The LysR-Type Transcription Regulator YhjC Promotes the Systemic Infection of Salmonella Typhimurium in Mice
Abstract
1. Introduction
2. Results
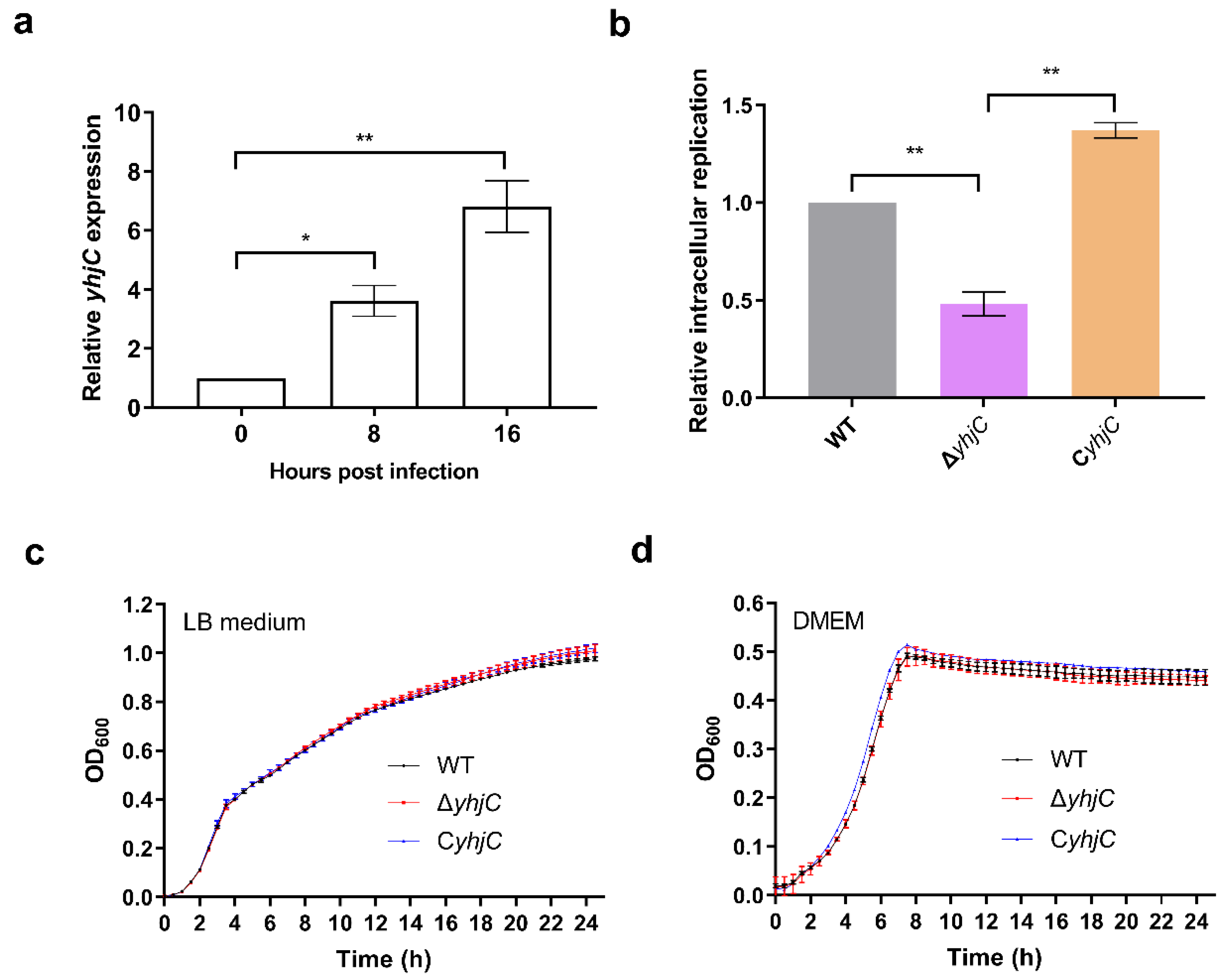
2.1. The Deletion of yhjC Reduced the Replication of S. Typhimurium in RAW 264.7 Cells
2.2. The Deletion of yhjC Reduced the Virulence of S. Typhimurium to Mice
2.3. The Transcriptional Regulation Function of YhjC in S. Typhimurium
2.4. YhjC Directly Activates spvD and zraP
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains and Plasmids
4.2. Determination of Bacterial Growth Curves
4.3. Intracellular Replication Experiment
4.4. Mouse Infection Experiment
4.5. RNA Extraction and Purification
4.6. Reverse Transcription and Real-Time Quantitative PCR
4.7. Library Construction and Transcriptome Sequencing Analysis
4.8. Immunofluorescence Microscopy
4.9. Purification of Recombinant Proteins
4.10. Electrophoretic Mobility Shift Assay
4.11. Statistical Analysis and Plotting
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Dhanoa, A.; Fatt, Q.K. Non-typhoidal Salmonella bacteraemia: Epidemiology, clinical characteristics and its’ association with severe immunosuppression. Ann. Clin. Microbiol. Antimicrob. 2009, 8, 15. [Google Scholar] [CrossRef] [PubMed]
- Neupane, D.; Dulal, H.; Song, J. Enteric Fever Diagnosis: Current Challenges and Future Directions. Pathogens 2021, 10, 410. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Wang, P.; Song, X.; Zhang, H.; Ma, S.; Wang, J.; Li, W.; Lv, R.; Liu, X.; Ma, S.; et al. Salmonella Typhimurium reprograms macrophage metabolism via T3SS effector SopE2 to promote intracellular replication and virulence. Nat. Commun. 2021, 12, 879. [Google Scholar] [CrossRef] [PubMed]
- Ly, K.T.; Casanova, J.E. Abelson Tyrosine Kinase Facilitates Salmonella enterica Serovar Typhimurium Entry into Epithelial Cells. Infect. Immun. 2009, 77, 60–69. [Google Scholar] [CrossRef]
- Bao, H.; Wang, S.; Zhao, J.-H.; Liu, S.-L. Salmonella secretion systems: Differential roles in pathogen-host interactions. Microbiol. Res. 2020, 241, 126591. [Google Scholar] [CrossRef]
- Griffin, A.J.; McSorley, S.J. Development of protective immunity to Salmonella, a mucosal pathogen with a systemic agenda. Mucosal Immunol. 2011, 4, 371–382. [Google Scholar] [CrossRef]
- Ruby, T.; McLaughlin, L.; Gopinath, S.; Monack, D. Salmonella’s long-term relationship with its host. FEMS Microbiol. Rev. 2012, 36, 600–615. [Google Scholar] [CrossRef]
- Salcedo, S.P.; Noursadeghi, M.; Cohen, J.; Holden, D.W. Intracellular replication of Salmonella typhimurium strains in specific subsets of splenic macrophages in vivo. Cell. Microbiol. 2001, 3, 587–597. [Google Scholar] [CrossRef]
- Mastroeni, P.; Grant, A.J. Spread of Salmonella enterica in the body during systemic infection: Unravelling host and pathogen determinants. Expert Rev. Mol. Med. 2011, 13, e12. [Google Scholar] [CrossRef]
- Lahiri, A.; Eswarappa, S.M.; Chakravortty, D.; Das, P. Altering the balance between pathogen containing vacuoles and lysosomes: A lesson from Salmonella. Virulence 2010, 1, 325–329. [Google Scholar] [CrossRef]
- Yang, F.; Sheng, X.; Huang, X.; Zhang, Y. Interactions between Salmonella and host macrophages—Dissecting NF-κB signaling pathway responses. Microb. Pathog. 2021, 154, 104846. [Google Scholar] [CrossRef] [PubMed]
- Yin, C.; Liu, Z.; Xian, H.; Jiao, Y.; Yuan, Y.; Li, Y.; Li, Q.; Jiao, X. AvrA Exerts Inhibition of NF-κB Pathway in Its Naïve Salmonella Serotype through Suppression of p-JNK and Beclin-1 Molecules. Int. J. Mol. Sci. 2020, 21, 6063. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Kamanova, J.; Lara-Tejero, M.; Galán, J.E. A Family of Salmonella Type III Secretion Effector Proteins Selectively Targets the NF-κB Signaling Pathway to Preserve Host Homeostasis. PLOS Pathog. 2016, 12, e1005484. [Google Scholar] [CrossRef] [PubMed]
- Rolhion, N.; Furniss, R.C.D.; Grabe, G.; Ryan, A.; Liu, M.; Matthews, S.A.; Holden, D.W. Inhibition of Nuclear Transport of NF-ĸB p65 by the Salmonella Type III Secretion System Effector SpvD. PLOS Pathog. 2016, 12, e1005653. [Google Scholar] [CrossRef] [PubMed]
- Günster, R.A.; Matthews, S.A.; Holden, D.W.; Thurston, T.L.M. SseK1 and SseK3 Type III Secretion System Effectors Inhibit NF-κB Signaling and Necroptotic Cell Death in Salmonella-Infected Macrophages. Infect. Immun. 2017, 85, e00010–e00017. [Google Scholar] [CrossRef] [PubMed]
- Steeb, B.; Claudi, B.; Burton, N.A.; Tienz, P.; Schmidt, A.; Farhan, H.; Mazé, A.; Bumann, D. Parallel Exploitation of Diverse Host Nutrients Enhances Salmonella Virulence. PLOS Pathog. 2013, 9, e1003301. [Google Scholar] [CrossRef]
- Deng, Q.; Yang, S.; Sun, L.; Dong, K.; Li, Y.; Wu, S.; Huang, R. Salmonella effector SpvB aggravates dysregulation of systemic iron metabolism via modulating the hepcidin−ferroportin axis. Gut Microbes 2021, 13, 1–18. [Google Scholar] [CrossRef]
- Mey, A.R.; Gómez-Garzón, C.; Payne, S.M. Iron Transport and Metabolism in Escherichia, Shigella, and Salmonella. EcoSal Plus 2021, 9, eESP00342020. [Google Scholar] [CrossRef]
- Crouch, M.-L.V.; Castor, M.; Karlinsey, J.E.; Kalhorn, T.; Fang, F.C. Biosynthesis and IroC-dependent export of the siderophore salmochelin are essential for virulence of Salmonella enterica serovar Typhimurium. Mol. Microbiol. 2008, 67, 971–983. [Google Scholar] [CrossRef]
- Cunrath, O.; Bumann, D. Host resistance factor SLC11A1 restricts Salmonella growth through magnesium deprivation. Science 2019, 366, 995–999. [Google Scholar] [CrossRef]
- Choi, J.; Groisman, E.A. Acidic pH sensing in the bacterial cytoplasm is required for Salmonella virulence. Mol. Microbiol. 2016, 101, 1024–1038. [Google Scholar] [CrossRef] [PubMed]
- Wu, A.; Tymoszuk, P.; Haschka, D.; Heeke, S.; Dichtl, S.; Petzer, V.; Seifert, M.; Hilbe, R.; Sopper, S.; Talasz, H.; et al. Salmonella Utilizes Zinc To Subvert Antimicrobial Host Defense of Macrophages via Modulation of NF-κB Signaling. Infect. Immun. 2017, 85. [Google Scholar] [CrossRef] [PubMed]
- Álvarez, R.; Neumann, G.; Frávega, J.; Díaz, F.; Tejías, C.; Collao, B.; Fuentes, J.A.; Paredes-Sabja, D.; Calderón, I.L.; Gil, F. CysB-dependent upregulation of the Salmonella Typhimurium cysJIH operon in response to antimicrobial compounds that induce oxidative stress. Biochem. Biophys. Res. Commun. 2015, 458, 46–51. [Google Scholar] [CrossRef]
- Espinosa, E.; Casadesús, J. Regulation of Salmonella enterica pathogenicity island 1 (SPI-1) by the LysR-type regulator LeuO. Mol. Microbiol. 2014, 91, 1057–1069. [Google Scholar] [CrossRef]
- Fernández-Mora, M.; Puente, J.L.; Calva, E. OmpR and LeuO Positively Regulate the Salmonella enterica Serovar Typhi ompS2 Porin Gene. J. Bacteriol. 2004, 186, 2909–2920. [Google Scholar] [CrossRef] [PubMed]
- Lim, S.; Kim, M.; Choi, J.; Ryu, S. A mutation in tdcA attenuates the virulence of Salmonella enterica serovar Typhimurium. Mol. Cells 2010, 29, 509–517. [Google Scholar] [CrossRef] [PubMed]
- Dillon, S.C.; Espinosa, E.; Hokamp, K.; Ussery, D.W.; Casadesús, J.; Dorman, C.J. LeuO is a global regulator of gene expression inSalmonella entericaserovar Typhimurium. Mol. Microbiol. 2012, 85, 1072–1089. [Google Scholar] [CrossRef] [PubMed]
- Saliba, A.-E.; Li, L.; Westermann, A.J.; Appenzeller, S.; Stapels, D.A.C.; Schulte, L.N.; Helaine, S.; Vogel, J. Single-cell RNA-seq ties macrophage polarization to growth rate of intracellular Salmonella. Nat. Microbiol. 2016, 2, 16206. [Google Scholar] [CrossRef]
- Grabe, G.J.; Zhang, Y.; Przydacz, M.; Rolhion, N.; Yang, Y.; Pruneda, J.N.; Komander, D.; Holden, D.W.; Hare, S.A. The Salmonella Effector SpvD Is a Cysteine Hydrolase with a Serovar-specific Polymorphism Influencing Catalytic Activity, Suppression of Immune Responses, and Bacterial Virulence. J. Biol. Chem. 2016, 291, 25853–25863. [Google Scholar] [CrossRef]
- Appia-Ayme, C.; Hall, A.; Patrick, E.; Rajadurai, S.; Clarke, T.A.; Rowley, G. ZraP is a periplasmic molecular chaperone and a repressor of the zinc-responsive two-component regulator ZraSR. Biochem. J. 2012, 442, 85–93. [Google Scholar] [CrossRef] [PubMed]
- Alix, E.; Blanc-Potard, A.-B. Peptide-assisted degradation of the Salmonella MgtC virulence factor. EMBO J. 2008, 27, 546–557. [Google Scholar] [CrossRef] [PubMed]
- Ogasawara, H.; Ishizuka, T.; Hotta, S.; Aoki, M.; Shimada, T.; Ishihama, A. Novel regulators of the csgD gene encoding the master regulator of biofilm formation in Escherichia coli K-12. Microbiology 2020, 166, 880–890. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Jiang, L.; Liu, X.; Guo, R.; Ma, S.; Wang, J.; Ma, S.; Li, S.; Li, H. YhjC is a novel transcriptional regulator required for Shigella flexneri virulence. Virulence 2021, 12, 1661–1671. [Google Scholar] [CrossRef] [PubMed]
- Behnsen, J.; Perez-Lopez, A.; Nuccio, S.-P.; Raffatellu, M. Exploiting host immunity: The Salmonella paradigm. Trends Immunol. 2015, 36, 112–120. [Google Scholar] [CrossRef]
- Nagy, T.A.; Moreland, S.M.; Detweiler, C.S. Salmonella acquires ferrous iron from haemophagocytic macrophages. Mol. Microbiol. 2014, 93, 1314–1326. [Google Scholar] [CrossRef]
- Zhu, M.; Valdebenito, M.; Winkelmann, G.; Hantke, K. Functions of the siderophore esterases IroD and IroE in iron-salmochelin utilization. Microbiology 2005, 151, 2363–2372. [Google Scholar] [CrossRef]
- Lin, H.; Fischbach, M.A.; Liu, D.R.; Walsh, C.T. In Vitro Characterization of Salmochelin and Enterobactin Trilactone Hydrolases IroD, IroE, and Fes. J. Am. Chem. Soc. 2005, 127, 11075–11084. [Google Scholar] [CrossRef]
- Choi, E.; Lee, K.-Y.; Shin, D. The MgtR regulatory peptide negatively controls expression of the MgtA Mg2+ transporter in Salmonella enterica serovar Typhimurium. Biochem. Biophys. Res. Commun. 2012, 417, 318–323. [Google Scholar] [CrossRef] [PubMed]
- Jean-Francois, F.L.; Dai, J.; Yu, L.; Myrick, A.; Rubin, E.; Fajer, P.G.; Song, L.; Zhou, H.-X.; Cross, T.A. Binding of MgtR, a Salmonella Transmembrane Regulatory Peptide, to MgtC, a Mycobacterium tuberculosis Virulence Factor: A Structural Study. J. Mol. Biol. 2014, 426, 436–446. [Google Scholar] [CrossRef]
- Roudier, C.; Fierer, J.; Guiney, D.G. Characterization of translation termination mutations in the spv operon of the Salmonella virulence plasmid pSDL2. J. Bacteriol. 1992, 174, 6418–6423. [Google Scholar] [CrossRef]
- Guiney, D.G.M.; Fierer, J.M. The Role of the spv Genes in Salmonella Pathogenesis. Front. Microbiol. 2011, 2, 129. [Google Scholar] [CrossRef]
- Krause, M.; Fang, F.C.; Guiney, D.G. Regulation of plasmid virulence gene expression in Salmonella dublin involves an unusual operon structure. J. Bacteriol. 1992, 174, 4482–4489. [Google Scholar] [CrossRef] [PubMed]
- Parsek, M.R.; Ye, R.W.; Pun, P.; Chakrabarty, A.M. Critical nucleotides in the interaction of a LysR-type regulator with its target promoter region. catBC promoter activation by CatR. J. Biol. Chem. 1994, 269, 11279–11284. [Google Scholar] [PubMed]
- Lim, D.; Kim, K.; Song, M.; Jeong, J.-H.; Chang, J.H.; Kim, S.R.; Hong, C.-W.; Im, S.-S.; Park, S.-H.; Lee, J.C.; et al. Transcriptional regulation of Salmochelin glucosyltransferase by Fur in Salmonella. Biochem. Biophys. Res. Commun. 2020, 529, 70–76. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Li, L.; Yan, X.; Wu, P.; Zhang, T.; Fan, Y.; Ma, S.; Wang, X.; Jiang, L. Nitrate Utilization Promotes Systemic Infection of Salmonella Typhimurium in Mice. Int. J. Mol. Sci. 2022, 23, 7220. [Google Scholar] [CrossRef] [PubMed]
- Ma, S.; Jiang, L.; Wang, J.; Liu, X.; Li, W.; Ma, S.; Feng, L. Downregulation of a novel flagellar synthesis regulator AsiR promotes intracellular replication and systemic pathogenicity of Salmonella Typhimurium. Virulence 2021, 12, 298–311. [Google Scholar] [CrossRef]
- Zhang, H.; Song, X.; Lv, R.; Liu, X.; Wang, P.; Jiang, L. The LysR-type transcriptional regulator STM0030 contributes to Salmonella Typhimurium growth in macrophages and virulence in mice. J. Basic Microbiol. 2019, 59, 1143–1153. [Google Scholar] [CrossRef]
- Song, X.; Zhang, H.; Liu, X.; Yuan, J.; Wang, P.; Lv, R.; Yang, B.; Huang, D.; Jiang, L. The putative transcriptional regulator STM14_3563 facilitates Salmonella Typhimurium pathogenicity by activating virulence-related genes. Int. Microbiol. 2019, 23, 381–390. [Google Scholar] [CrossRef]
- Jiang, L.; Wang, P.; Li, X.; Lv, R.; Wang, L.; Yang, B.; Huang, D.; Feng, L.; Liu, B. PagR mediates the precise regulation of Salmonella pathogenicity island 2 gene expression in response to magnesium and phosphate signals in Salmonella Typhimurium. Cell. Microbiol. 2019, 22, e13125. [Google Scholar] [CrossRef]
- Zhang, H.; Song, X.; Wang, P.; Lv, R.; Ma, S.; Jiang, L. YaeB, Expressed in Response to the Acidic pH in Macrophages, Promotes Intracellular Replication and Virulence of Salmonella Typhimurium. Int. J. Mol. Sci. 2019, 20, 4339. [Google Scholar] [CrossRef]
- Riggs, P. Expression and Purification of Recombinant Proteins by Fusion to Maltose-Binding Protein. Mol. Biotechnol. 2000, 15, 51–63. [Google Scholar] [CrossRef] [PubMed]





| Gene Name | Log2FC | Product |
|---|---|---|
| ProP | −3.532 | MFS transporter |
| STM14_RS03435 | −2.113 | YkgB family protein |
| ssrS | −2.085 | 6S RNA |
| zraP | −1.739 | zinc resistance sensor/chaperone ZraP |
| rplV | −1.694 | 50S ribosomal protein L22 |
| STM14_RS15610 | −1.618 | SDR family oxidoreductase |
| ygbJ | −1.593 | NAD(P)-dependent oxidoreductase |
| ygbM | −1.542 | HPr family phosphocarrier protein |
| spvD | −1.438 | SPI-2 type III secretion system effector cysteine hydrolase SpvD |
| lhgO | −1.277 | L-2-hydroxyglutarate oxidase |
| STM14_RS14715 | −1.242 | hypothetical protein |
| iroD | −1.221 | esterase family protein |
| iroE | −1.220 | alpha/beta hydrolase |
| ygbL | −1.208 | aldolase |
| hypC | −1.205 | hydrogenase 3 maturation protein HypC |
| csiD | −1.187 | carbon starvation induced protein CsiD |
| Sbp | −1.173 | sulfate ABC transporter substrate-binding protein |
| nrdF | −1.126 | class 1b ribonucleoside-diphosphate reductase subunit beta |
| ygbK | −1.096 | 3-oxo-tetronate kinase |
| nadB | −1.074 | L-aspartate oxidase |
| iroC | −1.063 | ABC transporter ATP-binding protein |
| rplC | −1.031 | 50S ribosomal protein L3 |
| STM14_RS16725 | −1.016 | polysaccharide deacetylase |
| rplD | −1.008 | 50S ribosomal protein L4 |
| ompW | 1.038 | outer membrane protein OmpW |
| lysA | 1.111 | diaminopimelate decarboxylase |
| ygiW | 1.115 | YgiW/YdeI family stress tolerance OB fold protein |
| mgtR | 1.131 | protein MgtR |
| fljB | 1.137 | FliC/FljB family flagellin |
| STM14_RS06850 | 1.165 | DUF2441 domain-containing protein |
| fdoI | 1.202 | formate dehydrogenase cytochrome b556 subunit |
| mgtA | 1.207 | magnesium-translocating P-type ATPase |
| narG | 1.252 | nitrate reductase subunit alpha |
| grcA | 1.277 | autonomous glycyl radical cofactor GrcA |
| fdxH | 1.312 | formate dehydrogenase subunit beta |
| fliI | 1.354 | flagellum-specific ATP synthase FliI |
| STM14_1795 | 1.705 | acid shock protein |
| STM14_RS12610 | 1.829 | MFS transporter |
| adiY | 2.469 | helix-turn-helix domain-containing protein |
| Targets | Primer Sequences (5′–3′) | |
|---|---|---|
| Primers used for the establishment of mutant strains | ||
| ΔyhjC | F | AGAAAATCAATAAAACCAGCCGCTTCAGCGTCACTGTTTCAATAAGAACAATAATAGAGCCTGATG GTGTAGGCTGGAGCTGCTTC |
| R | GGTAGTCCTTCATCATTAATAGAATATGTCAGTATAGCTTCTCCCCGGTGGATGCTGAAAATGCGG CATATGAATATCCTCCTTAGTTC | |
| Primers used for RT-qPCR analysis | ||
| 16S rRNA | F | ACTGGCAGGCTTGAGTCTTGTAGA |
| R | GGCACAACCTCCAAGTAGACATCG | |
| spvD | F | GAGAGTTTCTGGTAGTGCGTCATCCC |
| R | CGCTGTCTAATCCCACTGTAGGAGAG | |
| zraP | F | GTGGCAACAGGGAGGTAGCCC |
| R | ACTGGCGGTCAGTAGCGCGT | |
| ssrS | F | CCGCAGGCTGTAACCCTTGAAC |
| R | GAGATGTTTGCAAGCGGGCC | |
| ygbJ | F | CGGCGTCGTGGATGCGTTA |
| R | CAGCACAGGCTTCAGGCGGG | |
| iroC | F | GGTCAATAACGGCGGCACGG |
| R | CGTCACCCTGGTCAGCGAGG | |
| rplV | F | TCGCCTTGTTGCTGACCTGATTC |
| R | CGCTTCATGCTCGGGCCTT | |
| mgtR | F | CGCTCACCCGATAAAATCATCGC |
| R | CGATTTGCCAGAGGGCTAAACAC | |
| STM14_1795 | F | GTTGTTGCCGCTGCAATGGG |
| R | TGGTGCTGGGTGGTCTGGGT | |
| Primers for the construction of recombinant plasmids | ||
| pMAL-c5x-yhjC | F | CATGCCATGGGC ATGGATAAAATATATGCAATGAAATTGT |
| R | CGGGATCC CTACTCTGCGGCCTCTTTAATG | |
| pWSK129-yhjC | F | CGGGATCC AAATAGCCAATCCGCCTGAAA |
| R | GGAATTC AAATAGCCAATCCGCCTGAAA | |
| pWSK129-spvD | F | CGGGATCC TTTACGTGAGGAACCGTTTTATCG |
| R | CCGCTCGAG TCAATCGTGTTTTTCATCATAAGCC | |
| Primers used for the amplification of DNA sequences used in EMSA experiments | ||
| spvD | F | TTTACGTGAGGAACCGTTTTATCG |
| R | CTTTAATTCTCTTGACTTCATTTGAATCA | |
| mgtR | F | CTGGTTTATTGAGGGTCTGCTCT |
| R | TTAGAAAACGATTTGCCAGAGG | |
| zraP | F | CTCAACCACGTTGCAGCGG |
| R | CATTGTTGCCCCAGTGATGC | |
| iroC | F | TGGCCAGGGTGCCGAC |
| R | CTACGATGACAATGATGCTCAGTG | |
| 16S rDNA | F | AAATTGAAGAGTTTGATCATGGCTC |
| R | GCATGGCTGCATCAGGCTT | |
| Reaction Stages | Temperatures (°C) | Duration | Cycle Number |
|---|---|---|---|
| Activation of Uracil-DNA Glycosylase | 50 | 2 min | No |
| Initial denaturation | 95 | 2 min | No |
| Denaturation | 95 | 15 s | 40 |
| Annealing/extension | 60 | 1 min |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, W.; Ma, S.; Yan, X.; Wang, X.; Li, H.; Jiang, L. The LysR-Type Transcription Regulator YhjC Promotes the Systemic Infection of Salmonella Typhimurium in Mice. Int. J. Mol. Sci. 2023, 24, 1302. https://doi.org/10.3390/ijms24021302
Li W, Ma S, Yan X, Wang X, Li H, Jiang L. The LysR-Type Transcription Regulator YhjC Promotes the Systemic Infection of Salmonella Typhimurium in Mice. International Journal of Molecular Sciences. 2023; 24(2):1302. https://doi.org/10.3390/ijms24021302
Chicago/Turabian StyleLi, Wanwu, Shuai Ma, Xiaolin Yan, Xinyue Wang, Huiying Li, and Lingyan Jiang. 2023. "The LysR-Type Transcription Regulator YhjC Promotes the Systemic Infection of Salmonella Typhimurium in Mice" International Journal of Molecular Sciences 24, no. 2: 1302. https://doi.org/10.3390/ijms24021302
APA StyleLi, W., Ma, S., Yan, X., Wang, X., Li, H., & Jiang, L. (2023). The LysR-Type Transcription Regulator YhjC Promotes the Systemic Infection of Salmonella Typhimurium in Mice. International Journal of Molecular Sciences, 24(2), 1302. https://doi.org/10.3390/ijms24021302






