Participates in and Regulates the Formation of Other Cell Junctions
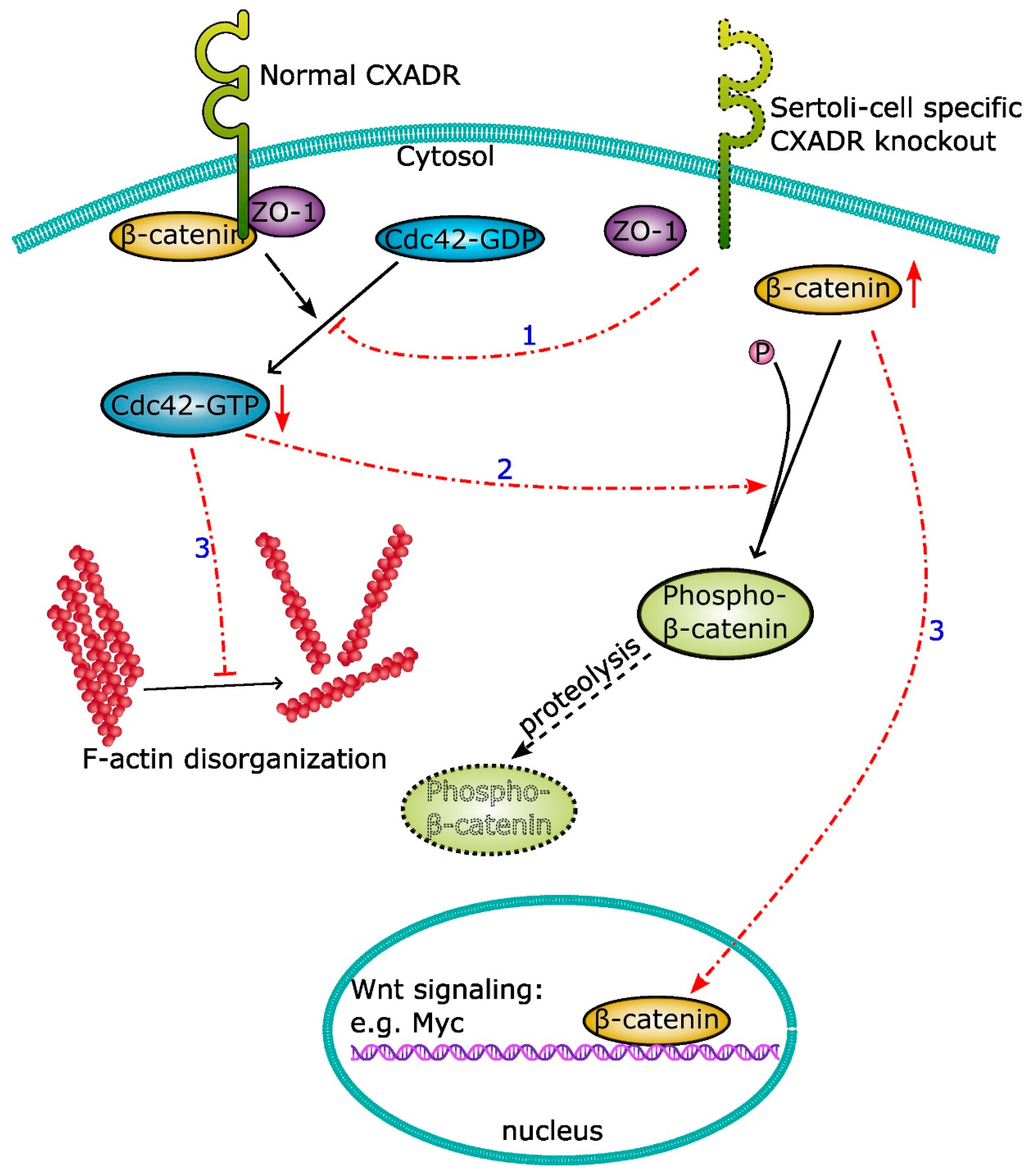
5.3. An Emerging Role of CXADR as a Signaling Mediator
6. Regulation of CXADR
6.1. Ectodomain Shedding and Intramembrane Proteolysis
6.2. Cytokines
6.3. Components of Cell Junctions
6.4. MicroRNAs and E3 Ligase
7. Future Directions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Mruk, D.D.; Cheng, C.Y. The Mammalian Blood-Testis Barrier: Its Biology and Regulation. Endocr. Rev. 2015, 36, 564–591. [Google Scholar] [CrossRef] [PubMed]
- Mruk, D.D.; Cheng, C.Y. Cell-cell interactions at the ectoplasmic specialization in the testis. Trends Endocrinol. Metab. 2004, 15, 439–447. [Google Scholar] [CrossRef] [PubMed]
- Stanton, P.G. Regulation of the blood-testis barrier. Semin. Cell Dev. Biol. 2016, 59, 166–173. [Google Scholar] [CrossRef]
- Bergelson, J.M.; Cunningham, J.A.; Droguett, G.; Kurt-Jones, E.A.; Krithivas, A.; Hong, J.S.; Horwitz, M.S.; Crowell, R.L.; Finberg, R.W. Isolation of a common receptor for Coxsackie B viruses and adenoviruses 2 and 5. Science 1997, 275, 1320–1323. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.Q.; Mruk, D.D.; Lee, W.M.; Cheng, C.Y. Coxsackie and adenovirus receptor (CAR) is a product of Sertoli and germ cells in rat testes which is localized at the Sertoli-Sertoli and Sertoli-germ cell interface. Exp. Cell Res. 2007, 313, 1373–1392. [Google Scholar] [CrossRef] [Green Version]
- Peters, A.H.; Drumm, J.; Ferrell, C.; Roth, D.A.; Roth, D.M.; McCaman, M.; Novak, P.L.; Friedman, J.; Engler, R.; Braun, R.E. Absence of germline infection in male mice following intraventricular injection of adenovirus. Mol. Ther. 2001, 4, 603–613. [Google Scholar] [CrossRef]
- Pong, R.C.; Lai, Y.J.; Chen, H.; Okegawa, T.; Frenkel, E.; Sagalowsky, A.; Hsieh, J.T. Epigenetic regulation of coxsackie and adenovirus receptor (CAR) gene promoter in urogenital cancer cells. Cancer Res. 2003, 63, 8680–8686. [Google Scholar]
- Thoelen, I.; Magnusson, C.; Tagerud, S.; Polacek, C.; Lindberg, M.; Van Ranst, M. Identification of alternative splice products encoded by the human coxsackie-adenovirus receptor gene. Biochem. Biophys. Res. Commun. 2001, 287, 216–222. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.W.; Ghosh, R.; Finberg, R.W.; Bergelson, J.M. Structure and chromosomal localization of the murine coxsackievirus and adenovirus receptor gene. DNA Cell Biol. 2003, 22, 253–259. [Google Scholar] [CrossRef]
- Cunningham, F.; Allen, J.E.; Allen, J.; Alvarez-Jarreta, J.; Amode, M.R.; Armean, I.M.; Austine-Orimoloye, O.; Azov, A.G.; Barnes, I.; Bennett, R.; et al. Ensembl 2022. Nucleic Acids Res. 2022, 50, D988–D995. [Google Scholar] [CrossRef]
- Andersson, B.; Tomko, R.P.; Edwards, K.; Mirza, M.; Darban, H.; Öncü, D.; Sonnhammer, E.; Sollerbrant, K.; Philipson, L. Putative regulatory domains in the human and mouse CVADR genes. Gene Funct. Dis. 2000, 1, 82–86. [Google Scholar] [CrossRef]
- Freimuth, P.; Springer, K.; Berard, C.; Hainfeld, J.; Bewley, M.; Flanagan, J. Coxsackievirus and adenovirus receptor amino-terminal immunoglobulin V-related domain binds adenovirus type 2 and fiber knob from adenovirus type 12. J. Virol. 1999, 73, 1392–1398. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bewley, M.C.; Springer, K.; Zhang, Y.B.; Freimuth, P.; Flanagan, J.M. Structural analysis of the mechanism of adenovirus binding to its human cellular receptor, CAR. Science 1999, 286, 1579–1583. [Google Scholar] [CrossRef] [Green Version]
- van Raaij, M.J.; Chouin, E.; van der Zandt, H.; Bergelson, J.M.; Cusack, S. Dimeric structure of the coxsackievirus and adenovirus receptor D1 domain at 1.7 A resolution. Structure 2000, 8, 1147–1155. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Philipson, L.; Pettersson, R.F. The coxsackie-adenovirus receptor—A new receptor in the immunoglobulin family involved in cell adhesion. Curr. Top. Microbiol. Immunol. 2004, 273, 87–111. [Google Scholar] [CrossRef] [PubMed]
- Honda, T.; Saitoh, H.; Masuko, M.; Katagiri-Abe, T.; Tominaga, K.; Kozakai, I.; Kobayashi, K.; Kumanishi, T.; Watanabe, Y.G.; Odani, S.; et al. The coxsackievirus-adenovirus receptor protein as a cell adhesion molecule in the developing mouse brain. Brain Res. Mol. Brain Res. 2000, 77, 19–28. [Google Scholar] [CrossRef]
- Jiang, S.; Caffrey, M. Solution structure of the coxsackievirus and adenovirus receptor domain 2. Protein Sci. 2007, 16, 539–542. [Google Scholar] [CrossRef]
- Patzke, C.; Max, K.E.; Behlke, J.; Schreiber, J.; Schmidt, H.; Dorner, A.A.; Kroger, S.; Henning, M.; Otto, A.; Heinemann, U.; et al. The coxsackievirus-adenovirus receptor reveals complex homophilic and heterophilic interactions on neural cells. J. Neurosci. 2010, 30, 2897–2910. [Google Scholar] [CrossRef] [Green Version]
- van’t Hof, W.; Crystal, R.G. Fatty acid modification of the coxsackievirus and adenovirus receptor. J. Virol. 2002, 76, 6382–6386. [Google Scholar] [CrossRef] [Green Version]
- Excoffon, K.J.; Hruska-Hageman, A.; Klotz, M.; Traver, G.L.; Zabner, J. A role for the PDZ-binding domain of the coxsackie B virus and adenovirus receptor (CAR) in cell adhesion and growth. J. Cell Sci. 2004, 117, 4401–4409. [Google Scholar] [CrossRef] [Green Version]
- Yan, R.; Sharma, P.; Kolawole, A.O.; Martin, S.C.; Readler, J.M.; Kotha, P.L.; Hostetler, H.A.; Excoffon, K.J. The PDZ3 domain of the cellular scaffolding protein MAGI-1 interacts with the Coxsackievirus and adenovirus receptor (CAR). Int. J. Biochem. Cell Biol. 2015, 61, 29–34. [Google Scholar] [CrossRef] [PubMed]
- Sollerbrant, K.; Raschperger, E.; Mirza, M.; Engstrom, U.; Philipson, L.; Ljungdahl, P.O.; Pettersson, R.F. The Coxsackievirus and adenovirus receptor (CAR) forms a complex with the PDZ domain-containing protein ligand-of-numb protein-X (LNX). J. Biol. Chem. 2003, 278, 7439–7444. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Excoffon, K.J.; Gansemer, N.D.; Mobily, M.E.; Karp, P.H.; Parekh, K.R.; Zabner, J. Isoform-specific regulation and localization of the coxsackie and adenovirus receptor in human airway epithelia. PLoS ONE 2010, 5, e9909. [Google Scholar] [CrossRef]
- Alghamri, M.S.; Sharma, P.; Williamson, T.L.; Readler, J.M.; Yan, R.; Rider, S.D., Jr.; Hostetler, H.A.; Cool, D.R.; Kolawole, A.O.; Excoffon, K. MAGI-1 PDZ2 Domain Blockade Averts Adenovirus Infection via Enhanced Proteolysis of the Apical Coxsackievirus and Adenovirus Receptor. J. Virol. 2021, 95, e0004621. [Google Scholar] [CrossRef] [PubMed]
- Cohen, C.J.; Gaetz, J.; Ohman, T.; Bergelson, J.M. Multiple regions within the coxsackievirus and adenovirus receptor cytoplasmic domain are required for basolateral sorting. J. Biol. Chem. 2001, 276, 25392–25398. [Google Scholar] [CrossRef] [Green Version]
- Asher, D.R.; Cerny, A.M.; Weiler, S.R.; Horner, J.W.; Keeler, M.L.; Neptune, M.A.; Jones, S.N.; Bronson, R.T.; Depinho, R.A.; Finberg, R.W. Coxsackievirus and adenovirus receptor is essential for cardiomyocyte development. Genesis 2005, 42, 77–85. [Google Scholar] [CrossRef]
- Dorner, A.A.; Wegmann, F.; Butz, S.; Wolburg-Buchholz, K.; Wolburg, H.; Mack, A.; Nasdala, I.; August, B.; Westermann, J.; Rathjen, F.G.; et al. Coxsackievirus-adenovirus receptor (CAR) is essential for early embryonic cardiac development. J. Cell Sci. 2005, 118, 3509–3521. [Google Scholar] [CrossRef] [Green Version]
- Sultana, T.; Hou, M.; Stukenborg, J.B.; Tohonen, V.; Inzunza, J.; Chagin, A.S.; Sollerbrant, K. Mice depleted of the coxsackievirus and adenovirus receptor display normal spermatogenesis and an intact blood-testis barrier. Reproduction 2014, 147, 875–883. [Google Scholar] [CrossRef]
- Marsman, R.F.; Bezzina, C.R.; Freiberg, F.; Verkerk, A.O.; Adriaens, M.E.; Podliesna, S.; Chen, C.; Purfurst, B.; Spallek, B.; Koopmann, T.T.; et al. Coxsackie and adenovirus receptor is a modifier of cardiac conduction and arrhythmia vulnerability in the setting of myocardial ischemia. J. Am. Coll. Cardiol. 2014, 63, 549–559. [Google Scholar] [CrossRef] [Green Version]
- Sharma, V.; Perry, D.J.; Eghtesady, P. Role of coxsackie-adenovirus receptor in cardiac development and pathogenesis of congenital heart disease. Birth Defects Res. 2021, 113, 535–545. [Google Scholar] [CrossRef]
- Chen, J.W.; Zhou, B.; Yu, Q.C.; Shin, S.J.; Jiao, K.; Schneider, M.D.; Baldwin, H.S.; Bergelson, J.M. Cardiomyocyte-specific deletion of the coxsackievirus and adenovirus receptor results in hyperplasia of the embryonic left ventricle and abnormalities of sinuatrial valves. Circ. Res. 2006, 98, 923–930. [Google Scholar] [CrossRef]
- O’Gorman, S.; Dagenais, N.A.; Qian, M.; Marchuk, Y. Protamine-Cre recombinase transgenes efficiently recombine target sequences in the male germ line of mice, but not in embryonic stem cells. Proc. Natl. Acad. Sci. USA 1997, 94, 14602–14607. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiao, K.; Kulessa, H.; Tompkins, K.; Zhou, Y.; Batts, L.; Baldwin, H.S.; Hogan, B.L. An essential role of Bmp4 in the atrioventricular septation of the mouse heart. Genes Dev. 2003, 17, 2362–2367. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Agah, R.; Frenkel, P.A.; French, B.A.; Michael, L.H.; Overbeek, P.A.; Schneider, M.D. Gene recombination in postmitotic cells. Targeted expression of Cre recombinase provokes cardiac-restricted, site-specific rearrangement in adult ventricular muscle in vivo. J. Clin. Investig. 1997, 100, 169–179. [Google Scholar] [CrossRef] [Green Version]
- Lim, B.K.; Xiong, D.; Dorner, A.; Youn, T.J.; Yung, A.; Liu, T.I.; Gu, Y.; Dalton, N.D.; Wright, A.T.; Evans, S.M.; et al. Coxsackievirus and adenovirus receptor (CAR) mediates atrioventricular-node function and connexin 45 localization in the murine heart. J. Clin. Investig. 2008, 118, 2758–2770. [Google Scholar] [CrossRef] [Green Version]
- Lisewski, U.; Shi, Y.; Wrackmeyer, U.; Fischer, R.; Chen, C.; Schirdewan, A.; Juttner, R.; Rathjen, F.; Poller, W.; Radke, M.H.; et al. The tight junction protein CAR regulates cardiac conduction and cell-cell communication. J. Exp. Med. 2008, 205, 2369–2379. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sohal, D.S.; Nghiem, M.; Crackower, M.A.; Witt, S.A.; Kimball, T.R.; Tymitz, K.M.; Penninger, J.M.; Molkentin, J.D. Temporally regulated and tissue-specific gene manipulations in the adult and embryonic heart using a tamoxifen-inducible Cre protein. Circ. Res. 2001, 89, 20–25. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Chen, C.; Lisewski, U.; Wrackmeyer, U.; Radke, M.; Westermann, D.; Sauter, M.; Tschope, C.; Poller, W.; Klingel, K.; et al. Cardiac deletion of the Coxsackievirus-adenovirus receptor abolishes Coxsackievirus B3 infection and prevents myocarditis in vivo. J. Am. Coll. Cardiol. 2009, 53, 1219–1226. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hayashi, S.; McMahon, A.P. Efficient recombination in diverse tissues by a tamoxifen-inducible form of Cre: A tool for temporally regulated gene activation/inactivation in the mouse. Dev. Biol. 2002, 244, 305–318. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pazirandeh, A.; Sultana, T.; Mirza, M.; Rozell, B.; Hultenby, K.; Wallis, K.; Vennstrom, B.; Davis, B.; Arner, A.; Heuchel, R.; et al. Multiple phenotypes in adult mice following inactivation of the Coxsackievirus and Adenovirus Receptor (Car) gene. PLoS ONE 2011, 6, e20203. [Google Scholar] [CrossRef] [Green Version]
- Mirza, M.; Pang, M.F.; Zaini, M.A.; Haiko, P.; Tammela, T.; Alitalo, K.; Philipson, L.; Fuxe, J.; Sollerbrant, K. Essential role of the coxsackie- and adenovirus receptor (CAR) in development of the lymphatic system in mice. PLoS ONE 2012, 7, e37523. [Google Scholar] [CrossRef] [PubMed]
- Schell, C.; Kretz, O.; Bregenzer, A.; Rogg, M.; Helmstadter, M.; Lisewski, U.; Gotthardt, M.; Tharaux, P.L.; Huber, T.B.; Grahammer, F. Podocyte-Specific Deletion of Murine CXADR Does Not Impair Podocyte Development, Function or Stress Response. PLoS ONE 2015, 10, e0129424. [Google Scholar] [CrossRef] [Green Version]
- Moeller, M.J.; Sanden, S.K.; Soofi, A.; Wiggins, R.C.; Holzman, L.B. Podocyte-specific expression of cre recombinase in transgenic mice. Genesis 2003, 35, 39–42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Outhwaite, J.E.; Patel, J.; Simmons, D.G. Secondary Placental Defects in Cxadr Mutant Mice. Front. Physiol. 2019, 10, 622. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, B.; Zhou, B.; Wang, Y.; Cheng, H.L.; Hang, C.T.; Pu, W.T.; Chang, C.P.; Zhou, B. Inducible cardiomyocyte-specific gene disruption directed by the rat Tnnt2 promoter in the mouse. Genesis 2010, 48, 63–72. [Google Scholar] [CrossRef] [Green Version]
- Hayashi, S.; Lewis, P.; Pevny, L.; McMahon, A.P. Efficient gene modulation in mouse epiblast using a Sox2Cre transgenic mouse strain. Mech. Dev. 2002, 119 (Suppl. 1), S97–S101. [Google Scholar] [CrossRef]
- Huang, K.; Ru, B.; Zhang, Y.; Chan, W.L.; Chow, S.C.; Zhang, J.; Lo, C.; Lui, W.Y. Sertoli cell-specific coxsackievirus and adenovirus receptor regulates cell adhesion and gene transcription via beta-catenin inactivation and Cdc42 activation. FASEB J. 2019, 33, 7588–7602. [Google Scholar] [CrossRef] [Green Version]
- Sadate-Ngatchou, P.I.; Payne, C.J.; Dearth, A.T.; Braun, R.E. Cre recombinase activity specific to postnatal, premeiotic male germ cells in transgenic mice. Genesis 2008, 46, 738–742. [Google Scholar] [CrossRef] [Green Version]
- Lecureuil, C.; Fontaine, I.; Crepieux, P.; Guillou, F. Sertoli and granulosa cell-specific Cre recombinase activity in transgenic mice. Genesis 2002, 33, 114–118. [Google Scholar] [CrossRef]
- Caruso, L.; Yuen, S.; Smith, J.; Husain, M.; Opavsky, M.A. Cardiomyocyte-targeted overexpression of the coxsackie-adenovirus receptor causes a cardiomyopathy in association with beta-catenin signaling. J. Mol. Cell Cardiol. 2010, 48, 1194–1205. [Google Scholar] [CrossRef]
- Ito, M.; Kodama, M.; Masuko, M.; Yamaura, M.; Fuse, K.; Uesugi, Y.; Hirono, S.; Okura, Y.; Kato, K.; Hotta, Y.; et al. Expression of coxsackievirus and adenovirus receptor in hearts of rats with experimental autoimmune myocarditis. Circ. Res. 2000, 86, 275–280. [Google Scholar] [CrossRef] [PubMed]
- Noutsias, M.; Fechner, H.; de Jonge, H.; Wang, X.; Dekkers, D.; Houtsmuller, A.B.; Pauschinger, M.; Bergelson, J.; Warraich, R.; Yacoub, M.; et al. Human coxsackie-adenovirus receptor is colocalized with integrins alpha(v)beta(3) and alpha(v)beta(5) on the cardiomyocyte sarcolemma and upregulated in dilated cardiomyopathy: Implications for cardiotropic viral infections. Circulation 2001, 104, 275–280. [Google Scholar] [CrossRef] [Green Version]
- Sasse, A.; Wallich, M.; Ding, Z.; Goedecke, A.; Schrader, J. Coxsackie-and-adenovirus receptor mRNA expression in human heart failure. J. Gene Med. 2003, 5, 876–882. [Google Scholar] [CrossRef] [PubMed]
- Fechner, H.; Noutsias, M.; Tschoepe, C.; Hinze, K.; Wang, X.; Escher, F.; Pauschinger, M.; Dekkers, D.; Vetter, R.; Paul, M.; et al. Induction of coxsackievirus-adenovirus-receptor expression during myocardial tissue formation and remodeling: Identification of a cell-to-cell contact-dependent regulatory mechanism. Circulation 2003, 107, 876–882. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garcia-Becerril, G.E.; Cruz-Montalvo, A.E.; De La Cruz, M.A.; Ares, M.A.; Moreno-Ruiz, L.A.; Garcia-Chequer, A.J.; Maldonado-Bernal, C.; Gomez-Jimenez, L.M.; Flores-Garcia, C.A.; Garrido-Garduno, M.H.; et al. Differential expression of coxsackievirus and adenovirus receptor in endomyocardial tissue of patients with myocarditis. Mol. Med. Rep. 2019, 20, 2189–2198. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaur, T.; Mishra, B.; Saikia, U.N.; Sharma, M.; Bahl, A.; Ratho, R.K. Expression of coxsackievirus and adenovirus receptor and its cellular localization in myocardial tissues of dilated cardiomyopathy. Exp. Clin. Cardiol. 2012, 17, 183–186. [Google Scholar]
- Tatrai, E.; Bedi, K.; Kovalszky, I.; Hartyanszky, I.; Laszik, A.; Acsady, G.; Sotonyi, P.; Hubay, M. No mutation but high mRNA expression of Coxsackie-Adenovirus Receptor was observed in both dilated and ischemic cardiomyopathy. Forensic. Sci. Int. 2011, 212, 47–50. [Google Scholar] [CrossRef]
- Gupalo, E.M.; Buryachkovskaya, L.I.; Chumachenko, P.V.; Mironova, N.A.; Narusov, O.Y.; Tereschenko, S.N.; Golitsyn, S.P.; Othman, M. Implication of inflammation on Coxsackie virus and Adenovirus receptor expression on cardiomyocytes and the role of platelets in patients with dilated cardiomyopathy. Cardiovasc. Pathol. 2022, 60, 107452. [Google Scholar] [CrossRef]
- Zussy, C.; Loustalot, F.; Junyent, F.; Gardoni, F.; Bories, C.; Valero, J.; Desarmenien, M.G.; Bernex, F.; Henaff, D.; Bayo-Puxan, N.; et al. Coxsackievirus Adenovirus Receptor Loss Impairs Adult Neurogenesis, Synapse Content, and Hippocampus Plasticity. J. Neurosci. 2016, 36, 9558–9571. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wehbi, A. Function of the Coxsackie and Adenovirus Receptor (CAR) in Adult Neurogenesis. Ph.D. Thesis, Université Montpellier, Montpellier, France, 2022. [Google Scholar]
- Wehbi, A.; Kremer, E.J.; Dopeso-Reyes, I.G. Location of the Cell Adhesion Molecule “Coxsackievirus and Adenovirus Receptor” in the Adult Mouse Brain. Front. Neuroanat. 2020, 14, 28. [Google Scholar] [CrossRef]
- Cohen, C.J.; Shieh, J.T.; Pickles, R.J.; Okegawa, T.; Hsieh, J.T.; Bergelson, J.M. The coxsackievirus and adenovirus receptor is a transmembrane component of the tight junction. Proc. Natl. Acad. Sci. USA 2001, 98, 15191–15196. [Google Scholar] [CrossRef] [Green Version]
- Raschperger, E.; Thyberg, J.; Pettersson, S.; Philipson, L.; Fuxe, J.; Pettersson, R.F. The coxsackie- and adenovirus receptor (CAR) is an in vivo marker for epithelial tight junctions, with a potential role in regulating permeability and tissue homeostasis. Exp. Cell Res. 2006, 312, 1566–1580. [Google Scholar] [CrossRef] [PubMed]
- Martinez, A.D.; Hayrapetyan, V.; Moreno, A.P.; Beyer, E.C. connexin 43 and connexin 45 form heteromeric gap junction channels in which individual components determine permeability and regulation. Circ. Res. 2002, 90, 1100–1107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lie, P.P.; Cheng, C.Y.; Mruk, D.D. Crosstalk between desmoglein-2/desmocollin-2/Src kinase and coxsackie and adenovirus receptor/ZO-1 protein complexes, regulates blood-testis barrier dynamics. Int. J. Biochem. Cell Biol. 2010, 42, 975–986. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, C.Y.; Mruk, D.D. The blood-testis barrier and its implications for male contraception. Pharmacol. Rev. 2012, 64, 16–64. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tamanini, A.; Nicolis, E.; Bonizzato, A.; Bezzerri, V.; Melotti, P.; Assael, B.M.; Cabrini, G. Interaction of adenovirus type 5 fiber with the coxsackievirus and adenovirus receptor activates inflammatory response in human respiratory cells. J. Virol. 2006, 80, 11241–11254. [Google Scholar] [CrossRef] [Green Version]
- Walters, R.W.; Freimuth, P.; Moninger, T.O.; Ganske, I.; Zabner, J.; Welsh, M.J. Adenovirus fiber disrupts CAR-mediated intercellular adhesion allowing virus escape. Cell 2002, 110, 789–799. [Google Scholar] [CrossRef] [Green Version]
- MacDonald, B.T.; Tamai, K.; He, X. Wnt/beta-catenin signaling: Components, mechanisms, and diseases. Dev. Cell 2009, 17, 9–26. [Google Scholar] [CrossRef] [Green Version]
- Nilchian, A.; Johansson, J.; Ghalali, A.; Asanin, S.T.; Santiago, A.; Rosencrantz, O.; Sollerbrant, K.; Vincent, C.T.; Sund, M.; Stenius, U.; et al. CXADR-Mediated Formation of an AKT Inhibitory Signalosome at Tight Junctions Controls Epithelial-Mesenchymal Plasticity in Breast Cancer. Cancer Res. 2019, 79, 47–60. [Google Scholar] [CrossRef] [Green Version]
- Morton, P.E.; Hicks, A.; Nastos, T.; Santis, G.; Parsons, M. CAR regulates epithelial cell junction stability through control of E-cadherin trafficking. Sci. Rep. 2013, 3, 2889. [Google Scholar] [CrossRef] [Green Version]
- Farmer, C.; Morton, P.E.; Snippe, M.; Santis, G.; Parsons, M. Coxsackie adenovirus receptor (CAR) regulates integrin function through activation of p44/42 MAPK. Exp. Cell Res. 2009, 315, 2637–2647. [Google Scholar] [CrossRef]
- Kashimura, T.; Kodama, M.; Hotta, Y.; Hosoya, J.; Yoshida, K.; Ozawa, T.; Watanabe, R.; Okura, Y.; Kato, K.; Hanawa, H.; et al. Spatiotemporal changes of coxsackievirus and adenovirus receptor in rat hearts during postnatal development and in cultured cardiomyocytes of neonatal rat. Virchows Arch. 2004, 444, 283–292. [Google Scholar] [CrossRef] [PubMed]
- Houri, N.; Huang, K.C.; Nalbantoglu, J. The Coxsackievirus and Adenovirus Receptor (CAR) undergoes ectodomain shedding and regulated intramembrane proteolysis (RIP). PLoS ONE 2013, 8, e73296. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lichtenthaler, S.F.; Lemberg, M.K.; Fluhrer, R. Proteolytic ectodomain shedding of membrane proteins in mammals-hardware, concepts, and recent developments. EMBO J. 2018, 37, e99456. [Google Scholar] [CrossRef] [PubMed]
- Herrmann, L.; Schelletter, L.; Hoffrogge, R.; Niehaus, K.; Rudolph, V.; Farr, M. Human Coxsackie- and adenovirus receptor is a putative target of neutrophil elastase-mediated shedding. Mol. Biol. Rep. 2022, 49, 3213–3223. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Barao, S.; Laga, M.; Bockstael, K.; Borgers, M.; Gijsen, H.; Annaert, W.; Moechars, D.; Mercken, M.; Gevaert, K.; et al. The neural cell adhesion molecules L1 and CHL1 are cleaved by BACE1 protease in vivo. J. Biol. Chem. 2012, 287, 25927–25940. [Google Scholar] [CrossRef] [Green Version]
- Morton, P.E.; Hicks, A.; Ortiz-Zapater, E.; Raghavan, S.; Pike, R.; Noble, A.; Woodfin, A.; Jenkins, G.; Rayner, E.; Santis, G.; et al. TNFalpha promotes CAR-dependent migration of leukocytes across epithelial monolayers. Sci. Rep. 2016, 6, 26321. [Google Scholar] [CrossRef] [Green Version]
- Gao, Y.; Lui, W.Y. Synergistic effect of interferon-gamma and tumor necrosis factor-alpha on coxsackievirus and adenovirus receptor expression: An explanation of cell sloughing during testicular inflammation in mice. Biol. Reprod. 2014, 90, 59. [Google Scholar] [CrossRef]
- Laura, R.P.; Ross, S.; Koeppen, H.; Lasky, L.A. MAGI-1: A widely expressed, alternatively spliced tight junction protein. Exp. Cell Res. 2002, 275, 155–170. [Google Scholar] [CrossRef]
- Kolawole, A.O.; Sharma, P.; Yan, R.; Lewis, K.J.; Xu, Z.; Hostetler, H.A.; Ashbourne Excoffon, K.J. The PDZ1 and PDZ3 domains of MAGI-1 regulate the eight-exon isoform of the coxsackievirus and adenovirus receptor. J. Virol. 2012, 86, 9244–9254. [Google Scholar] [CrossRef] [Green Version]
- Li, M.W.; Mruk, D.D.; Lee, W.M.; Cheng, C.Y. Connexin 43 and plakophilin-2 as a protein complex that regulates blood-testis barrier dynamics. Proc. Natl. Acad. Sci. USA 2009, 106, 10213–10218. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lam, W.Y.; Cheung, A.C.; Tung, C.K.; Yeung, A.C.; Ngai, K.L.; Lui, V.W.; Chan, P.K.; Tsui, S.K. miR-466 is putative negative regulator of Coxsackie virus and Adenovirus Receptor. FEBS Lett. 2015, 589, 246–254. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Wang, C.; Wei, J.; Zhu, Z.; Wang, X.; Sun, C. Ligand-of-Numb protein X1 controls the coxsackievirus B3-induced myocarditis via regulating the stability of coxsackievirus and adenovirus receptor. Genes Immun. 2022, 23, 42–46. [Google Scholar] [CrossRef] [PubMed]
- Siu, M.K.; Wong, C.H.; Lee, W.M.; Cheng, C.Y. Sertoli-germ cell anchoring junction dynamics in the testis are regulated by an interplay of lipid and protein kinases. J. Biol. Chem. 2005, 280, 25029–25047. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, K.; Pintilie, G.D.; Li, S.; Schmid, M.F.; Chiu, W. Resolving individual atoms of protein complex by cryo-electron microscopy. Cell Res. 2020, 30, 1136–1139. [Google Scholar] [CrossRef] [PubMed]
| Model | Exon | Time | Phenotypes | Reference |
|---|---|---|---|---|
| Cxadr−/− | 2 | E0 * | Embryonal lethal between E11.5 and E13.5 Thinner myocardial wall Cardiomyocytes degenerating Cardiomyocytes death | [26] |
| Cxadr+/− | 2 | E0 | Normal | [26] |
| Cxadr+/− | 1 | E0 | Normal | [27] |
| Cxadr−/− | 1 | E0 | Embryonal lethal between E11.5 and E13.5 Reduced density of myofibrils Disorganized orientation and bundling of myofibrils Formation of pericardial edema | [27] |
| Cxadr−/− | 2 | E0 | Embryonal lethal between E11.5 and E12.5, or die shortly after birth Regional over-proliferation of cardiomyocytes Hyperplasia of the left ventricle Abnormal junction between sinus venosus | [31] |
| Cxadrflox/flox; Protamine-Cre | 2 | Cxadr-null sperm is still fertile | [31,32] | |
| Cxadrflox/flox; TNT-Cre | 2 | E9.5 | Embryonal lethality | [31,33] |
| Cxadrflox/flox; α-MHC-Cre | 2 | E11.5 | Viable | [31,34] |
| Cxadrflox/flox; α-MHC-cre | 1 | Blockage of atrioventricular conduction in the adult heart Prolonged atrioventricular conduction in the embryonic heart Loss of connexin 45 Decreased β-catenin and ZO-1 amount and localization Cardiomyopathy | [34,35] | |
| Cxadr−/− | 2 | Embryonal lethal between E11.5 and E12.5 Hemorrhage Pericardial effusion | [35] | |
| Cxadrflox/flox; α-MHC-cre, Tamoxifen-Inducible | 1 | P2 months | Impaired electrical conductance from the atrium to ventricle Reduced expression of ZO-1 Reduced expression of connexin 45 Altered localization of connexin 43 | [36,37] |
| Cxadrflox/flox; α-MHC-cre, Tamoxifen-Inducible | 1 | P2 months | Prevent signs of inflammatory cardiomyopathy after CVB3 | [37,38] |
| Cxadrflox/flox; Cre-ERTM | 2 | P3 weeks | Dilated intestinal tract Atrophy of the exocrine pancreas Abnormal thymopoiesis | [39,40] |
| Cxadrflox/flox; Cre-ERTM | 2 | E12.5 | Lethal Subcutaneous edema Hemorrhage Embryonic lethality Dilated subcutaneous lymphatic vessels Abnormal structure with gaps and holes presents at lymphatic endothelial cell-cell junctions Erythrocyte leakage | [39,41] |
| Cxadrflox/flox; Cre-ERTM | 2 | E13.5 | Viable | [39,41] |
| Cxadr+/− | 1 | Slower ventricular conduction Increased arrhythmia susceptibility Reduced sodium current magnitude | [29] | |
| Cxadrflox/flox; Cre-ERTM | 2 | P4–6 weeks | Intact blood-testis barrier Uncompromised fertility No detectable phenotype up to the age of 8 months | [28,39] |
| Cxadrflox/flox; Cre-ERTM | 2 | P8 | Intact blood-testis barrier uncompromised fertility | [28,39] |
| Cxadrflox/flox; hNphs2-Cre | 1 | E14.0 | Normal podocyte development Normal stress response | [42,43] |
| Cxadrflox/flox; Tnnt2-Cre | 1 | E7.5 | Embryonic lethality by E12.5 Thinner placentas Decreased labyrinth depth | [44,45] |
| CxadrC210A/C210A-ENU | N.A. | Thinness of the labyrinth | [44] | |
| Cxadrflox/flox; Myh-6-Cre | 1 | Viable No obvious labyrinth defects | [34,44] | |
| Cxadrflox/flox; Sox2-Cre | 1 | E14.5 | Lethal between E11.5 and E12.5 Altered interheamal membrane architecture Reduced IHM branching Flatter placentas | [44,46] |
| Cxadrflox/flox; Stra8-iCre | 3 | P8 | No observable changes in reproductive functions; | [47,48] |
| Cxadrflox/flox; Amh-Cre | 3 | E14.5 | Reduced fertility with age Increased germ cell apoptosis Premature loss of elongated spermatids Compromised BTB function and apical ES structure Dysregulation of occludin and ZO-1 Altered β-catenin/Cdc42 signaling | [47,49] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.; Lui, W.-Y. CXADR: From an Essential Structural Component to a Vital Signaling Mediator in Spermatogenesis. Int. J. Mol. Sci. 2023, 24, 1288. https://doi.org/10.3390/ijms24021288
Zhang Y, Lui W-Y. CXADR: From an Essential Structural Component to a Vital Signaling Mediator in Spermatogenesis. International Journal of Molecular Sciences. 2023; 24(2):1288. https://doi.org/10.3390/ijms24021288
Chicago/Turabian StyleZhang, Yang, and Wing-Yee Lui. 2023. "CXADR: From an Essential Structural Component to a Vital Signaling Mediator in Spermatogenesis" International Journal of Molecular Sciences 24, no. 2: 1288. https://doi.org/10.3390/ijms24021288






