New Transcriptomic Biomarkers of 5-Fluorouracil Resistance
Abstract
1. Introduction
2. Results
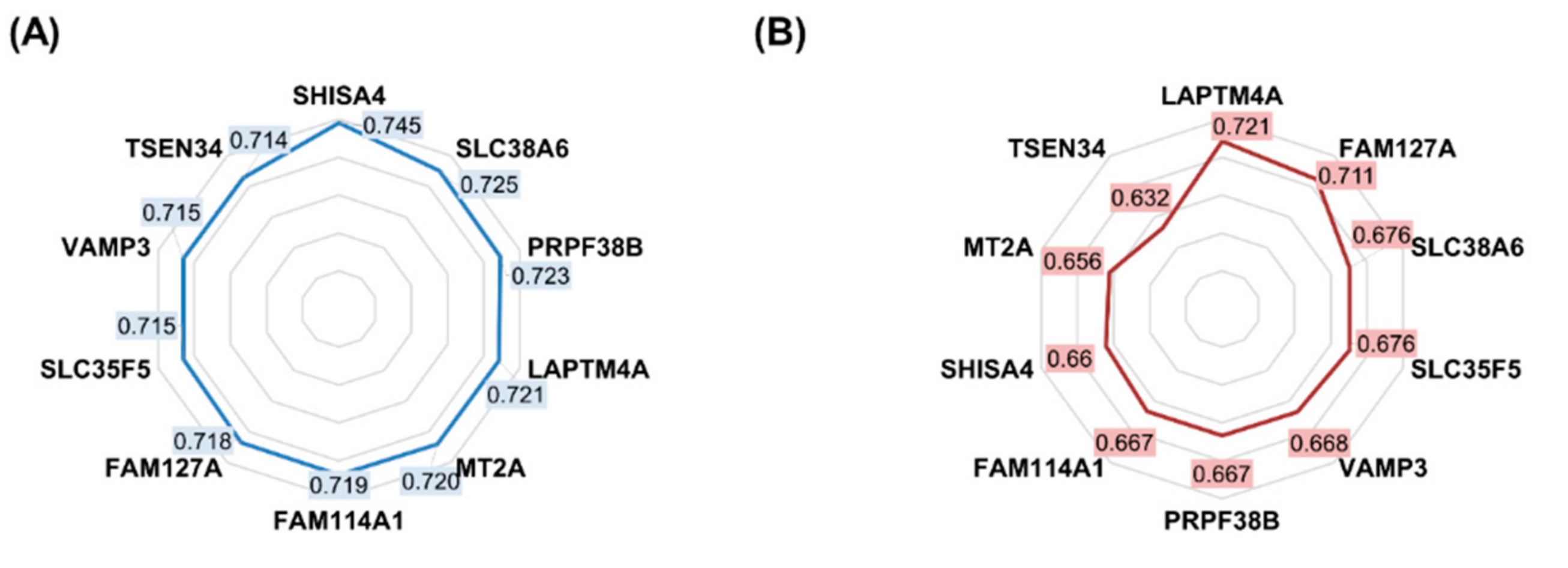
2.1. Features Significant in the In Vitro Dataset
2.2. Clinical Sample Database Construction
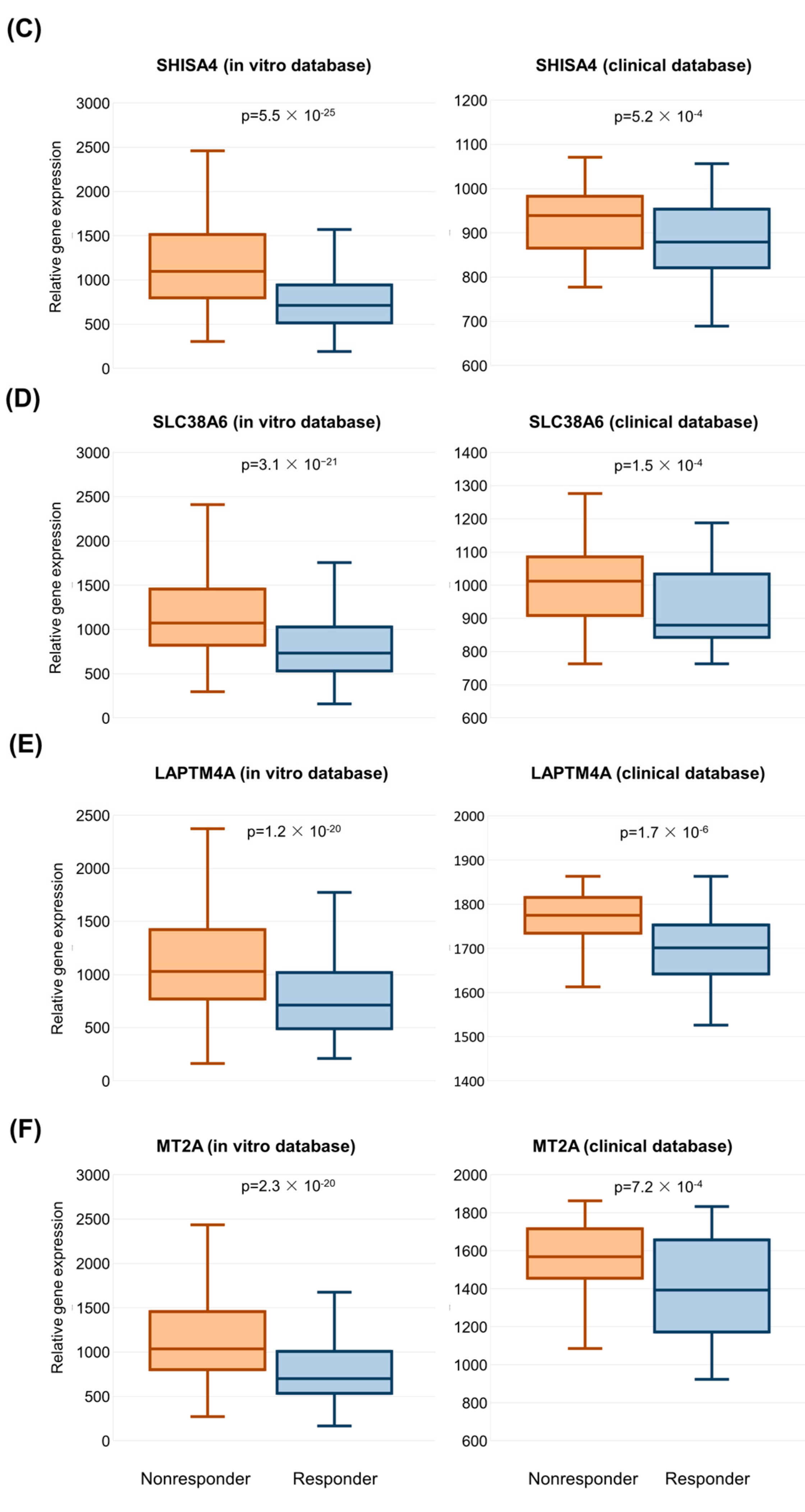
2.3. Validation of Significant Genes in the Clinical Sample Database
2.4. Functional Annotation of Validated Genes
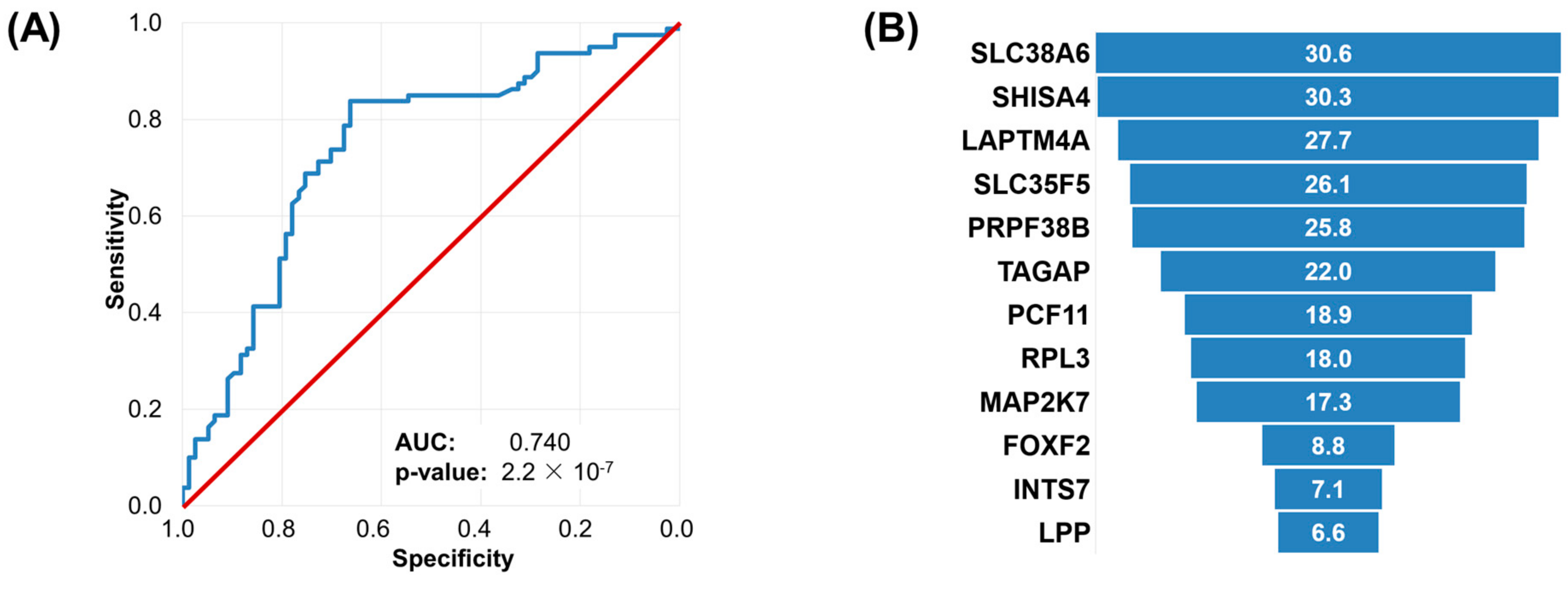
2.5. Machine Learning Approach to Set Up an Integrated Predictive Tool
3. Discussion
4. Materials and Methods
4.1. In Vitro Biomarker Discovery
4.2. Validation in a Dataset Comprising Clinical Samples
4.3. Machine Learning Approach to Set Up an Integrated Predictive Tool
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Mármol, I.; Sánchez-de-Diego, C.; Pradilla Dieste, A.; Cerrada, E.; Rodriguez Yoldi, M.J. Colorectal Carcinoma: A General Overview and Future Perspectives in Colorectal Cancer. Int. J. Mol. Sci. 2017, 18, 197. [Google Scholar] [CrossRef] [PubMed]
- Benson, A.B.; Venook, A.P.; Al-Hawary, M.M.; Arain, M.A.; Chen, Y.J.; Ciombor, K.K.; Cohen, S.; Cooper, H.S.; Deming, D.; Farkas, L.; et al. Colon Cancer, Version 2.2021, NCCN Clinical Practice Guidelines in Oncology. J. Natl. Compr. Cancer Netw. 2021, 19, 329–359. [Google Scholar] [CrossRef] [PubMed]
- Dekker, E.; Tanis, P.J.; Vleugels, J.L.A.; Kasi, P.M.; Wallace, M.B. Colorectal cancer. Lancet 2019, 394, 1467–1480. [Google Scholar] [CrossRef] [PubMed]
- Koncina, E.; Haan, S.; Rauh, S.; Letellier, E. Prognostic and Predictive Molecular Biomarkers for Colorectal Cancer: Updates and Challenges. Cancers 2020, 12, E319. [Google Scholar] [CrossRef]
- Fidai, S.S.; Sharma, A.E.; Johnson, D.N.; Segal, J.P.; Lastra, R.R. Dihydropyrimidine dehydrogenase deficiency as a cause of fatal 5-Fluorouracil toxicity. Autops. Case Rep. 2018, 8, e2018049. [Google Scholar] [CrossRef]
- Thorn, C.F.; Marsh, S.; Carrillo, M.W.; McLeod, H.L.; Klein, T.E.; Altman, R.B. PharmGKB summary: Fluoropyrimidine pathways. Pharmacogenet. Genom. 2011, 21, 237–242. [Google Scholar] [CrossRef]
- Longley, D.B.; Harkin, D.P.; Johnston, P.G. 5-Fluorouracil: Mechanisms of action and clinical strategies. Nat. Rev. Cancer 2003, 3, 330–338. [Google Scholar] [CrossRef]
- De Gramont, A.; Figer, A.; Seymour, M.; Homerin, M.; Hmissi, A.; Cassidy, J.; Boni, C.; Cortes-Funes, H.; Cervantes, A.; Freyer, G.; et al. Leucovorin and fluorouracil with or without oxaliplatin as first-line treatment in advanced colorectal cancer. J. Clin. Oncol. 2000, 18, 2938–2947. [Google Scholar] [CrossRef]
- Tsuji, S.; Midorikawa, Y.; Takahashi, T.; Yagi, K.; Takayama, T.; Yoshida, K.; Sugiyama, Y.; Aburatani, H. Potential responders to FOLFOX therapy for colorectal cancer by Random Forests analysis. Br. J. Cancer 2012, 106, 126–132. [Google Scholar] [CrossRef]
- Agostini, M.; Zangrando, A.; Pastrello, C.; D’Angelo, E.; Romano, G.; Giovannoni, R.; Giordan, M.; Maretto, I.; Bedin, C.; Zanon, C.; et al. A functional biological network centered on XRCC3: A new possible marker of chemoradiotherapy resistance in rectal cancer patients. Cancer Biol. Ther. 2015, 16, 1160–1171. [Google Scholar] [CrossRef]
- Del Rio, M.; Molina, F.; Bascoul-Mollevi, C.; Copois, V.; Bibeau, F.; Chalbos, P.; Bareil, C.; Kramar, A.; Salvetat, N.; Fraslon, C.; et al. Gene expression signature in advanced colorectal cancer patients select drugs and response for the use of leucovorin, fluorouracil, and irinotecan. J. Clin. Oncol. 2007, 25, 773–780. [Google Scholar] [CrossRef]
- Estevez-Garcia, P.; Rivera, F.; Molina-Pinelo, S.; Benavent, M.; Gómez, J.; Limón, M.L.; Pastor, M.D.; Martinez-Perez, J.; Paz-Ares, L.; Carnero, A.; et al. Gene expression profile predictive of response to chemotherapy in metastatic colorectal cancer. Oncotarget 2015, 6, 6151–6159. [Google Scholar] [CrossRef]
- Cherradi, S.; Martineau, P.; Gongora, C.; Del Rio, M. Claudin gene expression profiles and clinical value in colorectal tumors classified according to their molecular subtype. Cancer Manag. Res. 2019, 11, 1337–1348. [Google Scholar] [CrossRef]
- Tong, M.; Zheng, W.; Lu, X.; Ao, L.; Li, X.; Guan, Q.; Cai, H.; Li, M.; Yan, H.; Guo, Y.; et al. Identifying clinically relevant drug resistance genes in drug-induced resistant cancer cell lines and post-chemotherapy tissues. Oncotarget 2015, 6, 41216–41227. [Google Scholar] [CrossRef]
- Cherradi, S.; Ayrolles-Torro, A.; Vezzo-Vié, N.; Gueguinou, N.; Denis, V.; Combes, E.; Boissière, F.; Busson, M.; Canterel-Thouennon, L.; Mollevi, C.; et al. Antibody targeting of claudin-1 as a potential colorectal cancer therapy. J. Exp. Clin. Cancer Res. CR 2017, 36, 89. [Google Scholar] [CrossRef]
- Okita, A.; Takahashi, S.; Ouchi, K.; Inoue, M.; Watanabe, M.; Endo, M.; Honda, H.; Yamada, Y.; Ishioka, C. Consensus molecular subtypes classification of colorectal cancer as a predictive factor for chemotherapeutic efficacy against metastatic colorectal cancer. Oncotarget 2018, 9, 18698–18711. [Google Scholar] [CrossRef]
- Ji, D.; Song, C.; Li, Y.; Xia, J.; Wu, Y.; Jia, J.; Cui, X.; Yu, S.; Gu, J. Combination of radiotherapy and suppression of Tregs enhances abscopal antitumor effect and inhibits metastasis in rectal cancer. J. Immunother. Cancer 2020, 8, e000826. [Google Scholar] [CrossRef]
- Intarajak, T.; Udomchaiprasertkul, W.; Bunyoo, C.; Yimnoon, J.; Soonklang, K.; Wiriyaukaradecha, K.; Lamlertthon, W.; Sricharunrat, T.; Chaiwiriyawong, W.; Siriphongpreeda, B.; et al. Genetic Aberration Analysis in Thai Colorectal Adenoma and Early-Stage Adenocarcinoma Patients by Whole-Exome Sequencing. Cancers 2019, 11, 977. [Google Scholar] [CrossRef]
- He, L.; Zhu, H.; Zhou, S.; Wu, T.; Wu, H.; Yang, H.; Mao, H.; SekharKathera, C.; Janardhan, A.; Edick, A.; et al. Wnt pathway is involved in 5-FU drug resistance of colorectal cancer cells. Exp. Mol. Med. 2018, 50, 101. [Google Scholar] [CrossRef]
- Huang, L.; Li, L.; Cheng, B.; Xing, T. SLC38A6, regulated by EP300-mediated modifications of H3K27ac, promotes cell proliferation, glutamine metabolism and mitochondrial respiration in hepatocellular carcinoma. Carcinogenesis 2022, 43, 885–894. [Google Scholar] [CrossRef] [PubMed]
- Grabner, A.; Brast, S.; Sucic, S.; Bierer, S.; Hirsch, B.; Pavenstädt, H.; Sitte, H.H.; Schlatter, E.; Ciarimboli, G. LAPTM4A interacts with hOCT2 and regulates its endocytotic recruitment. Cell. Mol. Life Sci. 2011, 68, 4079–4090. [Google Scholar] [CrossRef] [PubMed]
- Tatsumi, S.; Matsuoka, H.; Hashimoto, Y.; Hatta, K.; Maeda, K.; Kamoshida, S. Organic cation transporter 2 and tumor budding as independent prognostic factors in metastatic colorectal cancer patients treated with oxaliplatin-based chemotherapy. Int. J. Clin. Exp. Pathol. 2014, 7, 204–212. [Google Scholar] [PubMed]
- Chen, F.; Wang, H.; Zhu, J.; Zhao, R.; Xue, P.; Zhang, Q.; Nelson, M.B.; Qu, W.; Feng, B.; Pi, J. Camptothecin suppresses NRF2–ARE activity and sensitises hepatocellular carcinoma cells to anticancer drugs. Br. J. Cancer 2017, 117, 1495–1506. [Google Scholar] [CrossRef] [PubMed]
- Pathak, G.P.; Shah, R.; Kennedy, B.E.; Murphy, J.P.; Clements, D.; Konda, P.; Giacomantonio, M.; Xu, Z.; Schlaepfer, I.R.; Gujar, S. RTN4 Knockdown Dysregulates the AKT Pathway, Destabilizes the Cytoskeleton, and Enhances Paclitaxel-Induced Cytotoxicity in Cancers. Mol. Ther. 2018, 26, 2019–2033. [Google Scholar] [CrossRef]
- Jie, W.; Andrade, K.C.; Lin, X.; Yang, X.; Yue, X.; Chang, J. Pathophysiological Functions of Rnd3/RhoE. Compr. Physiol. 2015, 6, 169–186. [Google Scholar]
- Yang, W.; Soares, J.; Greninger, P.; Edelman, E.J.; Lightfoot, H.; Forbes, S.; Bindal, N.; Beare, D.; Smith, J.A.; Thompson, I.R.; et al. Genomics of Drug Sensitivity in Cancer (GDSC): A resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Res. 2013, 41, D955–D961. [Google Scholar] [CrossRef]
- Tibor Fekete, J.; Győrffy, B. A unified platform enabling biomarker ranking and validation for 1562 drugs using transcriptomic data of 1250 cancer cell lines. Comput. Struct. Biotechnol. J. 2022, 20, 2885–2894. [Google Scholar] [CrossRef]
- Menyhart, O.; Weltz, B.; Győrffy, B. MultipleTesting.com: A tool for life science researchers for multiple hypothesis testing correction. PLoS ONE 2021, 16, e0245824. [Google Scholar] [CrossRef]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef]
- Gautier, L.; Cope, L.; Bolstad, B.M.; Irizarry, R.A. affy—Analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 2004, 20, 307–315. [Google Scholar] [CrossRef]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Li, Q.; Birkbak, N.J.; Gyorffy, B.; Szallasi, Z.; Eklund, A.C. Jetset: Selecting the optimal microarray probe set to represent a gene. BMC Bioinform. 2011, 12, 474. [Google Scholar] [CrossRef]
- Walsh, C.J.; Hu, P.; Batt, J.; Santos, C.C.D. Microarray Meta-Analysis and Cross-Platform Normalization: Integrative Genomics for Robust Biomarker Discovery. Microarrays 2015, 4, 389–406. [Google Scholar] [CrossRef]
- Leek, J.T.; Johnson, W.E.; Parker, H.S.; Jaffe, A.E.; Storey, J.D. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics 2012, 28, 882–883. [Google Scholar] [CrossRef]
- Liaw, A.; Wiener, M. Classification and Regression by randomForest. R News 2002, 2, 5. [Google Scholar]
- Kuhn, M. Building Predictive Models in R Using the caret Package. J. Stat. Softw. 2008, 28, 1–26. [Google Scholar] [CrossRef]

| (A) | ||
|---|---|---|
| Cell Line CCLE Name | Tumor Type | Normalized AUDRC |
| PSN1_PANCREAS | pancreas exocrine adenocarcinoma | 0.352 |
| CAL148_BREAST | breast ductal carcinoma | 0.353 |
| JHU011_UPPER_AERODIGESTIVE_TRACT | upper aerodigestive squamous carcinoma | 0.39 |
| PCI4B_UPPER_AERODIGESTIVE_TRACT | upper aerodigestive squamous carcinoma | 0.423 |
| CAL27_UPPER_AERODIGESTIVE_TRACT | upper aerodigestive squamous carcinoma | 0.439 |
| PA1_OVARY | ovary mixed germ cell | 0.452 |
| MRKNU1_BREAST | breast carcinoma | 0.46 |
| 22RV1_PROSTATE | prostate adenocarcinoma | 0.483 |
| HUPT4_PANCREAS | pancreas exocrine adenocarcinoma | 0.488 |
| A2780_OVARY | ovary endometrioid adenocarcinoma | 0.496 |
| (B) | ||
| Cell Line CCLE Name | Tumor Type | Normalized AUDRC |
| LN18_CENTRAL_NERVOUS_SYSTEM | glioblastoma | 0.991 |
| HEC1_ENDOMETRIUM | uterus endometrial adenocarcinoma | 0.99 |
| ASH3_THYROID | thyroid carcinoma | 0.988 |
| SCH_STOMACH | choriocarcinoma | 0.988 |
| CCFSTTG1_CENTRAL_NERVOUS_SYSTEM | astrocytoma | 0.988 |
| NCIH2444_LUNG | non-small cell lung cancer | 0.988 |
| HSC2_UPPER_AERODIGESTIVE_TRACT | upper aerodigestive squamous carcinoma | 0.988 |
| NCIH524_LUNG | small cell lung cancer | 0.988 |
| SW13_ADRENAL_CORTEX | adrenal cortex | 0.987 |
| FTC133_THYROID | thyroid carcinoma | 0.987 |
| Gene Symbol | Approved Name | ROC AUC | Mean Expression (Nonresponder) | Mean Expression (Responder) | Mean Fold Change | Mann–Whitney U Test p-Value |
|---|---|---|---|---|---|---|
| SHISA4 | shisa family member 4 | 0.745 | 1096.7 | 714.4 | 1.54 | 5.52 × 10−25 |
| SLC38A6 | solute carrier family 38 member 6 | 0.725 | 1072.0 | 735.3 | 1.46 | 3.07 × 10−21 |
| PRPF38B | pre-mRNA processing factor 38B | 0.723 | 753.0 | 1056.3 | 0.71 | 6.39 × 10−21 |
| LAPTM4A | lysosomal protein transmembrane 4 alpha | 0.721 | 1027.9 | 712.3 | 1.44 | 1.23 × 10−20 |
| MT2A | metallothionein 2A | 0.720 | 1038.1 | 699.6 | 1.48 | 2.34 × 10−20 |
| FAM114A1 | family with sequence similarity 114 member A1 | 0.719 | 1058.4 | 687.9 | 1.54 | 2.73 × 10−20 |
| FAM127A | retrotransposon Gag-like 8C | 0.718 | 1031.1 | 678.9 | 1.52 | 4.75 × 10−20 |
| SLC35F5 | solute carrier family 35 member F5 | 0.715 | 1091.4 | 732.7 | 1.49 | 1.17 × 10−19 |
| TSPAN4 | tetraspanin 4 | 0.715 | 1090.0 | 703.4 | 1.55 | 1.25 × 10−19 |
| VAMP3 | vesicle-associated membrane protein 3 | 0.715 | 1051.6 | 719.4 | 1.46 | 1.44 × 10−19 |
| Dataset (Reference) | Platform | Sample Size | Age in Years, (Mean ± SD) | 5FU/Capecitabine | Irinotecan | Bevacizumab | Oxaliplatin | Of These: 5FU/Capecitabine Monotherapy |
|---|---|---|---|---|---|---|---|---|
| GSE19860 | Affymetrix HGU133 Plus 2.0 Array | 29 | - | 29 | - | 12 | 29 | - |
| GSE19862 | Affymetrix HGU133 Plus 2.0 Array | 14 | - | - | - | 14 | - | - |
| GSE28702 [10] | Affymetrix HGU133 Plus 2.0 Array | 56 | - | 56 | - | - | 56 | - |
| GSE45404 [11] | Affymetrix HGU133 Plus 2.0 Array | 42 | 59.9 ± 11.20 | 42 | - | - | - | 42 |
| GSE49355 [12] | Affymetrix HGU133A Array | 20 | 59.3 ± 7.83 | 20 | 20 | - | - | - |
| GSE52735 [13] | Affymetrix HGU133 Plus 2.0 Array | 37 | - | 37 | - | - | 37 | - |
| GSE62080 [14] | Affymetrix HGU133 Plus 2.0 Array | 21 | - | 21 | 21 | - | - | - |
| GSE69657 [15] | Affymetrix HGU133 Plus 2.0 Array | 16 | 53.6 ± 12.88 | 16 | - | - | 16 | - |
| GSE72970 [16] | Affymetrix HGU133 Plus 2.0 Array | 124 | 61.7 ± 11.39 | 124 | 88 | 28 | 40 | - |
| GSE104645 [17] | Agilent-014850 Whole Human Genome Microarray 4x44K | 167 | - | 140 | 54 | 83 | 113 | - |
| GSE119409 [18] | Affymetrix HGU133 Plus 2.0 Array | 56 | 56.8 ± 11.17 | 56 | - | - | - | 56 |
| TCGA | Illumina HiSeq | 223 | 60.7 ± 12.04 | 210 | 38 | 43 | 147 | 59 |
| Total | 805 | 60.1 ± 11.69 | 751 | 221 | 180 | 438 | 157 | |
| Class | Gene Ontology ID | Gene Ontology Term | Gene Ratio | Adjusted p-Value | Top Five Genes included in the Term |
|---|---|---|---|---|---|
| Biological process | GO:0006979 | response to oxidative stress | 35/679 | 3.90 × 10−2 | NFE2L2 (0.619; 0.714); HNRNPD (0.645; 0.698); MGST1 (0.610; 0.696) EGLN1 (0.631; 0.695); STX4 (0.636; 0.689) |
| GO:0000302 | response to reactive oxygen species | 22/679 | 4.01 × 10−2 | NFE2L2 (0.619; 0.714); HNRNPD (0.645; 0.698); EGLN1 (0.631; 0.695); EEF2 (0.668; 0.689); PPP2CB (0.659; 0.689) | |
| GO:1903311 | regulation of mRNA metabolic process | 28/679 | 4.01 × 10−2 | SF1 (0.670; 0.731); GIGYF2 (0.624; 0.702); HNRNPD (0.645; 0.698); HNRNPC (0.621; 0.688); THRAP3 (0.636; 0.684) | |
| GO:0048762 | mesenchymal cell differentiation | 22/679 | 4.01 × 10−2 | PDCD6 (0.624; 0.718); TAPT1 (0.610; 0.708); CLASP2 (0.651; 0.697); RTN4 (0.675; 0.690); SDCBP (0.644; 0.680) | |
| Molecular function | GO:0003924 | GTPase activity | 28/693 | 9.79 × 10−3 | RND3 (0.701; 0.726); ARF4 (0.652; 0.703); KRAS (0.614; 0.694); EEF2 (0.668; 0.689); TUBB2A (0.673; 0.681) |
| GO:0005525 | GTP binding | 31/693 | 9.89 × 10−3 | RND3 (0.701; 0.726); ARL11 (0.630; 0.703); ARF4 (0.652; 0.703); KRAS (0.614; 0.694); EEF2 (0.668; 0.689) | |
| GO:0032550 | purine ribonucleoside binding | 31/693 | 9.89 × 10−3 | ||
| GO:0001883 | purine nucleoside binding | 31/693 | 9.89 × 10−3 | ||
| GO:0032549 | ribonucleoside binding | 31/693 | 1.10 × 10−2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fekete, J.T.; Győrffy, B. New Transcriptomic Biomarkers of 5-Fluorouracil Resistance. Int. J. Mol. Sci. 2023, 24, 1508. https://doi.org/10.3390/ijms24021508
Fekete JT, Győrffy B. New Transcriptomic Biomarkers of 5-Fluorouracil Resistance. International Journal of Molecular Sciences. 2023; 24(2):1508. https://doi.org/10.3390/ijms24021508
Chicago/Turabian StyleFekete, János Tibor, and Balázs Győrffy. 2023. "New Transcriptomic Biomarkers of 5-Fluorouracil Resistance" International Journal of Molecular Sciences 24, no. 2: 1508. https://doi.org/10.3390/ijms24021508
APA StyleFekete, J. T., & Győrffy, B. (2023). New Transcriptomic Biomarkers of 5-Fluorouracil Resistance. International Journal of Molecular Sciences, 24(2), 1508. https://doi.org/10.3390/ijms24021508






