Integrated Data Analysis Uncovers New COVID-19 Related Genes and Potential Drug Re-Purposing Candidates
Abstract
1. Introduction
1.1. The COVID-19 Pandemic
1.2. Network-Medicine Drug Re-Purposing Methods
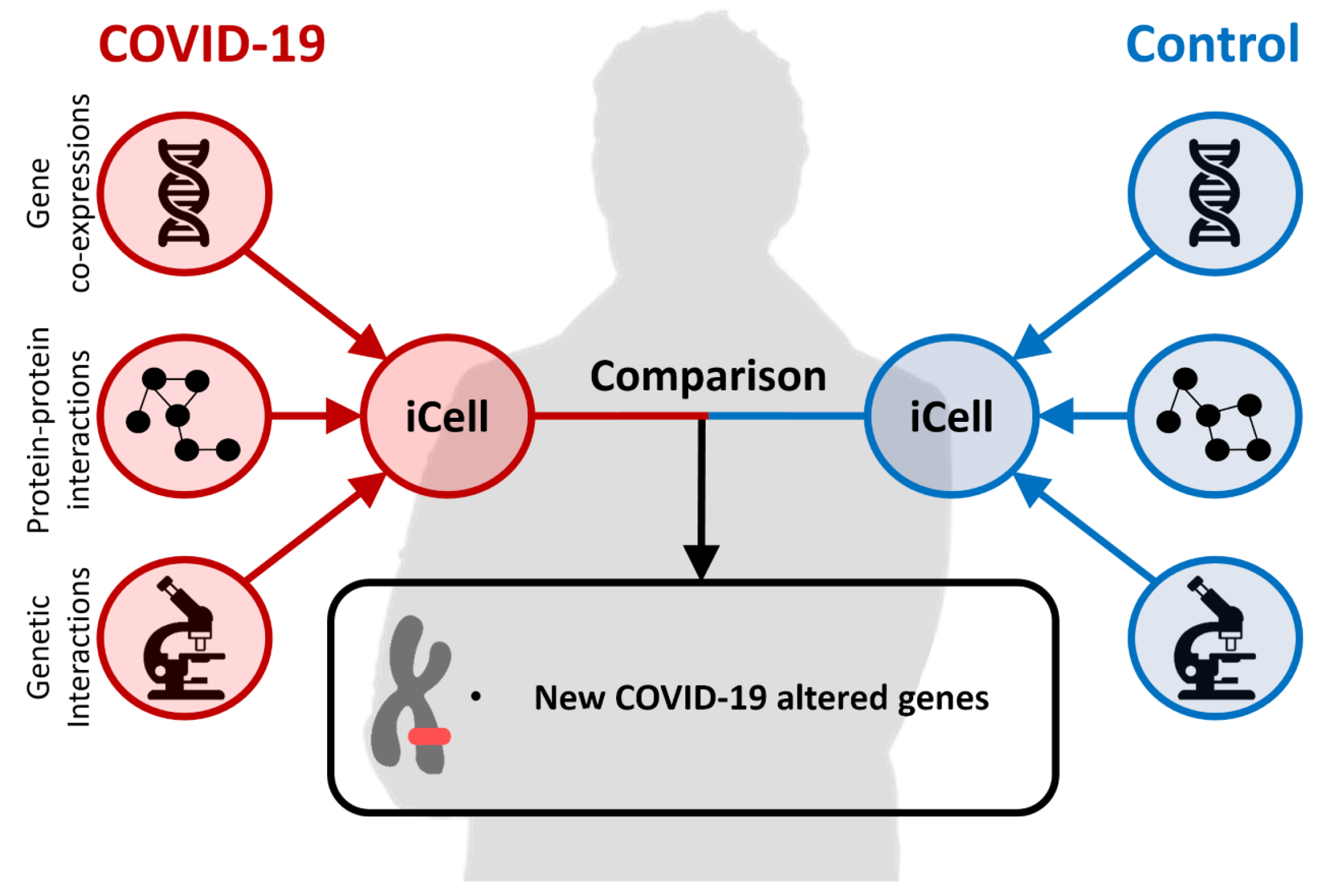
1.3. Comparative Data Integration with iCell
1.4. Contributions
2. Results
2.1. COVID-19 and Control iCells Are Biologically Coherent
2.2. Only iCells Are Intensely Rewired in COVID-19
2.3. Uncovering New COVID-19-Related Genes with iCells
2.4. Predicting Potential Drugs for Re-Purposing
3. Discussion
4. Materials and Methods
4.1. Creating Cell-Line and Tissue-Specific Molecular Interaction Networks
4.2. Gene Annotations
4.3. Differentially Expressed Genes from RNA-Seq Data
4.4. Drug Data
4.5. Creating Cell-Line and Tissue-Specific iCells
4.5.1. Clustering and Enrichment Analysis
4.5.2. Capturing the Wiring Patterns of Biological Networks
4.5.3. Predicting New Drug-Target Interactions
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| SARS-CoV-2 | severe acute respiratory syndrome-related coronavirus |
| PPI | Protein–protein interactions |
| COEX | Gene Co-Expressions |
| GI | Genetic Interactions |
| GO | Gene Ontology |
| DC | Drug Category |
| VHI | Viral–host interactions |
| DTI | Drug–target interactions |
| DCS | Drug–chemical similarity |
| SMILES | Simplified Molecular-Input Line-Entry System |
| DEGs | Differentially expressed genes |
| GO-BP | Gene Ontology Biological Process |
| RP | Reactome Pathways |
| NMTF | Non-Negative Matrix Tri-Factorization |
| MSNTF | Multiple Symmetric Non-negative Matrix Tri-Factorization |
| GNMTF | Graph Regularized Non-Negative Matrix Tri-Factorization |
| iCell | Integrated cell |
| SVD | Singular value decomposition |
References
- Zou, L.; Ruan, F.; Huang, M.; Liang, L.; Huang, H.; Hong, Z.; Yu, J.; Kang, M.; Song, Y.; Xia, J.; et al. SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N. Engl. J. Med. 2020, 382, 1177–1179. [Google Scholar] [CrossRef] [PubMed]
- Bai, Y.; Yao, L.; Wei, T.; Tian, F.; Jin, D.Y.; Chen, L.; Wang, M. Presumed Asymptomatic Carrier Transmission of COVID-19. JAMA 2020, 323, 1406–1407. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization (WHO) COVID-19 Dashboard. 2020. Available online: https://covid19.who.int/ (accessed on 1 November 2022).
- Hiscott, J.; Alexandridi, M.; Muscolini, M.; Tassone, E.; Palermo, E.; Soultsioti, M.; Zevini, A. The global impact of the coronavirus pandemic. Cytokine Growth Factor Rev. 2020, 53, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Ciotti, M.; Angeletti, S.; Minieri, M.; Giovannetti, M.; Benvenuto, D.; Pascarella, S.; Sagnelli, C.; Bianchi, M.; Bernardini, S.; Ciccozzi, M. COVID-19 Outbreak: An Overview. Chemotherapy 2020, 64, 215–223. [Google Scholar] [CrossRef] [PubMed]
- Pushpakom, S.; Iorio, F.; Eyers, P.A.; Escott, K.J.; Hopper, S.; Wells, A.; Doig, A.; Guilliams, T.; Latimer, J.; McNamee, C.; et al. Drug repurposing: Progress, challenges and recommendations. Nat. Rev. Drug Discov. 2019, 18, 41–58. [Google Scholar] [CrossRef]
- Galindez, G.; Matschinske, J.; Rose, T.D.; Sadegh, S.; Salgado-Albarrán, M.; Späth, J.; Baumbach, J.; Pauling, J.K. Lessons from the COVID-19 pandemic for advancing computational drug repurposing strategies. Nat. Comput. Sci. 2021, 1, 33–41. [Google Scholar] [CrossRef]
- Forni, G.; Mantovani, A. COVID-19 vaccines: Where we stand and challenges ahead. Cell Death Differ. 2021, 28, 626–639. [Google Scholar] [CrossRef]
- Dos Santos, W.G. Impact of virus genetic variability and host immunity for the success of COVID-19 vaccines. Biomed. Pharmacother. 2021, 136, 111272. [Google Scholar] [CrossRef]
- Liu, Y.; Liu, J.; Xia, H.; Zhang, X.; Fontes-Garfias, C.R.; Swanson, K.A.; Cai, H.; Sarkar, R.; Chen, W.; Cutler, M.; et al. Neutralizing activity of BNT162b2-elicited serum. N. Engl. J. Med. 2021, 384, 1466–1468. [Google Scholar]
- Wu, K.; Werner, A.P.; Koch, M.; Choi, A.; Narayanan, E.; Stewart-Jones, G.B.; Colpitts, T.; Bennett, H.; Boyoglu-Barnum, S.; Shi, W.; et al. Serum neutralizing activity elicited by mRNA-1273 vaccine. N. Engl. J. Med. 2021, 384, 1468–1470. [Google Scholar] [CrossRef]
- Ahlquist, P.; Noueiry, A.O.; Lee, W.M.; Kushner, D.B.; Dye, B.T. Host Factors in Positive-Strand RNA Virus Genome Replication. J. Virol. 2003, 77, 8181–8186. [Google Scholar] [CrossRef]
- Zhou, P.; Yang, X.L.; Wang, X.G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.R.; Zhu, Y.; Li, B.; Huang, C.L.; et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280. [Google Scholar] [CrossRef]
- Iwata-Yoshikawa, N.; Okamura, T.; Shimizu, Y.; Hasegawa, H.; Takeda, M.; Nagata, N. TMPRSS2 Contributes to Virus Spread and Immunopathology in the Airways of Murine Models after Coronavirus Infection. J. Virol. 2019, 93, 6. [Google Scholar] [CrossRef]
- Gordon, D.E.; Jang, G.M.; Bouhaddou, M.; Xu, J.; Obernier, K.; White, K.M.; O’Meara, M.J.; Rezelj, V.V.; Guo, J.Z.; Swaney, D.L.; et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature 2020, 583, 459–468. [Google Scholar] [CrossRef]
- Blanco-Melo, D.; Nilsson-Payant, B.E.; Liu, W.C.; Uhl, S.; Hoagland, D.; Møller, R.; Jordan, T.X.; Oishi, K.; Panis, M.; Sachs, D.; et al. Imbalanced Host Response to SARS-CoV-2 Drives Development of COVID-19. Cell 2020, 181, 1036–1045.e9. [Google Scholar] [CrossRef]
- Zhou, Y.; Hou, Y.; Shen, J.; Huang, Y.; Martin, W.; Cheng, F. Network-based drug repurposing for novel coronavirus 2019-nCoV/SARS-CoV-2. Cell Discov. 2020, 6, 14. [Google Scholar] [CrossRef]
- Sadegh, S.; Matschinske, J.; Blumenthal, D.B.; Galindez, G.; Kacprowski, T.; List, M.; Nasirigerdeh, R.; Oubounyt, M.; Pichlmair, A.; Rose, T.D.; et al. Exploring the SARS-CoV-2 virus–host-drug interactome for drug repurposing. Nat. Commun. 2020, 11, 1–9. [Google Scholar] [CrossRef]
- Morselli Gysi, D.; Do Valle, Í.; Zitnik, M.; Ameli, A.; Gan, X.; Varol, O.; Ghiassian, S.D.; Patten, J.; Davey, R.A.; Loscalzo, J.; et al. Network medicine framework for identifying drug-repurposing opportunities for COVID-19. Proc. Natl. Acad. Sci. USA 2021, 118, e2025581118. [Google Scholar] [CrossRef]
- Zambrana, C.; Xenos, A.; Böttcher, R.; Malod-Dognin, N.; Pržulj, N. Network neighbors of viral targets and differentially expressed genes in COVID-19 are drug target candidates. Sci. Rep. 2021, 11, 1–15. [Google Scholar] [CrossRef]
- Malod-Dognin, N.; Petschnigg, J.; Windels, S.F.; Povh, J.; Hemmingway, H.; Ketteler, R.; Pržulj, N. Towards a data-integrated cell. Nat. Commun. 2019, 10, 1–13. [Google Scholar] [CrossRef]
- Ding, C.; Li, T.; Peng, W.; Park, H. Orthogonal nonnegative matrix tri-factorizations for clustering. In Proceedings of the 12th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, Philadelphia, PA, USA, 20–23 August 2006; pp. 126–135. [Google Scholar]
- Kuleshov, M.V.; Stein, D.J.; Clarke, D.J.; Kropiwnicki, E.; Jagodnik, K.M.; Bartal, A.; Evangelista, J.E.; Hom, J.; Cheng, M.; Bailey, A.; et al. The COVID-19 drug and gene set library. Patterns 2020, 1, 100090. [Google Scholar] [CrossRef] [PubMed]
- Nchioua, R.; Kmiec, D.; Müller, J.A.; Conzelmann, C.; Groß, R.; Swanson, C.M.; Neil, S.J.D.; Stenger, S.; Sauter, D.; Münch, J.; et al. SARS-CoV-2 Is Restricted by Zinc Finger Antiviral Protein despite Preadaptation to the Low-CpG Environment in Humans. mBio 2020, 11, e01930-20. [Google Scholar] [CrossRef] [PubMed]
- Qin, S.; Xu, W.; Wang, C.; Jiang, S.; Dai, W.; Yang, Y.; Shen, J.; Jin, P.; Ma, F.; Xia, X. Analyzing master regulators and scRNA-seq of COVID-19 patients reveals an underlying anti-SARS-CoV-2 mechanism of ZNF proteins. Briefings Bioinform. 2021, 22, bbab118. [Google Scholar] [CrossRef]
- Beigel, J.H.; Tomashek, K.M.; Dodd, L.E.; Mehta, A.K.; Zingman, B.S.; Kalil, A.C.; Hohmann, E.; Chu, H.Y.; Luetkemeyer, A.; Kline, S.; et al. Remdesivir for the treatment of COVID-19. N. Engl. J. Med. 2020, 383, 1813–1826. [Google Scholar] [CrossRef] [PubMed]
- Oughtred, R.; Stark, C.; Breitkreutz, B.J.; Rust, J.; Boucher, L.; Chang, C.; Kolas, N.; O’Donnell, L.; Leung, G.; McAdam, R.; et al. The BioGRID interaction database: 2019 update. Nucleic Acids Res. 2019, 47, D529–D541. [Google Scholar] [CrossRef]
- Obayashi, T.; Kagaya, Y.; Aoki, Y.; Tadaka, S.; Kinoshita, K. COXPRESdb v7: A gene coexpression database for 11 animal species supported by 23 coexpression platforms for technical evaluation and evolutionary inference. Nucleic Acids Res. 2018, 47, D55–D62. [Google Scholar] [CrossRef]
- Guo, J.; Liu, H.; Zheng, J. SynLethDB: Synthetic lethality database toward discovery of selective and sensitive anticancer drug targets. Nucleic Acids Res. 2016, 44, D1011–D1017. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene Ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25. [Google Scholar] [CrossRef]
- Fabregat, A.; Jupe, S.; Matthews, L.; Sidiropoulos, K.; Gillespie, M.; Garapati, P.; Haw, R.; Jassal, B.; Korninger, F.; May, B.; et al. The reactome pathway knowledgebase. Nucleic Acids Res. 2017, 46, D649–D655. [Google Scholar] [CrossRef]
- Kodinariya, T.M.; Makwana, P.R. Review on determining number of Cluster in K-Means Clustering. Int. J. 2013, 1, 90–95. [Google Scholar]
- Supek, F.; Bošnjak, M.; Škunca, N.; Šmuc, T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS ONE 2011, 6, e21800. [Google Scholar] [CrossRef]
- Sinha, P.; Matthay, M.A.; Calfee, C.S. Is a “cytokine storm” relevant to COVID-19? JAMA Intern. Med. 2020, 180, 1152–1154. [Google Scholar] [CrossRef]
- Nidadavolu, L.S.; Walston, J.D. Underlying vulnerabilities to the cytokine storm and adverse COVID-19 outcomes in the aging immune system. J. Gerontol. Ser. 2021, 76, e13–e18. [Google Scholar] [CrossRef]
- Singh, B.; Ryan, H.; Kredo, T.; Chaplin, M.; Fletcher, T. Chloroquine or hydroxychloroquine for prevention and treatment of COVID-19. Cochrane Database Syst. Rev. 2021, 2. [Google Scholar] [CrossRef]
- Yaveroğlu, Ö.N.; Milenković, T.; Pržulj, N. Proper evaluation of alignment-free network comparison methods. Bioinformatics 2015, 31, 2697–2704. [Google Scholar] [CrossRef]
- McClain, M.T.; Constantine, F.J.; Henao, R.; Liu, Y.; Tsalik, E.L.; Burke, T.W.; Steinbrink, J.M.; Petzold, E.; Nicholson, B.P.; Rolfe, R.; et al. Dysregulated transcriptional responses to SARS-CoV-2 in the periphery. Nat. Commun. 2021, 12, 1–8. [Google Scholar] [CrossRef]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Menche, J.; Sharma, A.; Kitsak, M.; Ghiassian, S.D.; Vidal, M.; Loscalzo, J.; Barabási, A.L. Uncovering disease-disease relationships through the incomplete interactome. Science 2015, 347, 1257601. [Google Scholar] [CrossRef]
- Cassandri, M.; Smirnov, A.; Novelli, F.; Pitolli, C.; Agostini, M.; Malewicz, M.; Melino, G.; Raschellà, G. Zinc-finger proteins in health and disease. Cell Death Discov. 2017, 3, 1–12. [Google Scholar] [CrossRef]
- Seeßle, J.; Hippchen, T.; Schnitzler, P.; Gsenger, J.; Giese, T.; Merle, U. High rate of HSV-1 reactivation in invasively ventilated COVID-19 patients: Immunological findings. PLoS ONE 2021, 16, e0254129. [Google Scholar] [CrossRef] [PubMed]
- Bond, P. Ethnicity and the relationship between COVID-19 and the herpes simplex viruses. Med. Hypotheses 2021, 146, 110447. [Google Scholar] [CrossRef] [PubMed]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef]
- Hornuss, D.; Lange, B.; Schroeter, N.; Rieg, S.; Kern, W.V.; Wagner, D. Anosmia in COVID-19 patients. Clin. Microbiol. Infect. 2020, 26, 1426. [Google Scholar] [CrossRef] [PubMed]
- Abildúa, M.A.; Prieto, M.R.; Zabaleta, R.M.; Lucas, C.A.; López, C.P. Myopathy associated with severe SARS-CoV-2 infection. Neurología 2020, 35, 706. [Google Scholar] [CrossRef]
- Henry, B.M.; Lippi, G. Chronic kidney disease is associated with severe coronavirus disease 2019 (COVID-19) infection. Int. Urol. Nephrol. 2020, 52, 1193–1194. [Google Scholar] [CrossRef]
- De Erausquin, G.A.; Snyder, H.; Carrillo, M.; Hosseini, A.A.; Brugha, T.S.; Seshadri, S.; Consortium, C.S.C. The chronic neuropsychiatric sequelae of COVID-19: The need for a prospective study of viral impact on brain functioning. Alzheimer’s Dement. 2021, 17, 1056–1065. [Google Scholar] [CrossRef]
- Li, H.; Xiao, X.; Zhang, J.; Zafar, M.I.; Wu, C.; Long, Y.; Lu, W.; Pan, F.; Meng, T.; Zhao, K.; et al. Impaired spermatogenesis in COVID-19 patients. EClinicalMedicine 2020, 28, 100604. [Google Scholar] [CrossRef]
- Gligorijević, V.; Malod-Dognin, N.; Pržulj, N. Patient-specific data fusion for cancer stratification and personalised treatment. In Proceedings of the Biocomputing 2016: Proceedings of the Pacific Symposium, World Scientific, Kohala Coast, HI, USA, 4–8 January 2016; pp. 321–332. [Google Scholar]
- Okabe, K.; Yaku, K.; Tobe, K.; Nakagawa, T. Implications of altered NAD metabolism in metabolic disorders. J. Biomed. Sci. 2019, 26, 34. [Google Scholar] [CrossRef]
- Mueller, A.L.; McNamara, M.S.; Sinclair, D.A. Why does COVID-19 disproportionately affect older people? Aging 2020, 12, 9959–9981. [Google Scholar] [CrossRef]
- Miller, R.; Wentzel, A.; Richards, G. COVID-19: NAD+ deficiency may predispose the aged, obese and type2 diabetics to mortality through its effect on SIRT1 activity. Med. Hypotheses 2020, 144, 110044. [Google Scholar] [CrossRef]
- Mehta, P.; McAuley, D.F.; Brown, M.; Sanchez, E.; Tattersall, R.S.; Manson, J.J. COVID-19: Consider cytokine storm syndromes and immunosuppression. Lancet 2020, 395, 1033–1034. [Google Scholar] [CrossRef]
- Bogan-Brown, K.; Nkrumah-Elie, Y.; Ishtiaq, Y.; Redpath, P.; Shao, A. Potential efficacy of nutrient supplements for treatment or prevention of COVID-19. J. Diet. Suppl. 2021, 19, 336–365. [Google Scholar] [CrossRef]
- Krishna, S.; Bustamante, L.; Haynes, R.K.; Staines, H.M. Artemisinins: Their growing importance in medicine. Trends Pharmacol. Sci. 2008, 29, 520–527. [Google Scholar] [CrossRef]
- Wagner, S.D.; Yakovchuk, P.; Gilman, B.; Ponicsan, S.L.; Drullinger, L.F.; Kugel, J.F.; Goodrich, J.A. RNA polymerase II acts as an RNA-dependent RNA polymerase to extend and destabilize a non-coding RNA. EMBO J. 2013, 32, 781–790. [Google Scholar] [CrossRef]
- Gordon, C.J.; Tchesnokov, E.P.; Woolner, E.; Perry, J.K.; Feng, J.Y.; Porter, D.P.; Götte, M. Remdesivir is a direct-acting antiviral that inhibits RNA-dependent RNA polymerase from severe acute respiratory syndrome coronavirus 2 with high potency. J. Biol. Chem. 2020, 295, 6785–6797. [Google Scholar] [CrossRef]
- Yin, W.; Luan, X.; Li, Z.; Zhou, Z.; Wang, Q.; Gao, M.; Wang, X.; Zhou, F.; Shi, J.; You, E.; et al. Structural basis for inhibition of the SARS-CoV-2 RNA polymerase by suramin. Nat. Struct. Mol. Biol. 2021, 28, 319–325. [Google Scholar] [CrossRef]
- Wang, L.; Song, Y. Efficacy of zinc given as an adjunct to the treatment of severe pneumonia: A meta-analysis of randomized, double-blind and placebo-controlled trials. Clin. Respir. J. 2018, 12, 857–864. [Google Scholar] [CrossRef]
- Li, S.; Wu, B.; Ling, Y.; Guo, M.; Qin, B.; Ren, X.; Wang, C.; Yang, H.; Chen, L.; Liao, Y.; et al. Epigenetic landscapes of single-cell chromatin accessibility and transcriptomic immune profiles of T cells in COVID-19 patients. Front. Immunol. 2021, 12, 625881. [Google Scholar] [CrossRef]
- Porcu, E.; Sadler, M.C.; Lepik, K.; Auwerx, C.; Wood, A.R.; Weihs, A.; Sleiman, M.S.B.; Ribeiro, D.M.; Bandinelli, S.; Tanaka, T.; et al. Differentially expressed genes reflect disease-induced rather than disease-causing changes in the transcriptome. Nat. Commun. 2021, 12, 1–9. [Google Scholar] [CrossRef]
- Robinson, M.D.; Oshlack, A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010, 11, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Law, C.W.; Chen, Y.; Shi, W.; Smyth, G.K. voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014, 15, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Feunang, Y.D.; Guo, A.C.; Lo, E.J.; Marcu, A.; Grant, J.R.; Sajed, T.; Johnson, D.; Li, C.; Sayeeda, Z.; et al. DrugBank 5.0: A major update to the DrugBank database for 2018. Nucleic Acids Res. 2018, 46, D1074–D1082. [Google Scholar] [CrossRef] [PubMed]
- Nikolova, N.; Jaworska, J. Approaches to Measure Chemical Similarity—A Review. QSAR Comb. Sci. 2004, 22, 1006–1026. [Google Scholar] [CrossRef]
- Brunet, J.P.; Tamayo, P.; Golub, T.R.; Mesirov, J.P. Metagenes and molecular pattern discovery using matrix factorization. Proc. Natl. Acad. Sci. USA 2004, 101, 4164–4169. [Google Scholar] [CrossRef]
- Pržulj, N.; Corneil, D.G.; Jurisica, I. Modeling interactome: Scale-free or geometric? Bioinformatics 2004, 20, 3508–3515. [Google Scholar] [CrossRef]
- Pržulj, N. Biological network comparison using graphlet degree distribution. Bioinformatics 2007, 23, e177–e183. [Google Scholar] [CrossRef]
- Yaveroğlu, Ö.N.; Malod-Dognin, N.; Davis, D.; Levnajic, Z.; Janjic, V.; Karapandza, R.; Stojmirovic, A.; Pržulj, N. Revealing the hidden language of complex networks. Sci. Rep. 2014, 4, 4547. [Google Scholar] [CrossRef]
- Milenković, T.; Pržulj, N. Uncovering biological network function via graphlet degree signatures. Cancer Inform. 2008, 6, 257. [Google Scholar] [CrossRef]
- Qiao, H. New SVD based initialization strategy for non-negative matrix factorization. Pattern Recognit. Lett. 2015, 63, 71–77. [Google Scholar] [CrossRef]
- Kim, S.; Chen, J.; Cheng, T.; Gindulyte, A.; He, J.; He, S.; Li, Q.; Shoemaker, B.A.; Thiessen, P.A.; Yu, B.; et al. PubChem in 2021: New data content and improved web interfaces. Nucleic Acids Res. 2021, 49, D1388–D1395. [Google Scholar] [CrossRef]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The protein data bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef]
- Varadi, M.; Anyango, S.; Deshpande, M.; Nair, S.; Natassia, C.; Yordanova, G.; Yuan, D.; Stroe, O.; Wood, G.; Laydon, A.; et al. AlphaFold Protein Structure Database: Massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2022, 50, D439–D444. [Google Scholar] [CrossRef]


| PPI | COEX | GI | iCell | |||||
|---|---|---|---|---|---|---|---|---|
| #Node | #Edge | #Node | #Edge | #Node | #Edge | #Node | #Edge | |
| Infected A549 | 9623 | 178,828 | 9286 | 593,544 | 6968 | 22,418 | 9623 | 837,077 |
| Control A549 | 9592 | 177,728 | 9253 | 591,607 | 6970 | 22,903 | 9592 | 829,609 |
| Infected NHBE | 9391 | 174,892 | 9074 | 565,177 | 6933 | 22,027 | 9391 | 788,520 |
| Control NHBE | 9531 | 177,648 | 9204 | 585,361 | 7095 | 22,863 | 9531 | 822,374 |
| Infected CALU | 9434 | 175,830 | 9301 | 599,549 | 6957 | 20,505 | 9434 | 805,284 |
| Control CALU | 9149 | 169,229 | 9021 | 564,297 | 6536 | 18,391 | 9149 | 745,167 |
| Infected Patient | 5916 | 90,631 | 5845 | 241,213 | 3743 | 8978 | 5916 | 319,549 |
| Control Patient | 9552 | 168,284 | 9420 | 609,304 | 6739 | 20,143 | 9552 | 806,876 |
| Cell Line | Annotation Type | #Enriched in Control | #Enriched in Infected | Jaccard Similarity |
|---|---|---|---|---|
| GO-BP | 1193 | 1520 | 0.31 | |
| A549 | RP | 832 | 872 | 0.65 |
| GO-BP | 1201 | 1046 | 0.28 | |
| NHBE | RP | 752 | 852 | 0.57 |
| GO-BP | 1028 | 929 | 0.24 | |
| CALU | RP | 743 | 723 | 0.65 |
| GO-BP | 1145 | 828 | 0.21 | |
| Patient | RP | 862 | 618 | 0.53 |
| Cell Line | Rewirement of VHIs | Rewirement of DEGs | Rewirement of Background Genes |
|---|---|---|---|
| A549 | 0.037 | 0.064 (p value ) | 0.044 |
| NHBE | 0.038 | 0.063 (p value ) | 0.043 |
| CALU | 0.053 | 0.072 (p value ) | 0.057 |
| Patient | 0.073 | 0.088 (p value ) | 0.085 |
| Gene | External Validation (#Studies) | Diff. Exp. | Existing Drug (Drugbank) | Potential Drug for Re-Purposing | Binding Free Energy (kcal/mol) |
|---|---|---|---|---|---|
| ZNF35 | 8 | No | NADH | −9.8 | |
| RPSAP58 | 3 | No | NADH | - | |
| ZNF562 | 1 | No | NADH | −9.4 | |
| OLFM2 | 5 | No | FOSTAMATINIB | −9.6 | |
| CYB561 | 8 | No | ZINC CHLORIDE | - | |
| ZNF41 | 4 | No | FOSTAMATINIB | −8.5 | |
| LCMT2 | 5 | No | LEUCINE | N-FORMYLME THIONINE | - |
| CSTF2T | 3 | No | NADH | −10.8 | |
| NUP85 | 11 | No | CLADRIBINE | −7.2 | |
| REEP4 | 9 | No | FOSTAMATINIB | −9.3 | |
| ASRGL1 | 6 | No | ASPARTIC ACID ASPARTACIAL | NADH | -9.7 |
| ZFP62 | - | No | ARTENIMOL | −7.6 | |
| CBX5 | 10 | No | COPPER | ACETYLSALICYLIC ACID | - |
| KLHL9 | 7 | No | ARTENIMOL | −10.6 | |
| ZNF189 | 6 | No | FOSTAMATINIB | −9.9 | |
| ZNF597 | 4 | No | NADH | −10.8 | |
| H2AC20 | 7 | Yes | ARTENIMOL | −8.2 | |
| CSTF1 | 1 | No | FOSTAMATINIB | −13 | |
| ZNF507 | 9 | No | NADH | −8.6 | |
| ZNF286A | - | No | NADH | −10.7 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xenos, A.; Malod-Dognin, N.; Zambrana, C.; Pržulj, N. Integrated Data Analysis Uncovers New COVID-19 Related Genes and Potential Drug Re-Purposing Candidates. Int. J. Mol. Sci. 2023, 24, 1431. https://doi.org/10.3390/ijms24021431
Xenos A, Malod-Dognin N, Zambrana C, Pržulj N. Integrated Data Analysis Uncovers New COVID-19 Related Genes and Potential Drug Re-Purposing Candidates. International Journal of Molecular Sciences. 2023; 24(2):1431. https://doi.org/10.3390/ijms24021431
Chicago/Turabian StyleXenos, Alexandros, Noël Malod-Dognin, Carme Zambrana, and Nataša Pržulj. 2023. "Integrated Data Analysis Uncovers New COVID-19 Related Genes and Potential Drug Re-Purposing Candidates" International Journal of Molecular Sciences 24, no. 2: 1431. https://doi.org/10.3390/ijms24021431
APA StyleXenos, A., Malod-Dognin, N., Zambrana, C., & Pržulj, N. (2023). Integrated Data Analysis Uncovers New COVID-19 Related Genes and Potential Drug Re-Purposing Candidates. International Journal of Molecular Sciences, 24(2), 1431. https://doi.org/10.3390/ijms24021431






