Advances in Characterizing the Transport Systems of and Resistance to EntDD14, A Leaderless Two-Peptide Bacteriocin with Potent Inhibitory Activity
Abstract
:1. Introduction
2. Results
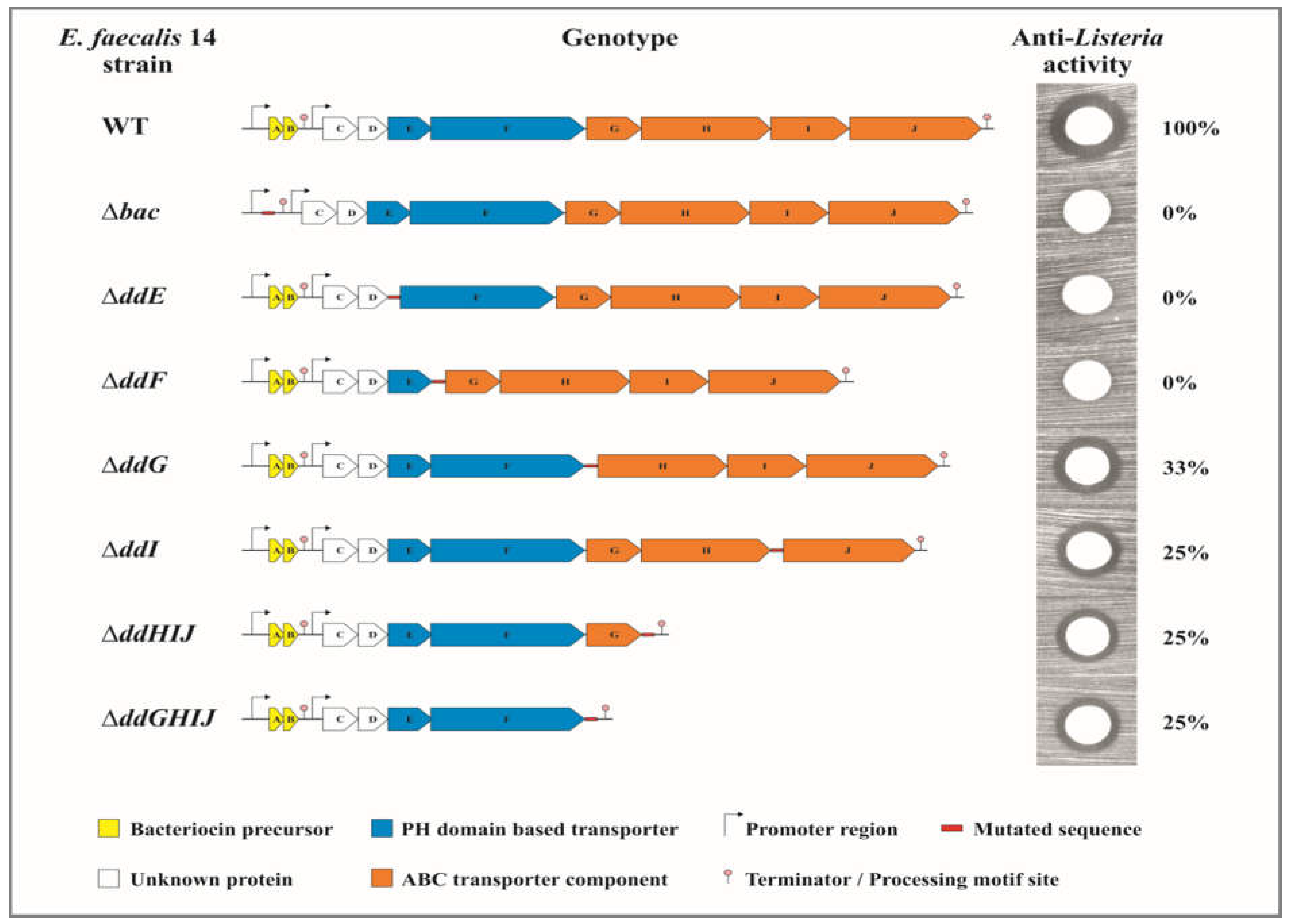
2.1. The EntDD14-ABC Transporter System Contains Four Proteins
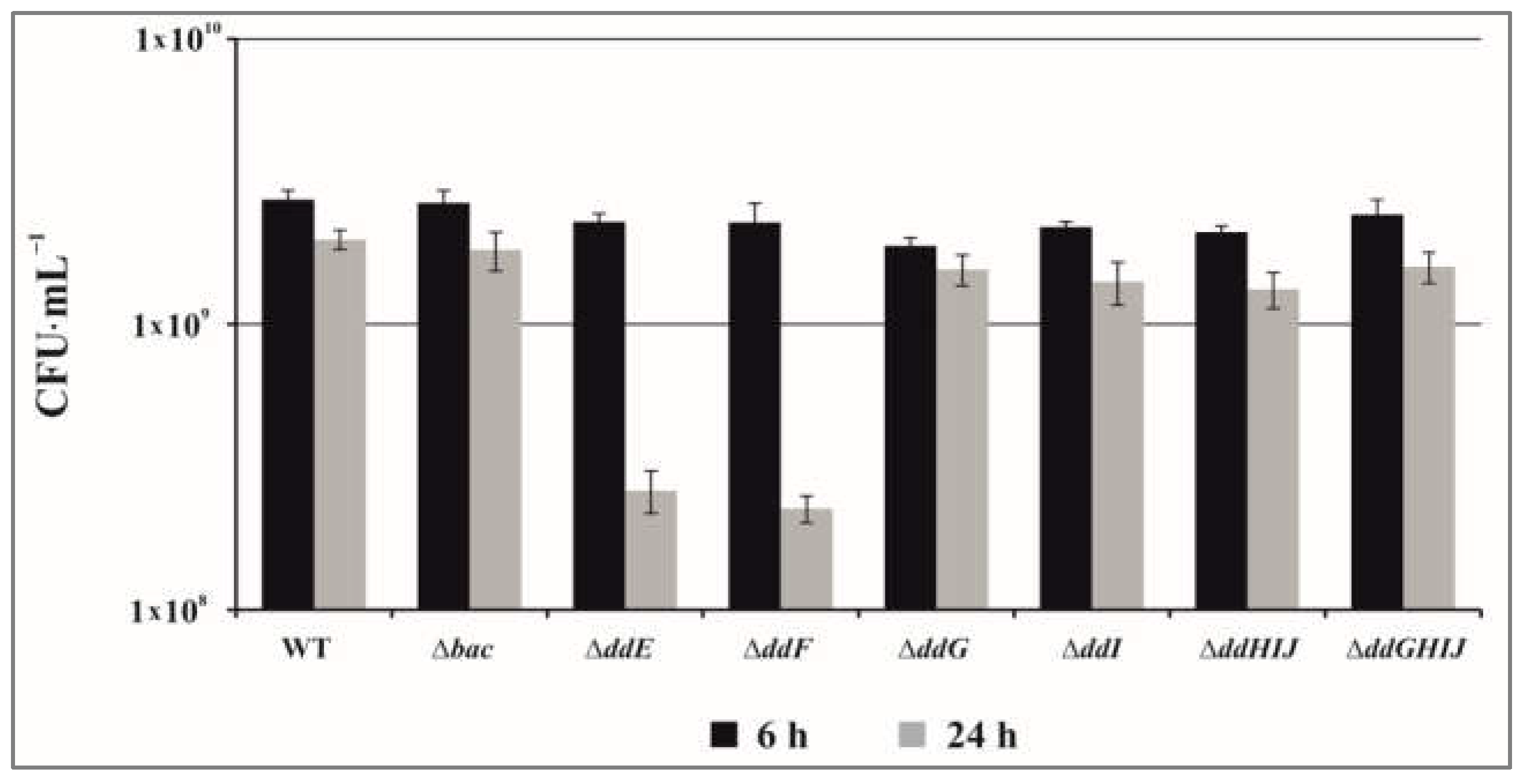
2.2. The Proteins DdF and DdE Are Essential for the Transport of EntDD14
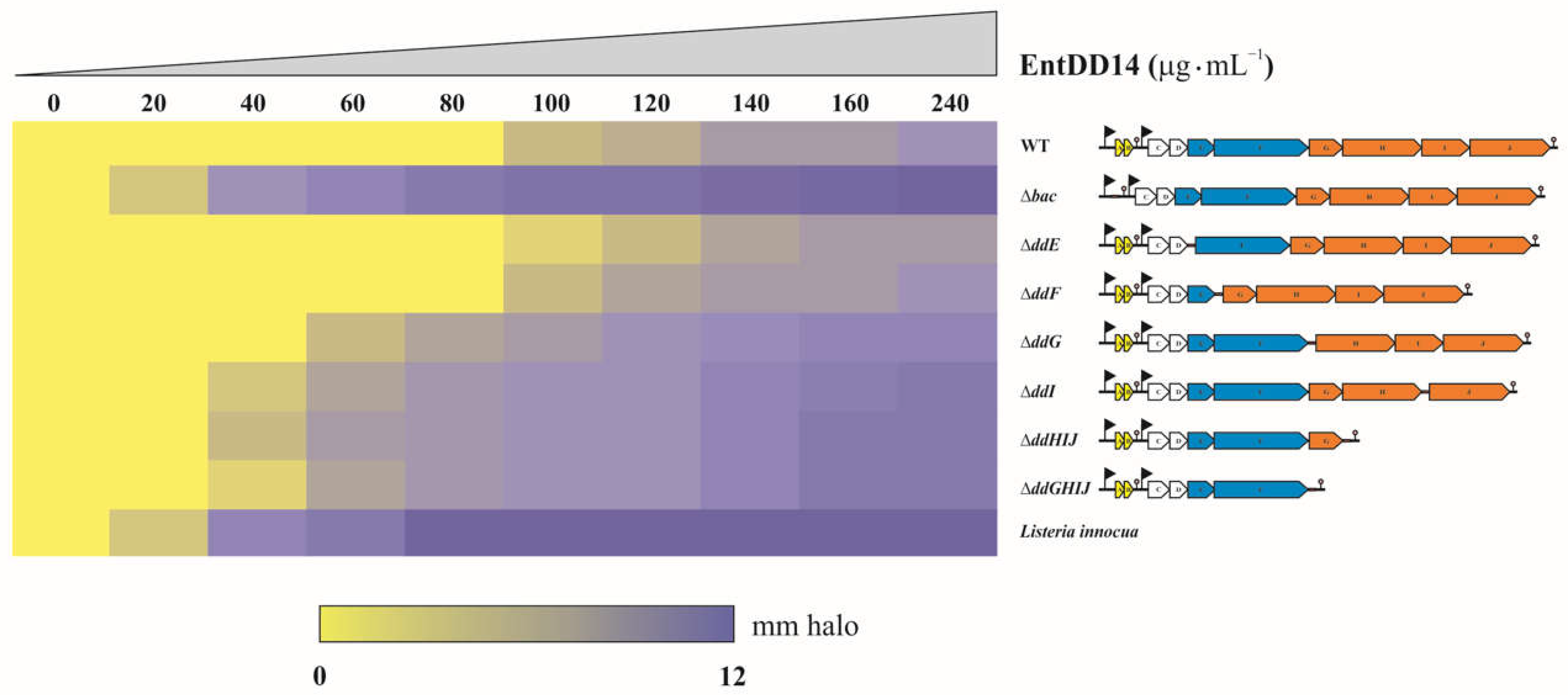
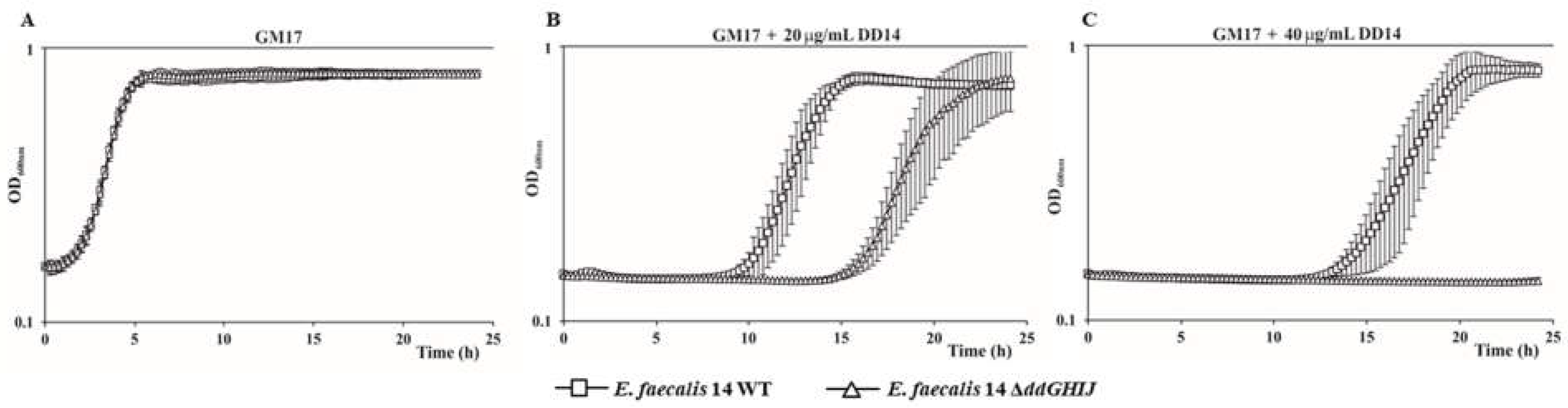
2.3. The ABC Transporter DdGHIJ Is Involved in the Resistance to Extracellular EntDD14
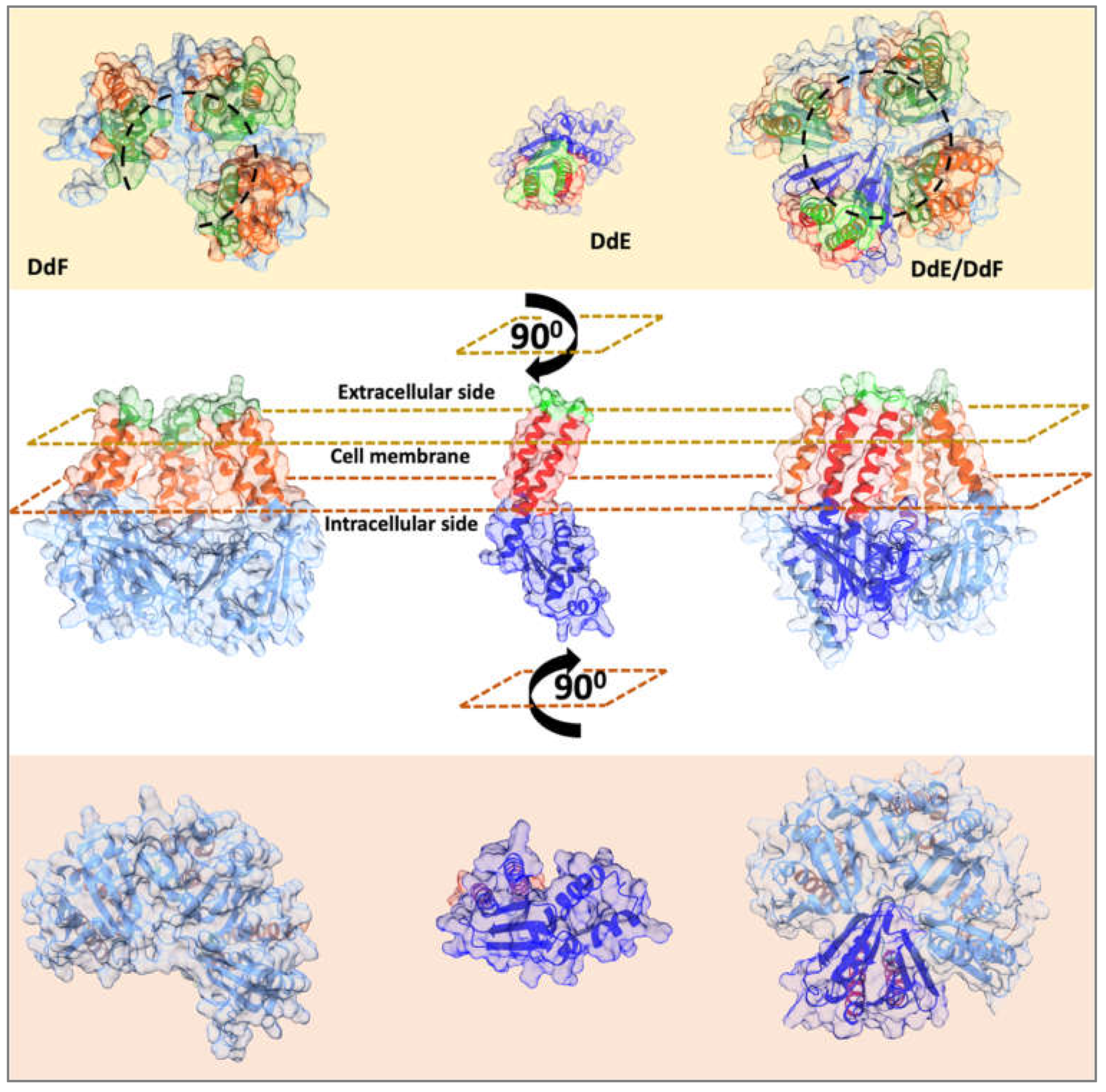
2.4. Predicted Structures of DdE, DdF, and DdE/DdF Show a Potential Transmembrane Channel
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains and Growth Conditions
4.2. Construction of the E. feacalis 14 Mutant Strains
4.3. Antimicrobial Activity against the Indicator Strain L. innocua ATCC 33090
4.4. Purification of Bacteriocin
4.5. Sensitivity of the WT and Engineered Variants to Extracellular EntDD14
4.6. Alphafold2 Structure Prediction of DdE, DdF and DdE/DdF Complex
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Drider, D.; Rebuffat, S. (Eds.) Prokaryotic Antimicrobial Peptides: From Genes to Applications; Springer: New York, NY, USA, 2011; ISBN 978-1-4419-7691-8. [Google Scholar]
- Flaherty, R.A.; Freed, S.D.; Lee, S.W. The Wide World of Ribosomally Encoded Bacterial Peptides. PLoS Pathog. 2014, 10, e1004221. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meade, E.; Slattery, M.A.; Garvey, M. Bacteriocins, Potent Antimicrobial Peptides and the Fight against Multi Drug Resistant Species: Resistance Is Futile? Antibiotics 2020, 9, 32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hammami, R.; Zouhir, A.; Le Lay, C.; Ben Hamida, J.; Fliss, I. BACTIBASE Second Release: A Database and Tool Platform for Bacteriocin Characterization. BMC Microbiol. 2010, 10, 22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Todorov, S.D.; de Melo Franco, B.D.G.; Tagg, J.R. Bacteriocins of Gram-Positive Bacteria Having Activity Spectra Extending beyond Closely-Related Species. Benef. Microbes 2019, 10, 315–328. [Google Scholar] [CrossRef]
- Zheng, S.; Sonomoto, K. Diversified Transporters and Pathways for Bacteriocin Secretion in Gram-Positive Bacteria. Appl. Microbiol. Biotechnol. 2018, 102, 4243–4253. [Google Scholar] [CrossRef]
- Ortega, M.A.; van der Donk, W.A. New Insights into the Biosynthetic Logic of Ribosomally Synthesized and Post-Translationally Modified Peptide Natural Products. Cell Chem. Biol. 2016, 23, 31–44. [Google Scholar] [CrossRef] [Green Version]
- Soltani, S.; Hammami, R.; Cotter, P.D.; Rebuffat, S.; Said, L.B.; Gaudreau, H.; Bédard, F.; Biron, E.; Drider, D.; Fliss, I. Bacteriocins as a New Generation of Antimicrobials: Toxicity Aspects and Regulations. FEMS Microbiol. Rev. 2021, 45, fuaa039. [Google Scholar] [CrossRef]
- Alvarez-Sieiro, P.; Montalbán-López, M.; Mu, D.; Kuipers, O.P. Bacteriocins of Lactic Acid Bacteria: Extending the Family. Appl. Microbiol. Biotechnol. 2016, 100, 2939–2951. [Google Scholar] [CrossRef] [Green Version]
- Pérez-Ramos, A.; Madi-Moussa, D.; Coucheney, F.; Drider, D. Current Knowledge of the Mode of Action and Immunity Mechanisms of LAB-Bacteriocins. Microorganisms 2021, 9, 2107. [Google Scholar] [CrossRef]
- Perez, R.H.; Zendo, T.; Sonomoto, K. Circular and Leaderless Bacteriocins: Biosynthesis, Mode of Action, Applications, and Prospects. Front. Microbiol. 2018, 9, 2085. [Google Scholar] [CrossRef]
- Cintas, L.M.; Casaus, P.; Holo, H.; Hernandez, P.E.; Nes, I.F.; Håvarstein, L.S. Enterocins L50A and L50B, Two Novel Bacteriocins from Enterococcus Faecium L50, Are Related to Staphylococcal Hemolysins. J. Bacteriol. 1998, 180, 1988–1994. [Google Scholar] [CrossRef] [Green Version]
- Drider, D.; Fimland, G.; Héchard, Y.; McMullen, L.M.; Prévost, H. The Continuing Story of Class IIa Bacteriocins. Microbiol. Mol. Biol. Rev. 2006, 70, 564–582. [Google Scholar] [CrossRef] [Green Version]
- Beis, K.; Rebuffat, S. Multifaceted ABC Transporters Associated to Microcin and Bacteriocin Export. Res. Microbiol. 2019, 170, 399–406. [Google Scholar] [CrossRef]
- Cintas, L.M.; Casaus, P.; Håvarstein, L.S.; Hernández, P.E.; Nes, I.F. Biochemical and Genetic Characterization of Enterocin P, a Novel Sec-Dependent Bacteriocin from Enterococcus faecium P13 with a Broad Antimicrobial Spectrum. Appl. Environ. Microbiol. 1997, 63, 4321–4330. [Google Scholar] [CrossRef] [Green Version]
- Netz, D.J.; Sahl, H.G.; Marcelino, R.; dos Santos Nascimento, J.; de Oliveira, S.S.; Soares, M.B.; do Carmo de Freire Bastos, M.; Marcolino, R. Molecular Characterisation of Aureocin A70, a Multi-Peptide Bacteriocin Isolated from Staphylococcus aureus. J. Mol. Biol. 2001, 311, 939–949. [Google Scholar] [CrossRef]
- Lagedroste, M.; Reiners, J.; Smits, S.H.J.; Schmitt, L. Impact of the Nisin Modification Machinery on the Transport Kinetics of NisT. Sci. Rep. 2020, 10, 12295. [Google Scholar] [CrossRef]
- Cotter, P.D.; Hill, C.; Ross, R.P. Bacteriocins: Developing Innate Immunity for Food. Nat. Rev. Microbiol. 2005, 3, 777–788. [Google Scholar] [CrossRef]
- Barraza, D.E.; Ríos Colombo, N.S.; Galván, A.E.; Acuña, L.; Minahk, C.J.; Bellomio, A.; Chalón, M.C. New Insights into Enterocin CRL35: Mechanism of Action and Immunity Revealed by Heterologous Expression in Escherichia coli. Mol. Microbiol. 2017, 105, 922–933. [Google Scholar] [CrossRef] [Green Version]
- Hacker, C.; Christ, N.A.; Duchardt-Ferner, E.; Korn, S.; Göbl, C.; Berninger, L.; Düsterhus, S.; Hellmich, U.A.; Madl, T.; Kötter, P.; et al. The Solution Structure of the Lantibiotic Immunity Protein NisI and Its Interactions with Nisin. J. Biol. Chem. 2015, 290, 28869–28886. [Google Scholar] [CrossRef] [Green Version]
- Ra, R.; Beerthuyzen, M.M.; de Vos, W.M.; Saris, P.E.J.; Kuipers, O.P. Effects of Gene Disruptions in the Nisin Gene Cluster of Lactococcus lactis on Nisin Production and Producer Immunity. Microbiology 1999, 145 Pt 5, 1227–1233. [Google Scholar] [CrossRef]
- Diaz, M.; Valdivia, E.; Martínez-Bueno, M.; Fernández, M.; Soler-González, A.S.; Ramírez-Rodrigo, H.; Maqueda, M. Characterization of a New Operon, as-48EFGH, from the as-48 Gene Cluster Involved in Immunity to Enterocin AS-48. Appl. Environ. Microbiol. 2003, 69, 1229–1236. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caly, D.L.; Chevalier, M.; Flahaut, C.; Cudennec, B.; Al Atya, A.K.; Chataigné, G.; D’Inca, R.; Auclair, E.; Drider, D. The Safe Enterocin DD14 Is a Leaderless Two-Peptide Bacteriocin with Anti-Clostridium perfringens Activity. Int. J. Antimicrob. Agents 2017, 49, 282–289. [Google Scholar] [CrossRef] [PubMed]
- Al Atya, A.K.; Drider-Hadiouche, K.; Ravallec, R.; Silvain, A.; Vachee, A.; Drider, D. Probiotic Potential of Enterococcus faecalis Strains Isolated from Meconium. Front. Microbiol. 2015, 6, 227. [Google Scholar] [CrossRef]
- Ladjouzi, R.; Lucau-Danila, A.; Benachour, A.; Drider, D. A Leaderless Two-Peptide Bacteriocin, Enterocin DD14, Is Involved in Its Own Self-Immunity: Evidence and Insights. Front. Bioeng. Biotechnol. 2020, 8, 644. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Ramos, A.; Ladjouzi, R.; Benachour, A.; Drider, D. Evidence for the Involvement of Pleckstrin Homology Domain-Containing Proteins in the Transport of Enterocin DD14 (EntDD14); a Leaderless Two-Peptide Bacteriocin. Int. J. Mol. Sci. 2021, 22, 12877. [Google Scholar] [CrossRef]
- Dos Santos Nascimento, J.; Coelho, M.L.V.; Ceotto, H.; Potter, A.; Fleming, L.R.; Salehian, Z.; Nes, I.F.; do Carmo de Freire Bastos, M. Genes Involved in Immunity to and Secretion of Aureocin A53, an Atypical Class II Bacteriocin Produced by Staphylococcus aureus A53. J. Bacteriol. 2012, 194, 875–883. [Google Scholar] [CrossRef] [Green Version]
- Iwatani, S.; Horikiri, Y.; Zendo, T.; Nakayama, J.; Sonomoto, K. Bifunctional Gene Cluster LnqBCDEF Mediates Bacteriocin Production and Immunity with Differential Genetic Requirements. Appl. Environ. Microbiol. 2013, 79, 2446–2449. [Google Scholar] [CrossRef] [Green Version]
- Rashid, R.; Veleba, M.; Kline, K.A. Focal Targeting of the Bacterial Envelope by Antimicrobial Peptides. Front. Cell Dev. Biol. 2016, 4, 55. [Google Scholar] [CrossRef] [Green Version]
- Rahman, S.; Mchaourab, H.S. ATP-Dependent Interactions of a Cargo Protein with the Transmembrane Domain of a Polypeptide Processing and Secretion ABC Transporter. J. Biol. Chem. 2020, 295, 14678–14685. [Google Scholar] [CrossRef]
- Hiron, A.; Falord, M.; Valle, J.; Débarbouillé, M.; Msadek, T. Bacitracin and Nisin Resistance in Staphylococcus aureus: A Novel Pathway Involving the BraS/BraR Two-Component System (SA2417/SA2418) and Both the BraD/BraE and VraD/VraE ABC Transporters. Mol. Microbiol. 2011, 81, 602–622. [Google Scholar] [CrossRef]
- Hinsa, S.M.; Espinosa-Urgel, M.; Ramos, J.L.; O’Toole, G.A. Transition from Reversible to Irreversible Attachment during Biofilm Formation by Pseudomonas fluorescens WCS365 Requires an ABC Transporter and a Large Secreted Protein. Mol. Microbiol. 2003, 49, 905–918. [Google Scholar] [CrossRef]
- Davidson, A.L.; Dassa, E.; Orelle, C.; Chen, J. Structure, Function, and Evolution of Bacterial ATP-Binding Cassette Systems. Microbiol. Mol. Biol. Rev. 2008, 72, 317–364. [Google Scholar] [CrossRef] [Green Version]
- Shimamura, H.; Gouda, H.; Nagai, K.; Hirose, T.; Ichioka, M.; Furuya, Y.; Kobayashi, Y.; Hirono, S.; Sunazuka, T.; Omura, S. Structure Determination and Total Synthesis of Bottromycin A2: A Potent Antibiotic against MRSA and VRE. Angew. Chem. Int. Ed. Engl. 2009, 48, 914–917. [Google Scholar] [CrossRef]
- Thurlow, L.R.; Thomas, V.C.; Hancock, L.E. Capsular Polysaccharide Production in Enterococcus faecalis and Contribution of CpsF to Capsule Serospecificity. J. Bacteriol. 2009, 191, 6203–6210. [Google Scholar] [CrossRef] [Green Version]
- Abriouel, H.; Valdivia, E.; Martínez-Bueno, M.; Maqueda, M.; Gálvez, A. A Simple Method for Semi-Preparative-Scale Production and Recovery of Enterocin AS-48 Derived from Enterococcus faecalis Subsp. liquefaciens A-48-32. J. Microbiol. Methods 2003, 55, 599–605. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly Accurate Protein Structure Prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Hornak, V.; Abel, R.; Okur, A.; Strockbine, B.; Roitberg, A.; Simmerling, C. Comparison of Multiple Amber Force Fields and Development of Improved Protein Backbone Parameters. Proteins 2006, 65, 712–725. [Google Scholar] [CrossRef] [Green Version]
- Eastman, P.; Swails, J.; Chodera, J.D.; McGibbon, R.T.; Zhao, Y.; Beauchamp, K.A.; Wang, L.-P.; Simmonett, A.C.; Harrigan, M.P.; Stern, C.D.; et al. OpenMM 7: Rapid Development of High Performance Algorithms for Molecular Dynamics. PLoS Comput. Biol. 2017, 13, e1005659. [Google Scholar] [CrossRef] [Green Version]
- Available online: https://www.biorxiv.org/content/10.1101/2021.10.04.463034v1 (accessed on 14 November 2022).
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Meng, E.C.; Couch, G.S.; Croll, T.I.; Morris, J.H.; Ferrin, T.E. UCSF ChimeraX: Structure Visualization for Researchers, Educators, and Developers. Protein Sci. 2021, 30, 70–82. [Google Scholar] [CrossRef]
- Available online: https://www.biorxiv.org/content/10.1101/2022.04.08.487609v1 (accessed on 14 November 2022).
| Bacteria | Plasmids | Resistance | Characteristics | Reference |
|---|---|---|---|---|
| Escherichia coli | ||||
| XL1-Blue | - | - | Plasmid-free type strain used for plasmid cloning | Agilent Technologies |
| XL1-Blue [plT06] | pLT06 | CmR | Source of the conditioned replicative pLT06 plasmid used for mutant strategies | [25] |
| XL1-Blue [pLT06:ΔddG] | pLT06:ΔddG | CmR | Derivative of pLT06 by cloning of a 2158 pb DNA fragment harboring flanked regions of ddG gene | This study |
| XL1-Blue [pLT06:ΔddHIJ] | pLT06:ΔddHIJ | CmR | Derivative of pLT06 by cloning of a 2052 pb DNA fragment harboring flanked regions of ddHIJ genes | This study |
| XL1-Blue [pLT06:ΔddGHIJ] | pLT06:ΔddGHIJ | CmR | Derivative of pLT06 by cloning of a 2134 pb DNA fragment harboring flanked regions of ddGHIJ genes | This study |
| Enterococcus faecalis | ||||
| 14 | - | - | Natural strain isolated from meconium | [24] |
| 14 Δbac | - | - | Deletion mutant strain of ddAB genes | [25] |
| 14 ΔddE | - | - | Deletion mutant strain of ddE gene | [26] |
| 14 ΔddF | - | - | Deletion mutant strain of ddF gene | [26] |
| 14 ΔddG | - | - | Deletion mutant strain of ddG gene | This study |
| 14 ΔddI | - | - | Deletion mutant strain of ddI gene | [25] |
| 14 ΔddHIJ | - | - | Deletion mutant strain of ddHIJ genes | This study |
| 14 ΔddGHIJ | - | - | Deletion mutant strain of ddGHIJ genes | This study |
| Listeria innocua | ||||
| ATCC 33090 | - | - | [25] | |
| Oligonucleotide | Sequence 3′-5′ | Utilization | Amplicon Size (pb) |
|---|---|---|---|
| ddG 1F-PstI | ATTAAACTGCAGTTGAATTCACTCAATCATTTT | Amplification of ddG upstream fragment | 1.133 |
| ddG 2R-Stop | CTATCACTAGGATCCTTAGACTTACTACGATACGTCTGTTTGTA | ||
| ddG 3F-Stop | TAAGTCTAAGGATCCTAGTGATAGGATATAGGAGAAGATAATGAGT | Amplification of ddG downstream fragment | 1.049 |
| ddG 4R-NcoI | ATTAAACCATGGGAACACTGATTTGGACAT | ||
| ddG 5F | AGGAAAATGTTGATTTGGTGTTT | Outer primer; verification of the plasmid integration | - |
| ddG 6R | CTAGAGATTGGGTTTGTTCTTCC | ||
| ddHIJ 1F-PstI | ATTAAACTGCAGAAATATGCTTTTTCCTTACA | Amplification of ddHIJ upstream fragment | 1.051 |
| ddHIJ 2R-Stop | CTATCACTAGGATCCTTAGACTTACTCATTATCTTCTCCTATATCT | ||
| ddHIJ 3F-Stop | TAAGTCTAAGGATCCTAGTGATAGGTAAAGGCCAAAGAATTAGA | Amplification of ddHIJ downstream fragment | 1.025 |
| ddHIJ 4R-NcoI | ATTAAACCATGGAATTTTATCCCAAAGAAAGT | ||
| ddHIJ 5F | ATCAGAATGTTTTCATGCGTT | Outer primer; verification of the plasmid integration | - |
| ddHIJ 6R | AAGTTAATGGTGATACTTCACAA | ||
| oriF | CAATAATCGCATCCGATTGCA | Cloning verification in pLT06 plasmid | - |
| Ks05R | CCTATTATACCATATTTTGGAC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pérez-Ramos, A.; Ladjouzi, R.; Mihasan, M.; Teiar, R.; Benachour, A.; Drider, D. Advances in Characterizing the Transport Systems of and Resistance to EntDD14, A Leaderless Two-Peptide Bacteriocin with Potent Inhibitory Activity. Int. J. Mol. Sci. 2023, 24, 1517. https://doi.org/10.3390/ijms24021517
Pérez-Ramos A, Ladjouzi R, Mihasan M, Teiar R, Benachour A, Drider D. Advances in Characterizing the Transport Systems of and Resistance to EntDD14, A Leaderless Two-Peptide Bacteriocin with Potent Inhibitory Activity. International Journal of Molecular Sciences. 2023; 24(2):1517. https://doi.org/10.3390/ijms24021517
Chicago/Turabian StylePérez-Ramos, Adrián, Rabia Ladjouzi, Marius Mihasan, Radja Teiar, Abdellah Benachour, and Djamel Drider. 2023. "Advances in Characterizing the Transport Systems of and Resistance to EntDD14, A Leaderless Two-Peptide Bacteriocin with Potent Inhibitory Activity" International Journal of Molecular Sciences 24, no. 2: 1517. https://doi.org/10.3390/ijms24021517





