Bypassing Mendel’s First Law: Transmission Ratio Distortion in Mammals
Abstract
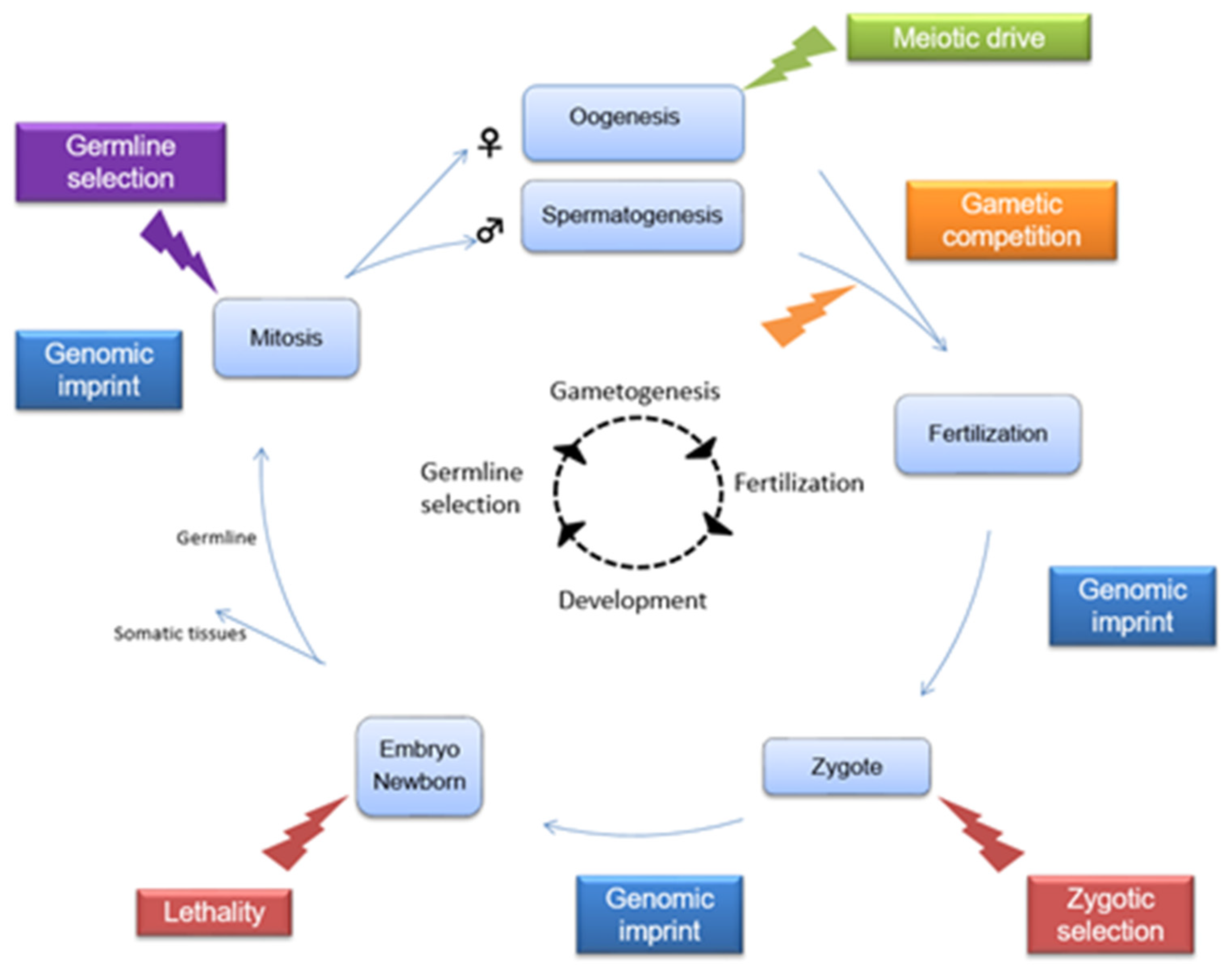
:1. Introduction
2. Transmission Biases Reported in Mice
2.1. Asymmetric Meiosis
2.1.1. Polarity during Female Meiosis
2.1.2. Meiotic Drive in the First Meiotic Division
Centromeric Drive
XO Mice
2.1.3. Meiotic Drive in the Second Meiotic Division
R2d2
Ovum Mutant
HSR
2.2. Symmetric Meiosis
2.2.1. T-Haplotype
2.2.2. Sex Chromosome Drive and Sly/Slx Genes
2.2.3. Other Male Transmission Distortion Systems in Mice
3. Transmission Biases Reported in Other Mammals
4. Transmission Biases Reported in Human and in Pathologies
4.1. Robertsonian Translocations
4.2. TRDs in Pathologies Caused by Point Mutations
4.3. TRDs in Pathologies Caused by Trinucleotide Expansions
4.3.1. Machado–Joseph Disease/Spinocerebellar Ataxia Type 3
4.3.2. Myotonic Dystrophy Type 1
4.3.3. ARX
5. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wyttenbach, A.; Borodin, P.; Hausser, J. Meiotic Drive Favors Robertsonian Metacentric Chromosomes in the Common Shrew (Sorex Araneus, Insectivora, Mammalia). Cytogenet. Cell Genet. 1998, 83, 199–206. [Google Scholar] [CrossRef] [PubMed]
- De Villena, F.P.-M.; de la Casa-Esperon, E.; Briscoe, T.L.; Sapienza, C. A Genetic Test to Determine the Origin of Maternal Transmission Ratio Distortion. Meiotic Drive at the Mouse Om Locus. Genetics 2000, 154, 333–342. [Google Scholar] [CrossRef] [PubMed]
- De Villena, F.P.-M.; Sapienza, C. Transmission Ratio Distortion in Offspring of Heterozygous Female Carriers of Robertsonian Translocations. Hum. Genet. 2001, 108, 31–36. [Google Scholar] [CrossRef] [PubMed]
- Vázquez-Gómez, M.; de Hijas-Villalba, M.M.; Varona, L.; Ibañez-Escriche, N.; Rosas, J.P.; Negro, S.; Noguera, J.L.; Casellas, J. Maternal Transmission Ratio Distortion in Two Iberian Pig Varieties. Genes 2020, 11, 1050. [Google Scholar] [CrossRef]
- Amaral, A.; Herrmann, B.G. RAC1 Controls Progressive Movement and Competitiveness of Mammalian Spermatozoa. PLoS Genet. 2021, 17, e1009308. [Google Scholar] [CrossRef] [PubMed]
- Hastings, I.M. Germline Selection: Population Genetic Aspects of the Sexual/Asexual Life Cycle. Genetics 1991, 129, 1167–1176. [Google Scholar] [CrossRef]
- Crow, J.F. Upsetting the Dogma: Germline Selection in Human Males. PLoS Genet. 2012, 8, e1002535. [Google Scholar] [CrossRef] [Green Version]
- Sandler, L.; Novitski, E. Meiotic Drive as an Evolutionary Force. Am. Nat. 1957, 91, 105–110. [Google Scholar] [CrossRef]
- de Villena, F.P.-M.; Sapienza, C. Nonrandom Segregation during Meiosis: The Unfairness of Females. Mamm. Genome 2001, 12, 331–339. [Google Scholar] [CrossRef]
- Bravo Núñez, M.A.; Nuckolls, N.L.; Zanders, S.E. Genetic Villains: Killer Meiotic Drivers. Trends Genet. 2018, 34, 424–433. [Google Scholar] [CrossRef]
- Lorenzen, M.D.; Gnirke, A.; Margolis, J.; Garnes, J.; Campbell, M.; Stuart, J.J.; Aggarwal, R.; Richards, S.; Park, Y.; Beeman, R.W. The maternal-effect, selfish genetic element Medea is associated with a composite Tc1 transposon. Proc. Natl. Acad. Sci. USA 2008, 105, 10085–10089. [Google Scholar] [CrossRef] [Green Version]
- Zöllner, S.; Wen, X.; Hanchard, N.A.; Herbert, M.A.; Ober, C.; Pritchard, J.K. Evidence for Extensive Transmission Distortion in the Human Genome. Am. J. Hum. Genet. 2004, 74, 62–72. [Google Scholar] [CrossRef] [Green Version]
- Moore, C.S. Postnatal Lethality and Cardiac Anomalies in the Ts65Dn Down Syndrome Mouse Model. Mamm. Genome 2006, 17, 1005–1012. [Google Scholar] [CrossRef] [PubMed]
- Moreno-Garcia, M.A.; Pupavac, M.; Rosenblatt, D.S.; Tremblay, M.L.; Jerome-Majewska, L.A. The Mmachc Gene Is Required for Pre-Implantation Embryogenesis in the Mouse. Mol. Genet. Metab. 2014, 112, 198–204. [Google Scholar] [CrossRef] [PubMed]
- Casellas, J.; Id-Lahoucine, S.; Cánovas, A. Discriminating between Allele- and Genotype-Specific Transmission Ratio Distortion. Anim. Genet. 2020, 51, 847–854. [Google Scholar] [CrossRef] [PubMed]
- Naumova, A.K.; Greenwood, C.M.; Morgan, K. Imprinting and Deviation from Mendelian Transmission Ratios. Genome 2001, 44, 311–320. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Andrade, M.F.; Labialle, S.; Moussette, S.; Geneau, G.; Sinnett, D.; Belisle, A.; Greenwood, C.M.T.; Naumova, A.K. Parental Effect of DNA (Cytosine-5) Methyltransferase 1 on Grandparental-Origin-Dependent Transmission Ratio Distortion in Mouse Crosses and Human Families. Genetics 2008, 178, 35–45. [Google Scholar] [CrossRef] [Green Version]
- Huang, L.O.; Infante-Rivard, C.; Labbe, A. Analysis of Case-Parent Trios for Imprinting Effect Using a Loglinear Model with Adjustment for Sex-of-Parent-Specific Transmission Ratio Distortion. Hum. Genet. 2017, 136, 951–961. [Google Scholar] [CrossRef]
- Iwata-Otsubo, A.; Dawicki-McKenna, J.M.; Akera, T.; Falk, S.J.; Chmátal, L.; Yang, K.; Sullivan, B.A.; Schultz, R.M.; Lampson, M.A.; Black, B.E. Expanded Satellite Repeats Amplify a Discrete CENP-A Nucleosome Assembly Site on Chromosomes That Drive in Female Meiosis. Curr. Biol. 2017, 27, 2365–2373.e8. [Google Scholar] [CrossRef]
- Akera, T.; Chmátal, L.; Trimm, E.; Yang, K.; Aonbangkhen, C.; Chenoweth, D.M.; Janke, C.; Schultz, R.M.; Lampson, M.A. Spindle Asymmetry Drives Non-Mendelian Chromosome Segregation. Science 2017, 358, 668–672. [Google Scholar] [CrossRef]
- Wu, T.; Lane, S.I.R.; Morgan, S.L.; Jones, K.T. Spindle Tubulin and MTOC Asymmetries May Explain Meiotic Drive in Oocytes. Nat. Commun. 2018, 9, 2952. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dumont, J.; Petri, S.; Pellegrin, F.; Terret, M.-E.; Bohnsack, M.T.; Rassinier, P.; Georget, V.; Kalab, P.; Gruss, O.J.; Verlhac, M.-H. A Centriole- and RanGTP-Independent Spindle Assembly Pathway in Meiosis I of Vertebrate Oocytes. J. Cell. Biol. 2007, 176, 295–305. [Google Scholar] [CrossRef] [Green Version]
- Schuh, M.; Ellenberg, J. Self-Organization of MTOCs Replaces Centrosome Function during Acentrosomal Spindle Assembly in Live Mouse Oocytes. Cell 2007, 130, 484–498. [Google Scholar] [CrossRef] [Green Version]
- Verlhac, M.H.; Lefebvre, C.; Guillaud, P.; Rassinier, P.; Maro, B. Asymmetric Division in Mouse Oocytes: With or without Mos. Curr. Biol. 2000, 10, 1303–1306. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akera, T.; Trimm, E.; Lampson, M.A. Molecular Strategies of Meiotic Cheating by Selfish Centromeres. Cell 2019, 178, 1132–1144.e10. [Google Scholar] [CrossRef]
- Henikoff, S.; Ahmad, K.; Malik, H.S. The Centromere Paradox: Stable Inheritance with Rapidly Evolving DNA. Science 2001, 293, 1098–1102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumon, T.; Ma, J.; Akins, R.B.; Stefanik, D.; Nordgren, C.E.; Kim, J.; Levine, M.T.; Lampson, M.A. Parallel Pathways for Recruiting Effector Proteins Determine Centromere Drive and Suppression. Cell 2021, 184, 4904–4918.e11. [Google Scholar] [CrossRef]
- LeMaire-Adkins, R.; Hunt, P.A. Nonrandom Segregation of the Mouse Univalent X Chromosome: Evidence of Spindle-Mediated Meiotic Drive. Genetics 2000, 156, 775–783. [Google Scholar] [CrossRef]
- Maro, B.; Johnson, M.H.; Pickering, S.J.; Flach, G. Changes in Actin Distribution during Fertilization of the Mouse Egg. J. Embryol. Exp. Morphol. 1984, 81, 211–237. [Google Scholar] [CrossRef]
- Dehapiot, B.; Clément, R.; Bourdais, A.; Carrière, V.; Huet, S.; Halet, G. RhoA- and Cdc42-Induced Antagonistic Forces Underlie Symmetry Breaking and Spindle Rotation in Mouse Oocytes. PLoS Biol. 2021, 19, e3001376. [Google Scholar] [CrossRef]
- Wang, H.; Li, Y.; Yang, J.; Duan, X.; Kalab, P.; Sun, S.X.; Li, R. Symmetry Breaking in Hydrodynamic Forces Drives Meiotic Spindle Rotation in Mammalian Oocytes. Sci. Adv. 2020, 6, eaaz5004. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Didion, J.P.; Morgan, A.P.; Clayshulte, A.M.-F.; Mcmullan, R.C.; Yadgary, L.; Petkov, P.M.; Bell, T.A.; Gatti, D.M.; Crowley, J.J.; Hua, K.; et al. A Multi-Megabase Copy Number Gain Causes Maternal Transmission Ratio Distortion on Mouse Chromosome 2. PLoS Genet. 2015, 11, e1004850. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Didion, J.P.; Morgan, A.P.; Yadgary, L.; Bell, T.A.; McMullan, R.C.; Ortiz de Solorzano, L.; Britton-Davidian, J.; Bult, C.J.; Campbell, K.J.; Castiglia, R.; et al. R2d2 Drives Selfish Sweeps in the House Mouse. Mol. Biol. Evol. 2016, 33, 1381–1395. [Google Scholar] [CrossRef] [Green Version]
- Wakasugi, N. Studies on Fertility of DDK Mice: Reciprocal Crosses between DDK and C57BL/6J Strains and Experimental Transplantation of the Ovary. J. Reprod. Fertil. 1973, 33, 283–291. [Google Scholar] [CrossRef] [Green Version]
- De Villena, F.P.-M.; Slamka, C.; Fonseca, M.; Naumova, A.K.; Paquette, J.; Pannunzio, P.; Smith, M.; Verner, A.; Morgan, K.; Sapienza, C. Transmission-Ratio Distortion through F1 Females at Chromosome 11 Loci Linked to Om in the Mouse DDK Syndrome. Genetics 1996, 142, 1299–1304. [Google Scholar] [CrossRef]
- De Villena, F.P.-M.; Naumova, A.K.; Verner, A.E.; Jin, W.H.; Sapienza, C. Confirmation of Maternal Transmission Ratio Distortion at Om and Direct Evidence That the Maternal and Paternal “DDK Syndrome” Genes Are Linked. Mamm. Genome 1997, 8, 642–646. [Google Scholar] [CrossRef]
- Wu, G.; Hao, L.; Han, Z.; Gao, S.; Latham, K.E.; de Villena, F.P.-M.; Sapienza, C. Maternal Transmission Ratio Distortion at the Mouse Om Locus Results from Meiotic Drive at the Second Meiotic Division. Genetics 2005, 170, 327–334. [Google Scholar] [CrossRef] [Green Version]
- Purmann, L.; Plass, C.; Grüneberg, M.; Winking, H.; Traut, W. A Long-Range Repeat Cluster in Chromosome 1 of the House Mouse, Mus Musculus, and Its Relation to a Germline Homogeneously Staining Region. Genomics 1992, 12, 80–88. [Google Scholar] [CrossRef] [PubMed]
- Agulnik, S.I.; Agulnik, A.I.; Ruvinsky, A.O. Meiotic Drive in Female Mice Heterozygous for the HSR Inserts on Chromosome 1. Genet. Res. 1990, 55, 97–100. [Google Scholar] [CrossRef]
- Ruvinsky, A. Meiotic drive in female mice: An essay. Mamm. Genome 1995, 6, 315–320. [Google Scholar] [CrossRef]
- Erickson, R.P. Haploid Gene Expresion versus Meiotic Drive: The Relevance of Intercellular Bridges during Spermatogenesis. Nat. New Biol. 1973, 243, 210–212. [Google Scholar] [CrossRef]
- Braun, R.E.; Behringer, R.R.; Peschon, J.J.; Brinster, R.L.; Palmiter, R.D. Genetically Haploid Spermatids Are Phenotypically Diploid. Nature 1989, 337, 373–376. [Google Scholar] [CrossRef] [Green Version]
- Ren, H.P.; Russell, L.D. Clonal Development of Interconnected Germ Cells in the Rat and Its Relationship to the Segmental and Subsegmental Organization of Spermatogenesis. Am. J. Anat. 1991, 192, 121–128. [Google Scholar] [CrossRef] [PubMed]
- Herrmann, B.G.; Bauer, H. The Mouse T-Haplotype: A Selfish Chromosome—Genetics, Molecular Mechanism, and Evolution. In Evolution of the House Mouse; Cambridge University Press: Cambridge, UK, 2012; pp. 297–314. [Google Scholar]
- Lyon, M.F. Transmission Ratio Distortion in Mouse T-Haplotypes Is Due to Multiple Distorter Genes Acting on a Responder Locus. Cell 1984, 37, 621–628. [Google Scholar] [CrossRef] [PubMed]
- Silver, L.M.; Olds-Clarke, P. Transmission Ratio Distortion of Mouse t Haplotypes Is Not a Consequence of Wild-Type Sperm Degeneration. Dev. Biol. 1984, 105, 250–252. [Google Scholar] [CrossRef] [PubMed]
- Olds-Clarke, P.; Johnson, L.R. T Haplotypes in the Mouse Compromise Sperm Flagellar Function. Dev. Biol. 1993, 155, 14–25. [Google Scholar] [CrossRef] [PubMed]
- Herrmann, B.G.; Koschorz, B.; Wertz, K.; McLaughlin, K.J.; Kispert, A. A Protein Kinase Encoded by the t Complex Responder Gene Causes Non-Mendelian Inheritance. Nature 1999, 402, 141–146. [Google Scholar] [CrossRef]
- Véron, N.; Bauer, H.; Weisse, A.Y.; Lüder, G.; Werber, M.; Herrmann, B.G. Retention of Gene Products in Syncytial Spermatids Promotes Non-Mendelian Inheritance as Revealed by the t Complex Responder. Genes Dev. 2009, 23, 2705–2710. [Google Scholar] [CrossRef] [Green Version]
- Charron, Y.; Willert, J.; Lipkowitz, B.; Kusecek, B.; Herrmann, B.G.; Bauer, H. Two Isoforms of the RAC-Specific Guanine Nucleotide Exchange Factor TIAM2 Act Oppositely on Transmission Ratio Distortion by the Mouse t-Haplotype. PLoS Genet. 2019, 15, e1007964. [Google Scholar] [CrossRef] [Green Version]
- Bauer, H.; Schindler, S.; Charron, Y.; Willert, J.; Kusecek, B.; Herrmann, B.G. The Nucleoside Diphosphate Kinase Gene Nme3 Acts as Quantitative Trait Locus Promoting Non-Mendelian Inheritance. PLoS Genet. 2012, 8, e1002567. [Google Scholar] [CrossRef]
- Bauer, H.; Willert, J.; Koschorz, B.; Herrmann, B.G. The t Complex-Encoded GTPase-Activating Protein Tagap1 Acts as a Transmission Ratio Distorter in Mice. Nat. Genet. 2005, 37, 969–973. [Google Scholar] [CrossRef] [PubMed]
- Bauer, H.; Véron, N.; Willert, J.; Herrmann, B.G. The T-Complex-Encoded Guanine Nucleotide Exchange Factor Fgd2 Reveals That Two Opposing Signaling Pathways Promote Transmission Ratio Distortion in the Mouse. Genes Dev. 2007, 21, 143–147. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ardlie, K.G. Putting the Brake on Drive: Meiotic Drive of t Haplotypes in Natural Populations of Mice. Trends Genet. 1998, 14, 189–193. [Google Scholar] [CrossRef] [PubMed]
- Winkler, L.; Lindholm, A.K. A Meiotic Driver Alters Sperm Form and Function in House Mice: A Possible Example of Spite. Chromosome Res. 2022, 30, 151–164. [Google Scholar] [CrossRef] [PubMed]
- Lyttle, T.W. Segregation Distorters. Annu. Rev. Genet. 1991, 25, 511–557. [Google Scholar] [CrossRef]
- Burt, A.; Trivers, R. Genes in Conflict: The Biology of Selfish Genetic Elements; Harvard University Press: Cambridge, UK, 2006. [Google Scholar]
- Fisher, R.A. The Genetical Theory of Natural Selection; Oxford University Press: Oxford, UK, 1930. [Google Scholar]
- Hamilton, W.D. Extraordinary Sex Ratios. A Sex-Ratio Theory for Sex Linkage and Inbreeding Has New Implications in Cytogenetics and Entomology. Science 1967, 156, 477–488. [Google Scholar] [CrossRef]
- Lindholm, A.K.; Dyer, K.A.; Firman, R.C.; Fishman, L.; Forstmeier, W.; Holman, L.; Johannesson, H.; Knief, U.; Kokko, H.; Larracuente, A.M.; et al. The Ecology and Evolutionary Dynamics of Meiotic Drive. Trends Ecol. Evol. 2016, 31, 315–326. [Google Scholar] [CrossRef]
- Courret, C.; Chang, C.-H.; Wei, K.H.-C.; Montchamp-Moreau, C.; Larracuente, A.M. Meiotic Drive Mechanisms: Lessons from Drosophila. Proc. Biol. Sci. 2019, 286, 20191430. [Google Scholar]
- Scavetta, R.J.; Tautz, D. Copy Number Changes of CNV Regions in Intersubspecific Crosses of the House Mouse. Mol. Biol. Evol. 2010, 27, 1845–1856. [Google Scholar] [CrossRef] [Green Version]
- Morgan, A.P.; de Villena, F.P.-M. Sequence and Structural Diversity of Mouse y Chromosomes. Mol. Biol. Evol. 2017, 34, 3186–3204. [Google Scholar] [CrossRef] [Green Version]
- Cocquet, J.; Ellis, P.J.I.; Mahadevaiah, S.K.; Affara, N.A.; Vaiman, D.; Burgoyne, P.S. A Genetic Basis for a Postmeiotic X versus Y Chromosome Intragenomic Conflict in the Mouse. PLoS Genet. 2012, 8, e1002900. [Google Scholar] [CrossRef]
- Cocquet, J.; Ellis, P.J.I.; Yamauchi, Y.; Mahadevaiah, S.K.; Affara, N.A.; Ward, M.A.; Burgoyne, P.S. The Multicopy Gene Sly Represses the Sex Chromosomes in the Male Mouse Germline after Meiosis. PLoS Biol. 2009, 7, e1000244. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cocquet, J.; Ellis, P.J.I.; Yamauchi, Y.; Riel, J.M.; Karacs, T.P.S.; Rattigan, A.; Ojarikre, O.A.; Affara, N.A.; Ward, M.A.; Burgoyne, P.S. Deficiency in the Multicopy Sycp3-like X-Linked Genes Slx and Slxl1 Causes Major Defects in Spermatid Differentiation. Mol. Biol. Cell 2010, 21, 3497–3505. [Google Scholar] [CrossRef] [Green Version]
- Kruger, A.N.; Brogley, M.A.; Huizinga, J.L.; Kidd, J.M.; de Rooij, D.G.; Hu, Y.-C.; Mueller, J.L. A Neofunctionalized X-Linked Ampliconic Gene Family Is Essential for Male Fertility and Equal Sex Ratio in Mice. Curr. Biol. 2019, 29, 3699–3706.e5. [Google Scholar] [CrossRef]
- Rathje, C.C.; Johnson, E.E.P.; Drage, D.; Patinioti, C.; Silvestri, G.; Affara, N.A.; Ialy-Radio, C.; Cocquet, J.; Skinner, B.M.; Ellis, P.J.I. Differential Sperm Motility Mediates the Sex Ratio Drive Shaping Mouse Sex Chromosome Evolution. Curr. Biol. 2019, 29, 3692–3698.e4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moretti, C.; Vaiman, D.; Tores, F.; Cocquet, J. Expression and Epigenomic Landscape of the Sex Chromosomes in Mouse Post-Meiotic Male Germ Cells. Epigenetics Chromatin 2016, 9, 47. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moretti, C.; Serrentino, M.-E.; Ialy-Radio, C.; Delessard, M.; Soboleva, T.A.; Tores, F.; Leduc, M.; Nitschké, P.; Drevet, J.R.; Tremethick, D.J.; et al. SLY Regulates Genes Involved in Chromatin Remodeling and Interacts with TBL1XR1 during Sperm Differentiation. Cell Death Differ. 2017, 24, 1029–1044. [Google Scholar] [CrossRef] [Green Version]
- Comptour, A.; Moretti, C.; Serrentino, M.-E.; Auer, J.; Ialy-Radio, C.; Ward, M.A.; Touré, A.; Vaiman, D.; Cocquet, J. SSTY Proteins Co-Localize with the Post-Meiotic Sex Chromatin and Interact with Regulators of Its Expression. FEBS J. 2014, 281, 1571–1584. [Google Scholar] [CrossRef] [Green Version]
- Moretti, C.; Blanco, M.; Ialy-Radio, C.; Serrentino, M.-E.; Gobé, C.; Friedman, R.; Battail, C.; Leduc, M.; Ward, M.A.; Vaiman, D.; et al. Battle of the Sex Chromosomes: Competition between X and Y Chromosome-Encoded Proteins for Partner Interaction and Chromatin Occupancy Drives Multicopy Gene Expression and Evolution in Muroid Rodents. Mol. Biol. Evol. 2020, 37, 3453–3468. [Google Scholar] [CrossRef]
- Chayko, C.A.; Martin-DeLeon, P.A. The Murine Rb(6.16) Translocation: Alterations in the Proportion of Alternate Sperm Segregants Effecting Fertilization in Vitro and in Vivo. Hum. Genet. 1992, 90, 79–85. [Google Scholar] [CrossRef]
- Aranha, I.P.; Martin-DeLeon, P.A. Mouse Chromosome 6 in Rb Translocations: Consequences in Singly and Doubly Heterozygous Males. Cytogenet. Cell Genet. 1995, 69, 253–259. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Deng, X.; Martin-DeLeon, P.A. Lack of Sharing of Spam1 (Ph-20) among Mouse Spermatids and Transmission Ratio Distortion. Biol. Reprod. 2001, 64, 1730–1738. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, H.; Shertok, S.; Miller, K.; Taylor, L.; Martin-Deleon, P.A. Sperm Dysfunction in the Rb(6.16)- and Rb(6.15)-Bearing Mice Revisited: Involvement of Hyalp1 and Hyal5. Mol. Reprod. Dev. 2005, 72, 404–410. [Google Scholar] [CrossRef] [PubMed]
- Martin-DeLeon, P.A.; Zhang, H.; Morales, C.R.; Zhao, Y.; Rulon, M.; Barnoski, B.L.; Chen, H.; Galileo, D.S. Spam I-Associated Transmission Ratio Distortion in Mice: Elucidating the Mechanism. Reprod. Biol. Endocrinol. 2005, 3, 32. [Google Scholar] [CrossRef] [Green Version]
- Umehara, T.; Tsujita, N.; Shimada, M. Activation of Toll-like Receptor 7/8 Encoded by the X Chromosome Alters Sperm Motility and Provides a Novel Simple Technology for Sexing Sperm. PLoS Biol. 2019, 17, e3000398. [Google Scholar] [CrossRef] [Green Version]
- Navarro-Costa, P.A.; Molaro, A.; Misra, C.S.; Meiklejohn, C.D.; Ellis, P.J. Sex and Suicide: The Curious Case of Toll-like Receptors. PLoS Biol. 2020, 18, e3000663. [Google Scholar] [CrossRef]
- Saunders, P.A.; Veyrunes, F. Unusual Mammalian Sex Determination Systems: A Cabinet of Curiosities. Genes 2021, 12, 1770. [Google Scholar] [CrossRef]
- Saunders, P.A.; Perez, J.; Ronce, O.; Veyrunes, F. Multiple Sex Chromosome Drivers in a Mammal with Three Sex Chromosomes. Curr. Biol. 2022, 32, 2001–2010.e3. [Google Scholar] [CrossRef]
- Ross, K.G.; Shoemaker, D. Unexpected Patterns of Segregation Distortion at a Selfish Supergene in the Fire Ant Solenopsis Invicta. BMC Genet. 2018, 19, 101. [Google Scholar] [CrossRef] [Green Version]
- Hamilton, P.T.; Hodson, C.N.; Curtis, C.I.; Perlman, S.J. Genetics and Genomics of an Unusual Selfish Sex Ratio Distortion in an Insect. Curr. Biol. 2018, 28, 3864–3870.e4. [Google Scholar] [CrossRef] [Green Version]
- Seymour, D.K.; Chae, E.; Arioz, B.I.; Koenig, D.; Weigel, D. Transmission Ratio Distortion Is Frequent in Arabidopsis Thaliana Controlled Crosses. Heredity 2019, 122, 294–304. [Google Scholar] [CrossRef] [PubMed]
- Meccariello, A.; Krsticevic, F.; Colonna, R.; Del Corsano, G.; Fasulo, B.; Papathanos, P.A.; Windbichler, N. Engineered Sex Ratio Distortion by X-Shredding in the Global Agricultural Pest Ceratitis Capitata. BMC Biol. 2021, 19, 78. [Google Scholar] [CrossRef]
- Knief, U.; Schielzeth, H.; Ellegren, H.; Kempenaers, B.; Forstmeier, W. A Prezygotic Transmission Distorter Acting Equally in Female and Male Zebra Finches Taeniopygia Guttata. Mol. Ecol. 2015, 24, 3846–3859. [Google Scholar] [CrossRef]
- Abdalla, E.A.; Id-Lahoucine, S.; Cánovas, A.; Casellas, J.; Schenkel, F.S.; Wood, B.J.; Baes, C.F. Discovering Lethal Alleles across the Turkey Genome Using a Transmission Ratio Distortion Approach. Anim. Genet. 2020, 51, 876–889. [Google Scholar] [CrossRef]
- Axelsson, E.; Albrechtsen, A.; van, A.P.; Li, L.; Megens, H.J.; Vereijken, A.L.J.; Crooijmans, R.P.M.A.; Groenen, M.A.M.; Ellegren, H.; Willerslev, E.; et al. Segregation Distortion in Chicken and the Evolutionary Consequences of Female Meiotic Drive in Birds. Heredity 2010, 105, 290–298. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Borowsky, R.; Cohen, D. Genomic Consequences of Ecological Speciation in Astyanax Cavefish. PLoS ONE 2013, 8, e79903. [Google Scholar] [CrossRef] [Green Version]
- Gagnaire, P.-A.; Normandeau, E.; Pavey, S.A.; Bernatchez, L. Mapping Phenotypic, Expression and Transmission Ratio Distortion QTL Using RAD Markers in the Lake Whitefish (Coregonus Clupeaformis). Mol. Ecol. 2013, 22, 3036–3048. [Google Scholar] [CrossRef]
- Roth, O.; Ebert, D.; Vizoso, D.B.; Bieger, A.; Lass, S. Male-Biased Sex-Ratio Distortion Caused by Octosporea Bayeri, a Vertically and Horizontally-Transmitted Parasite of Daphnia Magna. Int. J. Parasitol. 2008, 38, 969–979. [Google Scholar] [CrossRef] [PubMed]
- Fedyk, S.; Chetnicki, W. Preferential Segregation of Metacentric Chromosomes in Simple Robertsonian Heterozygotes of Sorex Araneus. Heredity 2007, 99, 545–552. [Google Scholar] [CrossRef] [PubMed]
- Wyttenbach, A.; Hawser, J. The Fixation of Metacentric Chromosomes during the Chromosomal Evolution in the Common Shrew (Sorex Araneus, Insectivora). Hereditas 1996, 125, 209–217. [Google Scholar] [CrossRef]
- Szyda, J.; Simianer, H.; Lien, S. Sex Ratio Distortion in Bovine Sperm Correlates to Recombination in the Pseudoautosomal Region. Genet. Res. 2000, 75, 53–59. [Google Scholar] [CrossRef] [PubMed]
- Murphy, A.M.; Meade, K.G.; Hayes, P.A.; Park, S.D.E.; Evans, A.C.O.; Lonergan, P.; MacHugh, D.E. Transmission Ratio Distortion at the Growth Hormone Gene (GH1) in Bovine Preimplantation Embryos: An in Vitro Culture-Induced Phenomenon? Mol. Reprod. Dev. 2008, 75, 715–722. [Google Scholar] [CrossRef] [PubMed]
- Casellas, J.; Cañas-Álvarez, J.J.; González-Rodríguez, A.; Puig-Oliveras, A.; Fina, M.; Piedrafita, J.; Molina, A.; Díaz, C.; Baró, J.A.; Varona, L. Bayesian Analysis of Parent-Specific Transmission Ratio Distortion in Seven Spanish Beef Cattle Breeds. Anim. Genet. 2017, 48, 93–96. [Google Scholar] [CrossRef] [PubMed]
- Id-Lahoucine, S.; Cánovas, A.; Jaton, C.; Miglior, F.; Fonseca, P.A.S.; Sargolzaei, M.; Miller, S.; Schenkel, F.S.; Medrano, J.F.; Casellas, J. Implementation of Bayesian Methods to Identify SNP and Haplotype Regions with Transmission Ratio Distortion across the Whole Genome: TRDscan v.1.0. J. Dairy Sci. 2019, 102, 3175–3188. [Google Scholar] [CrossRef] [Green Version]
- Philipsen, M.; Kristensen, B. Preliminary Evidence of Segregation Distortion in the SLA System. Anim. Blood Groups Biochem. Genet. 1985, 16, 125–133. [Google Scholar] [CrossRef]
- Jeon, J.T.; Carlborg, O.; Törnsten, A.; Giuffra, E.; Amarger, V.; Chardon, P.; Andersson-Eklund, L.; Andersson, K.; Hansson, I.; Lundström, K.; et al. A Paternally Expressed QTL Affecting Skeletal and Cardiac Muscle Mass in Pigs Maps to the IGF2 Locus. Nat. Genet. 1999, 21, 157–158. [Google Scholar] [CrossRef] [PubMed]
- Pinton, A.; Calgaro, A.; Bonnet, N.; Ferchaud, S.; Billoux, S.; Dudez, A.M.; Mary, N.; Massip, K.; Bonnet-Garnier, A.; Yerle, M.; et al. Influence of Sex on the Meiotic Segregation of a t(13;17) Robertsonian Translocation: A Case Study in the Pig. Hum. Reprod. 2009, 24, 2034–2043. [Google Scholar] [CrossRef] [Green Version]
- Casellas, J.; Manunza, A.; Mercader, A.; Quintanilla, R.; Amills, M. A Flexible Bayesian Model for Testing for Transmission Ratio Distortion. Genetics 2014, 198, 1357–1367. [Google Scholar] [CrossRef]
- Gòdia, M.; Casellas, J.; Ruiz-Herrera, A.; Rodríguez-Gil, J.E.; Castelló, A.; Sánchez, A.; Clop, A. Whole Genome Sequencing Identifies Allelic Ratio Distortion in Sperm Involving Genes Related to Spermatogenesis in a Swine Model. DNA Res. 2020, 27, dsaa019. [Google Scholar] [CrossRef]
- Weitkamp, L.R.; MacCluer, J.W.; Guttormsen, S.A.; King, R.H. Standardbred Stallion Gene Transmission for Twelve Protein Systems: Evidence for Selection in Trotters. Anim. Genet. 1988, 19, 317–330. [Google Scholar] [CrossRef]
- Bailey, E. Segregation Distortion within the Equine MHC. Analogy to a Mouse T/t-Complex Trait. Immunogenetics 1986, 24, 225–229. [Google Scholar] [CrossRef]
- Luigi-Sierra, M.G.; Casellas, J.; Martínez, A.; Vicente Delgado, J.; Fernández Álvarez, J.; Such, F.X.; Jordana, J.; Amills, M. Markers with Low GenTrain Scores Can Generate Spurious Signals in Genome-Wide Scans for Transmission Ratio Distortion. Anim. Genet. 2021, 52, 779–781. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Pemberton, J.M. Within-Trio Tests Provide Little Support for Post-Copulatory Selection on Major Histocompatibility Complex Haplotypes in a Free-Living Population. Proc. Biol. Sci. 2021, 288, 20202862. [Google Scholar] [CrossRef]
- Rexroad, C.; Vallet, J.; Matukumalli, L.K.; Reecy, J.; Bickhart, D.; Blackburn, H.; Boggess, M.; Cheng, H.; Clutter, A.; Cockett, N.; et al. Genome to Phenome: Improving Animal Health, Production, and Well-Being—A New USDA Blueprint for Animal Genome Research 2018-2027. Front. Genet. 2019, 10, 327. [Google Scholar] [CrossRef] [PubMed]
- Cothran, E.G.; MacCluer, J.W.; Weitkamp, L.R.; Bailey, E. Genetic Differentiation Associated with Gait within American Standardbred Horses. Anim. Genet. 1987, 18, 285–296. [Google Scholar] [CrossRef]
- Awdeh, Z.L.; Raum, D.; Yunis, E.J.; Alper, C.A. Extended HLA/Complement Allele Haplotypes: Evidence for T/t-like Complex in Man. Proc. Natl. Acad. Sci. USA 1983, 80, 259–263. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Casellas, J.; Gularte, R.J.; Farber, C.R.; Varona, L.; Mehrabian, M.; Schadt, E.E.; Lusis, A.J.; Attie, A.D.; Yandell, B.S.; Medrano, J.F. Genome Scans for Transmission Ratio Distortion Regions in Mice. Genetics 2012, 191, 247–259. [Google Scholar] [CrossRef] [Green Version]
- Wójcik, J.M.; Borodin, P.M.; Fedyk, S.; Fredga, K.; Hausser, J.; Mishta, A.; Orlov, V.N. The List of the Chromosome Races of the Common Shrew Sorex Araneus (Updated 2002). Mammalia 2003, 67, 169–178. [Google Scholar] [CrossRef]
- Kolano, A.; Brunet, S.; Silk, A.D.; Cleveland, D.W.; Verlhac, M.-H. Error-Prone Mammalian Female Meiosis from Silencing the Spindle Assembly Checkpoint without Normal Interkinetochore Tension. Proc. Natl. Acad. Sci. USA 2012, 109, E1858–E1867. [Google Scholar] [CrossRef] [Green Version]
- Hunt, P.A. Women in reproductive science: Errors and Insight: Intentional and Accidental Studies of Human Chromosome Abnormalities. Reproduction 2019, 158, F91–F99. [Google Scholar] [CrossRef] [Green Version]
- Lane, S.; Kauppi, L. Meiotic Spindle Assembly Checkpoint and Aneuploidy in Males versus Females. Cell. Mol. Life Sci. 2019, 76, 1135–1150. [Google Scholar] [CrossRef]
- Boué, A.; Gallano, P. A Collaborative Study of the Segregation of Inherited Chromosome Structural Rearrangements in 1356 Prenatal Diagnoses. Prenat. Diagn. 1984, 4, 45–67. [Google Scholar] [CrossRef] [PubMed]
- Daniel, A.; Hook, E.B.; Wulf, G. Risks of Unbalanced Progeny at Amniocentesis to Carriers of Chromosome Rearrangements: Data from United States and Canadian Laboratories. Am. J. Med. Genet. 1989, 33, 14–53. [Google Scholar] [CrossRef] [PubMed]
- Scriven, P.N.; Flinter, F.A.; Braude, P.R.; Ogilvie, C.M. Robertsonian Translocations—Reproductive Risks and Indications for Preimplantation Genetic Diagnosis. Hum. Reprod. 2001, 16, 2267–2273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Munné, S.; Escudero, T.; Sandalinas, M.; Sable, D.; Cohen, J. Gamete Segregation in Female Carriers of Robertsonian Translocations. Cytogenet. Cell Genet. 2000, 90, 303–308. [Google Scholar] [CrossRef]
- Morel, F.; Roux, C.; Bresson, J.-L. FISH Analysis of the Chromosomal Status of Spermatozoa from Three Men with 45,XY,Der(13;14)(Q10;Q10) Karyotype. Mol. Hum. Reprod. 2001, 7, 483–488. [Google Scholar] [CrossRef] [Green Version]
- Daniel, A. Distortion of Female Meiotic Segregation and Reduced Male Fertility in Human Robertsonian Translocations: Consistent with the Centromere Model of Co-Evolving Centromere DNA/Centromeric Histone (CENP-A). Am. J. Med. Genet. 2002, 111, 450–452. [Google Scholar] [CrossRef]
- Snanoudj, S.; Molin, A.; Colson, C.; Coudray, N.; Paulien, S.; Mittre, H.; Gérard, M.; Schaefer, E.; Goldenberg, A.; Bacchetta, J.; et al. Maternal Transmission Ratio Distortion of GNAS Loss-of-Function Mutations. J. Bone Miner. Res. 2020, 35, 913–919. [Google Scholar] [CrossRef]
- Chen, D.; Zhang, Y.; Yi, Q.; Huang, Y.; Hou, H.; Zhang, Y.; Hao, Q.; Cooke, H.J.; Li, L.; Sun, Q.; et al. Regulation of Asymmetrical Cytokinesis by CAMP during Meiosis I in Mouse Oocytes. PLoS ONE 2012, 7, e29735. [Google Scholar] [CrossRef] [Green Version]
- Rose, R.D.; Gilchrist, R.B.; Kelly, J.M.; Thompson, J.G.; Sutton-McDowall, M.L. Regulation of Sheep Oocyte Maturation Using CAMP Modulators. Theriogenology 2013, 79, 142–148. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Teng, Z.; Li, G.; Mu, X.; Wang, Z.; Feng, L.; Niu, W.; Huang, K.; Xiang, X.; Wang, C.; et al. Cyclic AMP in Oocytes Controls Meiotic Prophase I and Primordial Folliculogenesis in the Perinatal Mouse Ovary. Development 2015, 142, 343–351. [Google Scholar] [CrossRef]
- Conti, M.; Hsieh, M.; Zamah, A.M.; Oh, J.S. Novel Signaling Mechanisms in the Ovary during Oocyte Maturation and Ovulation. Mol. Cell. Endocrinol. 2012, 356, 65–73. [Google Scholar] [CrossRef] [Green Version]
- Mantovani, G.; Ballare, E.; Giammona, E.; Beck-Peccoz, P.; Spada, A. The Gsalpha Gene: Predominant Maternal Origin of Transcription in Human Thyroid Gland and Gonads. J. Clin. Endocrinol. Metab. 2002, 87, 4736–4740. [Google Scholar] [CrossRef] [Green Version]
- Imboden, M.; Swan, H.; Denjoy, I.; Van Langen, I.M.; Latinen-Forsblom, P.J.; Napolitano, C.; Fressart, V.; Breithardt, G.; Berthet, M.; Priori, S.; et al. Female Predominance and Transmission Distortion in the Long-QT Syndrome. N. Engl. J. Med. 2006, 355, 2744–2751. [Google Scholar] [CrossRef] [Green Version]
- Itoh, H.; Berthet, M.; Fressart, V.; Denjoy, I.; Maugenre, S.; Klug, D.; Mizusawa, Y.; Makiyama, T.; Hofman, N.; Stallmeyer, B.; et al. Asymmetry of Parental Origin in Long QT Syndrome: Preferential Maternal Transmission of KCNQ1 Variants Linked to Channel Dysfunction. Eur. J. Hum. Genet. 2016, 24, 1160–1166. [Google Scholar] [CrossRef] [Green Version]
- Kunz, L.; Roggors, C.; Mayerhofer, A. Ovarian Acetylcholine and Ovarian KCNQ Channels: Insights into Cellular Regulatory Systems of Steroidogenic Granulosa Cells. Life Sci. 2007, 80, 2195–2198. [Google Scholar] [CrossRef]
- Nolin, S.L.; Glicksman, A.; Tortora, N.; Allen, E.; Macpherson, J.; Mila, M.; Vianna-Morgante, A.M.; Sherman, S.L.; Dobkin, C.; Latham, G.J.; et al. Expansions and Contractions of the FMR1 CGG Repeat in 5,508 Transmissions of Normal, Intermediate, and Premutation Alleles. Am. J. Med. Genet. A 2019, 179, 1148–1156. [Google Scholar] [CrossRef] [Green Version]
- Souza, G.N.; Kersting, N.; Krum-Santos, A.C.; Santos, A.S.P.; Furtado, G.V.; Pacheco, D.; Gonçalves, T.A.; Saute, J.A.; Schuler-Faccini, L.; Mattos, E.P.; et al. Spinocerebellar Ataxia Type 3/Machado-Joseph Disease: Segregation Patterns and Factors Influencing Instability of Expanded CAG Transmissions. Clin. Genet. 2016, 90, 134–140. [Google Scholar] [CrossRef]
- Ikeuchi, T.; Igarashi, S.; Takiyama, Y.; Onodera, O.; Oyake, M.; Takano, H.; Koide, R.; Tanaka, H.; Tsuji, S. Non-Mendelian Transmission in Dentatorubral-Pallidoluysian Atrophy and Machado-Joseph Disease: The Mutant Allele Is Preferentially Transmitted in Male Meiosis. Am. J. Hum. Genet. 1996, 58, 730–733. [Google Scholar]
- Takiyama, Y.; Sakoe, K.; Soutome, M.; Namekawa, M.; Ogawa, T.; Nakano, I.; Igarashi, S.; Oyake, M.; Tanaka, H.; Tsuji, S.; et al. Single Sperm Analysis of the CAG Repeats in the Gene for Machado-Joseph Disease (MJD1): Evidence for Non-Mendelian Transmission of the MJD1 Gene and for the Effect of the Intragenic CGG/GGG Polymorphism on the Intergenerational Instability. Hum. Mol. Genet. 1997, 6, 1063–1068. [Google Scholar] [CrossRef] [Green Version]
- Grewal, R.P.; Cancel, G.; Leeflang, E.P.; Dürr, A.; McPeek, M.S.; Draghinas, D.; Yao, X.; Stevanin, G.; Alnot, M.O.; Brice, A.; et al. French Machado-Joseph Disease Patients Do Not Exhibit Gametic Segregation Distortion: A Sperm Typing Analysis. Hum. Mol. Genet. 1999, 8, 1779–1784. [Google Scholar] [CrossRef] [Green Version]
- Rubinsztein, D.C.; Leggo, J. Non-Mendelian Transmission at the Machado-Joseph Disease Locus in Normal Females: Preferential Transmission of Alleles with Smaller CAG Repeats. J. Med. Genet. 1997, 34, 234–236. [Google Scholar] [CrossRef]
- MacMillan, J.C.; Voisey, J.; Healey, S.C.; Martin, N.G. Mendelian Segregation of Normal CAG Trinucleotide Repeat Alleles at Three Autosomal Loci. J. Med. Genet. 1999, 36, 258–259. [Google Scholar]
- Bettencourt, C.; Fialho, R.N.; Santos, C.; Montiel, R.; Bruges-Armas, J.; Maciel, P.; Lima, M. Segregation Distortion of Wild-Type Alleles at the Machado-Joseph Disease Locus: A Study in Normal Families from the Azores Islands (Portugal). J. Hum. Genet. 2008, 53, 333–339. [Google Scholar] [CrossRef] [Green Version]
- Sena, L.S.; Dos Santos Pinheiro, J.; Saraiva-Pereira, M.L.; Jardim, L.B. Selective Forces Acting on Spinocerebellar Ataxia Type 3/Machado-Joseph Disease Recurrency: A Systematic Review and Meta-Analysis. Clin. Genet. 2021, 99, 347–358. [Google Scholar] [CrossRef]
- Depienne, C.; Mandel, J.-L. 30 Years of Repeat Expansion Disorders: What Have We Learned and What Are the Remaining Challenges? Am. J. Hum. Genet. 2021, 108, 764–785. [Google Scholar] [CrossRef]
- Barbé, L.; Lanni, S.; López-Castel, A.; Franck, S.; Spits, C.; Keymolen, K.; Seneca, S.; Tomé, S.; Miron, I.; Letourneau, J.; et al. CpG Methylation, a Parent-of-Origin Effect for Maternal-Biased Transmission of Congenital Myotonic Dystrophy. Am. J. Hum. Genet. 2017, 100, 488–505. [Google Scholar] [CrossRef] [Green Version]
- Shoubridge, C.; Gardner, A.; Schwartz, C.E.; Hackett, A.; Field, M.; Gecz, J. Is There a Mendelian Transmission Ratio Distortion of the c.429_452dup(24bp) Polyalanine Tract ARX Mutation? Eur. J. Hum. Genet. 2012, 20, 1311–1314. [Google Scholar] [CrossRef] [Green Version]
- Friocourt, G.; Parnavelas, J.G. Mutations in ARX Result in Several Defects Involving GABAergic Neurons. Front. Cell. Neurosci. 2010, 4, 4. [Google Scholar] [CrossRef] [Green Version]
- Curie, A.; CHU de Lyon-Hôpital Femme-Mère-Enfant, France; des Portes, V.; CHU de Lyon-Hôpital Femme-Mère-Enfant, France. Personal communication, 2012.
- Dubos, A.; Meziane, H.; Iacono, G.; Curie, A.; Riet, F.; Martin, C.; Loaëc, N.; Birling, M.-C.; Selloum, M.; Normand, E.; et al. A New Mouse Model of ARX Dup24 Recapitulates the Patients’ Behavioral and Fine Motor Alterations. Hum. Mol. Genet. 2018, 27, 2138–2153. [Google Scholar] [CrossRef] [Green Version]
- Friocourt, G.; Kanatani, S.; Tabata, H.; Yozu, M.; Takahashi, T.; Antypa, M.; Raguénès, O.; Chelly, J.; Férec, C.; Nakajima, K.; et al. Cell-Autonomous Roles of ARX in Cell Proliferation and Neuronal Migration during Corticogenesis. J. Neurosci. 2008, 28, 5794–5805. [Google Scholar] [CrossRef] [Green Version]
- Colasante, G.; Simonet, J.C.; Calogero, R.; Crispi, S.; Sessa, A.; Cho, G.; Golden, J.A.; Broccoli, V. ARX Regulates Cortical Intermediate Progenitor Cell Expansion and Upper Layer Neuron Formation through Repression of Cdkn1c. Cereb. Cortex 2015, 25, 322–335. [Google Scholar] [CrossRef] [Green Version]
- Arora, U.P.; Dumont, B.L. Meiotic Drive in House Mice: Mechanisms, Consequences, and Insights for Human Biology. Chromosome Res. 2022, 30, 165–186. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Friocourt, G.; Perrin, A.; Saunders, P.A.; Nikalayevich, E.; Voisset, C.; Coutton, C.; Martinez, G.; Morel, F. Bypassing Mendel’s First Law: Transmission Ratio Distortion in Mammals. Int. J. Mol. Sci. 2023, 24, 1600. https://doi.org/10.3390/ijms24021600
Friocourt G, Perrin A, Saunders PA, Nikalayevich E, Voisset C, Coutton C, Martinez G, Morel F. Bypassing Mendel’s First Law: Transmission Ratio Distortion in Mammals. International Journal of Molecular Sciences. 2023; 24(2):1600. https://doi.org/10.3390/ijms24021600
Chicago/Turabian StyleFriocourt, Gaëlle, Aurore Perrin, Paul A. Saunders, Elvira Nikalayevich, Cécile Voisset, Charles Coutton, Guillaume Martinez, and Frédéric Morel. 2023. "Bypassing Mendel’s First Law: Transmission Ratio Distortion in Mammals" International Journal of Molecular Sciences 24, no. 2: 1600. https://doi.org/10.3390/ijms24021600




