Phenotype Switching and the Melanoma Microenvironment; Impact on Immunotherapy and Drug Resistance
Abstract
1. Introduction
2. Understanding Heterogeneity in the Melanoma Tumor Microenvironment
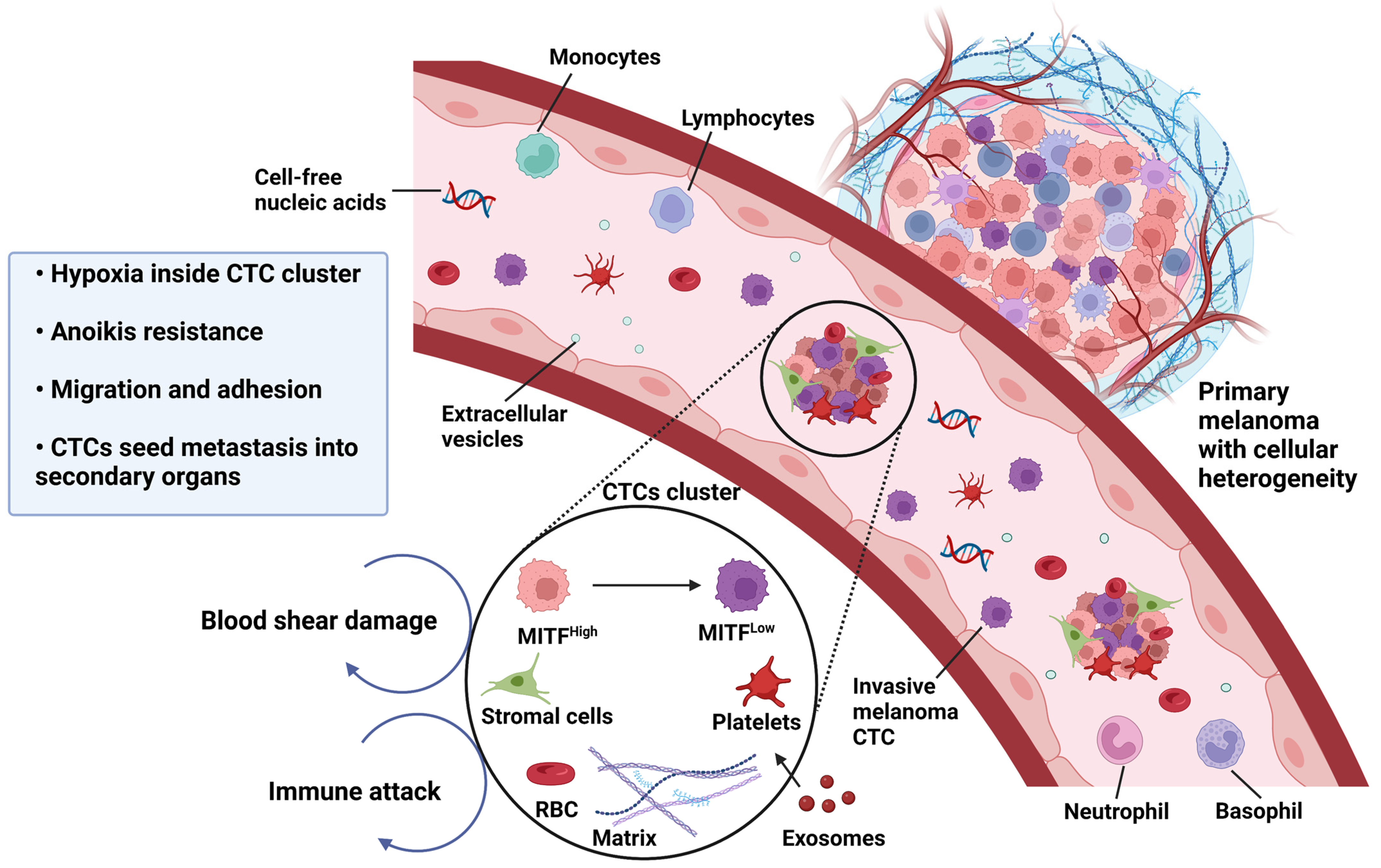
3. Melanoma Phenotypic Plasticity, Melanoma Cell Invasion and Metastasis
4. EMT-like Phenotype Switching in Melanoma
5. MITF and Phenotype Switching in Melanoma
6. The Rheostat Model of MITF
6.1. Phenotype Switching as a Driver of Metastasis
6.2. Phenotypic Plasticity as an Enabler of Melanoma Progression and Therapy Resistance
| Group | Drug Name | Target | Mechanism of Action | References |
|---|---|---|---|---|
| ICI therapy | Ipilimumab | CTLA4 | Primes anti-tumor immune response by inhibiting the cytotoxic T-lymphocyte-associated protein 4 (CTLA-4) signaling pathway. | [52] |
| Pembrolizumab/Nivolumab | PD-1 | Blocks the binding of PD-1 with its ligand PD-L1, in turn causing T cell activation and restoration of antitumor activity. | [53,54] | |
| Atezolizumab | PD-L1 | |||
| Targeted therapy | Dabrafenib/Vemurafenib/Encorafenib | BRAFv600E/K mutation | Acts on melanomas that have V600E or V600K mutations in the BRAF protein, as a potent inhibitor of BRAF through ATP competitive binding of the active conformation of BRAF kinase. | [55] |
| Trametinib/ Cobimetinib/Binimetinib | MEK | Interferes with abnormal BRAF signals to slow or stop out-of-control cell growth. | [56] | |
| Combination targeted therapy | Dabrafenib + Trametinib | BRAFV600E/K mutant, MEK | Inhibition of the BRAF-MEK pathway, and decreased tumor cell survival in BRAF-mutants. | [57,58,59] |
| Vemurafenib + Cobimetinib | ||||
| Encorafenib + Binimetinib | ||||
| Combination of ICI and targeted therapy | Pembrolizumab + Dabrafenib + Trametinib | PD1, BRAFV600E/K, MEK | Blocks programmed death 1 (PD-1) and enhances the durability of anti-tumor responses by combined inhibition of mutant BRAFV600 and MEK signaling pathways. | [60] |
| Atezolizumab + Vemurafenib + Cobimetinib | PD-L1, BRAFV600E/K, MEK | Combination BRAF plus MEK inhibitors and immune checkpoint inhibitor therapy in BRAFV600 mutation-positive advanced or metastatic melanomas. | [61] |
6.3. Inducers of EMT-like Phenotype Switching in Melanoma
7. Phenotypic Instability and the Tumor Microenvironment
8. Signaling, Dedifferentiation and Developmental Pathways Involved in Melanoma Phenotype Switching
| Pathways | Role in Melanoma Phenotype Switching |
|---|---|
| RAS/RAF/MEK/ERK pathway | Mutations in BRAF, NRAS, NF1 and KIT occur commonly during cutaneous melanoma carcinogenesis, which cause hyper-activation of the RAS/RAF/MEK/ERK pathway [103]. This pathway is responsible for regulating cancer cell proliferation, survival, and apoptosis, and is mainly controlled by phosphorylation and de-phosphorylation, involving phosphatases, GTP/GDP exchange proteins, adaptor proteins and scaffolding proteins [103,104]. For instance, the BRAFV600E mutation leads to elevated activation of MEK and ERK [105], which then facilitate changes in cell adhesion, cell cycle progression, cell migration, cell survival, differentiation, metabolism, proliferation and transcription [106,107,108,109,110]. Likewise, mutations in NRAS cause prolongation of the GTP binding, which activates RAF proteins, leading to enhanced signaling via the MAPK pathway. |
| Nuclear factor κ-B (NF-κB) signaling | Mutations in NRAS, for example, use CRAF to activate the MAPK pathway in melanoma [111,112]. Activation of ERK regulates melanoma cell survival, proliferation, resistance to apoptosis and metastasis though phosphorylation and activation of nuclear factor κ-B (NF-κB) transcription factor [113], which further induces Snail, promoting a mesenchymal phenotype in epithelial cells [114] and in melanoma [115]. |
| JAK/STAT pathway | The JAK/STAT pathway interacts with receptor tyrosine kinase (RTK) pathway signaling at multiple levels, leading to mutual activation. Further, the activation of RTK signaling causes activation of EGFR and downregulation of MAPK, which in turn phosphorylates STAT to promote JAK/STAT signaling pathway activation. In addition, RTK and PI3K pathways crosstalk with TGF-β signaling to interact with the JAK/STAT signaling cascades, which are activated upon cytokine stimulation [116]. |
| TGF-β signaling pathway | TGF-β is the most extensively studied inducer of EMT, with established roles in regulating ECM remodeling and in influencing cell phenotype [117,118]. TGF-β signals through SMAD3 to activate SNAI2/SLUG in a Rho-pathway dependent manner [119]. Enhanced TGF-β signaling is also involved in facilitating resistance to targeted inhibition of many oncogenic signaling pathways [120]. |
| WNT/β-catenin signaling pathway | The WNT/β-catenin signaling pathway is very important in embryonic development and in adult tissue homeostasis, cell migration, hematopoiesis and wound repair. In addition to the WNT/β-catenin canonical pathway, WNT glycoproteins can also activate WNT/Ca2+ pathways, involving activation of protein kinase C (PKC), and the WNT/planar polarity pathway, which involves activation of c-Jun N-terminal kinases (JNKs) [121,122]. WNT5A and protein kinase C (PKC) expression is associated with tumor aggressiveness, cell proliferation and invasiveness, and decreased differentiation. WNT5A/PKC also stimulates melanoma cell motility via induction of genes involved in the EMT program of carcinomas, including downregulation of E-cadherin [123]. |
| IFNγ | IFNγ plays an important role in reshaping the melanoma microenvironment, especially in relation to pro-survival and immune evasion effects involved in ultraviolet radiation-induced melanoma-genesis. IFN-γ pathways have a dual role in the anti-tumor immune response—producing an effective anti-tumor immune response by directing anti-proliferative and pro-apoptotic effects on tumor cells, enhancing the expression of MHC and other molecules to increase tumor neoantigen presentation, and by recruiting other immune cells [124]. Also, IFN-γ can cause immune escape by increasing the expression of PD-L1 on the surface of tumor cells [125]. IFN-γ is mainly produced by Th1 cells, and is critical for the immune response, and for sustained M1 macrophage bioactivity to eliminate neoplastic cells [126]. |
9. Balancing Melanoma Heterogeneity
10. Communication between Melanoma Cells in Different Phenotypic States
11. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ozretic, P.; Hanzic, N.; Proust, B.; Sabol, M.; Trnski, D.; Radic, M.; Musani, V.; Ciribilli, Y.; Milas, I.; Puljiz, Z.; et al. Expression profiles of p53/p73, NME and GLI families in metastatic melanoma tissue and cell lines. Sci. Rep. 2019, 9, 12470. [Google Scholar] [CrossRef] [PubMed]
- Lawrence, M.S.; Stojanov, P.; Polak, P.; Kryukov, G.V.; Cibulskis, K.; Sivachenko, A.; Carter, S.L.; Stewart, C.; Mermel, C.H.; Roberts, S.A.; et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 2013, 499, 214–218. [Google Scholar] [CrossRef]
- Arozarena, I.; Wellbrock, C. Phenotype plasticity as enabler of melanoma progression and therapy resistance. Nat. Rev. Cancer 2019, 19, 377–391. [Google Scholar] [CrossRef] [PubMed]
- Li, F.Z.; Dhillon, A.S.; Anderson, R.L.; McArthur, G.; Ferrao, P.T. Phenotype switching in melanoma: Implications for progression and therapy. Front. Oncol. 2015, 5, 31. [Google Scholar] [CrossRef] [PubMed]
- Satyamoorthy, K.; Herlyn, M. Cellular and molecular biology of human melanoma. Cancer Biol. Ther. 2002, 1, 14–17. [Google Scholar] [CrossRef] [PubMed]
- Damsky, W.E.; Theodosakis, N.; Bosenberg, M. Melanoma metastasis: New concepts and evolving paradigms. Oncogene 2014, 33, 2413–2422. [Google Scholar] [CrossRef]
- Clark, W.H., Jr.; Elder, D.E.; Guerry, D.t.; Epstein, M.N.; Greene, M.H.; Van Horn, M. A study of tumor progression: The precursor lesions of superficial spreading and nodular melanoma. Hum. Pathol. 1984, 15, 1147–1165. [Google Scholar] [CrossRef]
- Reuben, A.; Spencer, C.N.; Prieto, P.A.; Gopalakrishnan, V.; Reddy, S.M.; Miller, J.P.; Mao, X.; De Macedo, M.P.; Chen, J.; Song, X.; et al. Genomic and immune heterogeneity are associated with differential responses to therapy in melanoma. NPJ Genom. Med. 2017, 2, 10. [Google Scholar] [CrossRef]
- Prieto, P.A.; Yang, J.C.; Sherry, R.M.; Hughes, M.S.; Kammula, U.S.; White, D.E.; Levy, C.L.; Rosenberg, S.A.; Phan, G.Q. CTLA-4 blockade with ipilimumab: Long-term follow-up of 177 patients with metastatic melanoma. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2012, 18, 2039–2047. [Google Scholar] [CrossRef]
- Pedri, D.; Karras, P.; Landeloos, E.; Marine, J.C.; Rambow, F. Epithelial-to-mesenchymal-like transition events in melanoma. Febs. J. 2022, 289, 1352–1368. [Google Scholar] [CrossRef]
- Su, Y.; Bintz, M.; Yang, Y.; Robert, L.; Ng, A.H.C.; Liu, V.; Ribas, A.; Heath, J.R.; Wei, W. Phenotypic heterogeneity and evolution of melanoma cells associated with targeted therapy resistance. PLoS Comput. Biol. 2019, 15, e1007034. [Google Scholar] [CrossRef] [PubMed]
- Schadendorf, D.; Hodi, F.S.; Robert, C.; Weber, J.S.; Margolin, K.; Hamid, O.; Patt, D.; Chen, T.T.; Berman, D.M.; Wolchok, J.D. Pooled Analysis of Long-Term Survival Data From Phase II and Phase III Trials of Ipilimumab in Unresectable or Metastatic Melanoma. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2015, 33, 1889–1894. [Google Scholar] [CrossRef] [PubMed]
- Sabbatino, F.; Villani, V.; Yearley, J.H.; Deshpande, V.; Cai, L.; Konstantinidis, I.T.; Moon, C.; Nota, S.; Wang, Y.; Al-Sukaini, A.; et al. PD-L1 and HLA Class I Antigen Expression and Clinical Course of the Disease in Intrahepatic Cholangiocarcinoma. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2016, 22, 470–478. [Google Scholar] [CrossRef] [PubMed]
- Brandner, J.M.; Haass, N.K. Melanoma’s connections to the tumour microenvironment. Pathology 2013, 45, 443–452. [Google Scholar] [CrossRef] [PubMed]
- Marusyk, A.; Janiszewska, M.; Polyak, K. Intratumor Heterogeneity: The Rosetta Stone of Therapy Resistance. Cancer Cell 2020, 37, 471–484. [Google Scholar] [CrossRef]
- Haffner, M.C.; Zwart, W.; Roudier, M.P.; True, L.D.; Nelson, W.G.; Epstein, J.I.; De Marzo, A.M.; Nelson, P.S.; Yegnasubramanian, S. Genomic and phenotypic heterogeneity in prostate cancer. Nat. Rev. Urol. 2021, 18, 79–92. [Google Scholar] [CrossRef]
- Sauka-Spengler, T.; Bronner-Fraser, M. A gene regulatory network orchestrates neural crest formation. Nat. Rev. Mol. Cell Biol. 2008, 9, 557–568. [Google Scholar] [CrossRef]
- Konieczkowski, D.J.; Johannessen, C.M.; Abudayyeh, O.; Kim, J.W.; Cooper, Z.A.; Piris, A.; Frederick, D.T.; Barzily-Rokni, M.; Straussman, R.; Haq, R.; et al. A melanoma cell state distinction influences sensitivity to MAPK pathway inhibitors. Cancer Discov. 2014, 4, 816–827. [Google Scholar] [CrossRef]
- Hoek, K.S.; Eichhoff, O.M.; Schlegel, N.C.; Döbbeling, U.; Kobert, N.; Schaerer, L.; Hemmi, S.; Dummer, R. In vivo switching of human melanoma cells between proliferative and invasive states. Cancer Res. 2008, 68, 650–656. [Google Scholar] [CrossRef]
- Bailey, C.M.; Morrison, J.A.; Kulesa, P.M. Melanoma revives an embryonic migration program to promote plasticity and invasion. Pigment. Cell Melanoma Res. 2012, 25, 573–583. [Google Scholar] [CrossRef]
- Tuncer, E.; Calçada, R.R.; Zingg, D.; Varum, S.; Cheng, P.; Freiberger, S.N.; Deng, C.X.; Kleiter, I.; Levesque, M.P.; Dummer, R.; et al. SMAD signaling promotes melanoma metastasis independently of phenotype switching. J. Clin. Investig. 2019, 129, 2702–2716. [Google Scholar] [CrossRef] [PubMed]
- Hoek, K.S.; Goding, C.R. Cancer stem cells versus phenotype-switching in melanoma. Pigment. Cell Melanoma Res. 2010, 23, 746–759. [Google Scholar] [CrossRef]
- Goding, C.R.; Arnheiter, H. MITF-the first 25 years. Genes Dev. 2019, 33, 983–1007. [Google Scholar] [CrossRef] [PubMed]
- Kemper, K.; de Goeje, P.L.; Peeper, D.S.; van Amerongen, R. Phenotype switching: Tumor cell plasticity as a resistance mechanism and target for therapy. Cancer Res. 2014, 74, 5937–5941. [Google Scholar] [CrossRef]
- Thiery, J.P.; Acloque, H.; Huang, R.Y.; Nieto, M.A. Epithelial-mesenchymal transitions in development and disease. Cell 2009, 139, 871–890. [Google Scholar] [CrossRef]
- Tam, W.L.; Weinberg, R.A. The epigenetics of epithelial-mesenchymal plasticity in cancer. Nat. Med. 2013, 19, 1438–1449. [Google Scholar] [CrossRef] [PubMed]
- Baker, C.V.; Bronner-Fraser, M.; Le Douarin, N.M.; Teillet, M.A. Early- and late-migrating cranial neural crest cell populations have equivalent developmental potential in vivo. Development 1997, 124, 3077–3087. [Google Scholar] [CrossRef]
- De Craene, B.; Berx, G. Regulatory networks defining EMT during cancer initiation and progression. Nat. Rev. Cancer 2013, 13, 97–110. [Google Scholar] [CrossRef]
- Pastushenko, I.; Brisebarre, A.; Sifrim, A.; Fioramonti, M.; Revenco, T.; Boumahdi, S.; Van Keymeulen, A.; Brown, D.; Moers, V.; Lemaire, S.; et al. Identification of the tumour transition states occurring during EMT. Nature 2018, 556, 463–468. [Google Scholar] [CrossRef]
- Nieto, M.A.; Huang, R.Y.; Jackson, R.A.; Thiery, J.P. EMT: 2016. Cell 2016, 166, 21–45. [Google Scholar] [CrossRef]
- Ahn, A.; Chatterjee, A.; Eccles, M.R. The Slow Cycling Phenotype: A Growing Problem for Treatment Resistance in Melanoma. Mol. Cancer Ther. 2017, 16, 1002–1009. [Google Scholar] [CrossRef] [PubMed]
- Goding, C.R. Mitf from neural crest to melanoma: Signal transduction and transcription in the melanocyte lineage. Genes Dev. 2000, 14, 1712–1728. [Google Scholar] [CrossRef] [PubMed]
- Jeffs, A.R.; Glover, A.C.; Slobbe, L.J.; Wang, L.; He, S.; Hazlett, J.A.; Awasthi, A.; Woolley, A.G.; Marshall, E.S.; Joseph, W.R.; et al. A gene expression signature of invasive potential in metastatic melanoma cells. PLoS ONE 2009, 4, e8461. [Google Scholar] [CrossRef] [PubMed]
- Widmer, D.S.; Cheng, P.F.; Eichhoff, O.M.; Belloni, B.C.; Zipser, M.C.; Schlegel, N.C.; Javelaud, D.; Mauviel, A.; Dummer, R.; Hoek, K.S. Systematic classification of melanoma cells by phenotype-specific gene expression mapping. Pigment. Cell Melanoma Res. 2012, 25, 343–353. [Google Scholar] [CrossRef]
- Ferguson, J.; Smith, M.; Zudaire, I.; Wellbrock, C.; Arozarena, I. Glucose availability controls ATF4-mediated MITF suppression to drive melanoma cell growth. Oncotarget 2017, 8, 32946–32959. [Google Scholar] [CrossRef]
- Lee, K.E. Hypoxia as a regulator of tumor stroma and metastasis. Am. J. Physiol. Cell Physiol. 2023, 324, C10–C13. [Google Scholar] [CrossRef]
- Carreira, S.; Goodall, J.; Aksan, I.; La Rocca, S.A.; Galibert, M.D.; Denat, L.; Larue, L.; Goding, C.R. Mitf cooperates with Rb1 and activates p21Cip1 expression to regulate cell cycle progression. Nature 2005, 433, 764–769. [Google Scholar] [CrossRef]
- Carreira, S.; Goodall, J.; Denat, L.; Rodriguez, M.; Nuciforo, P.; Hoek, K.S.; Testori, A.; Larue, L.; Goding, C.R. Mitf regulation of Dia1 controls melanoma proliferation and invasiveness. Genes Dev. 2006, 20, 3426–3439. [Google Scholar] [CrossRef]
- Tsoi, J.; Robert, L.; Paraiso, K.; Galvan, C.; Sheu, K.M.; Lay, J.; Wong, D.J.L.; Atefi, M.; Shirazi, R.; Wang, X.; et al. Multi-stage Differentiation Defines Melanoma Subtypes with Differential Vulnerability to Drug-Induced Iron-Dependent Oxidative Stress. Cancer Cell 2018, 33, 890–904.e895. [Google Scholar] [CrossRef]
- Hoek, K.S.; Schlegel, N.C.; Brafford, P.; Sucker, A.; Ugurel, S.; Kumar, R.; Weber, B.L.; Nathanson, K.L.; Phillips, D.J.; Herlyn, M.; et al. Metastatic potential of melanomas defined by specific gene expression profiles with no BRAF signature. Pigment. Cell Res. 2006, 19, 290–302. [Google Scholar] [CrossRef]
- Cheli, Y.; Giuliano, S.; Botton, T.; Rocchi, S.; Hofman, V.; Hofman, P.; Bahadoran, P.; Bertolotto, C.; Ballotti, R. Mitf is the key molecular switch between mouse or human melanoma initiating cells and their differentiated progeny. Oncogene 2011, 30, 2307–2318. [Google Scholar] [CrossRef] [PubMed]
- Rambow, F.; Rogiers, A.; Marin-Bejar, O.; Aibar, S.; Femel, J.; Dewaele, M.; Karras, P.; Brown, D.; Chang, Y.H.; Debiec-Rychter, M.; et al. Toward Minimal Residual Disease-Directed Therapy in Melanoma. Cell 2018, 174, 843–855.e819. [Google Scholar] [CrossRef] [PubMed]
- Hao, Y.; Baker, D.; Ten Dijke, P. TGF-β-Mediated Epithelial-Mesenchymal Transition and Cancer Metastasis. Int. J. Mol. Sci. 2019, 20, 2767. [Google Scholar] [CrossRef] [PubMed]
- Tirosh, I.; Izar, B.; Prakadan, S.M.; Wadsworth, M.H., 2nd; Treacy, D.; Trombetta, J.J.; Rotem, A.; Rodman, C.; Lian, C.; Murphy, G.; et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science 2016, 352, 189–196. [Google Scholar] [CrossRef] [PubMed]
- Widmer, D.S.; Hoek, K.S.; Cheng, P.F.; Eichhoff, O.M.; Biedermann, T.; Raaijmakers, M.I.G.; Hemmi, S.; Dummer, R.; Levesque, M.P. Hypoxia contributes to melanoma heterogeneity by triggering HIF1α-dependent phenotype switching. J. Investig. Dermatol. 2013, 133, 2436–2443. [Google Scholar] [CrossRef]
- Chapman, A.; Fernandez del Ama, L.; Ferguson, J.; Kamarashev, J.; Wellbrock, C.; Hurlstone, A. Heterogeneous tumor subpopulations cooperate to drive invasion. Cell Rep. 2014, 8, 688–695. [Google Scholar] [CrossRef] [PubMed]
- Fallahi-Sichani, M.; Becker, V.; Izar, B.; Baker, G.J.; Lin, J.R.; Boswell, S.A.; Shah, P.; Rotem, A.; Garraway, L.A.; Sorger, P.K. Adaptive resistance of melanoma cells to RAF inhibition via reversible induction of a slowly dividing de-differentiated state. Mol. Syst. Biol. 2017, 13, 905. [Google Scholar] [CrossRef]
- Su, Y.; Wei, W.; Robert, L.; Xue, M.; Tsoi, J.; Garcia-Diaz, A.; Homet Moreno, B.; Kim, J.; Ng, R.H.; Lee, J.W.; et al. Single-cell analysis resolves the cell state transition and signaling dynamics associated with melanoma drug-induced resistance. Proc. Natl. Acad. Sci. USA 2017, 114, 13679–13684. [Google Scholar] [CrossRef]
- Shaffer, S.M.; Dunagin, M.C.; Torborg, S.R.; Torre, E.A.; Emert, B.; Krepler, C.; Beqiri, M.; Sproesser, K.; Brafford, P.A.; Xiao, M.; et al. Rare cell variability and drug-induced reprogramming as a mode of cancer drug resistance. Nature 2017, 546, 431–435. [Google Scholar] [CrossRef]
- Hossain, S.M.; Gimenez, G.; Stockwell, P.A.; Tsai, P.; Print, C.G.; Rys, J.; Cybulska-Stopa, B.; Ratajska, M.; Harazin-Lechowska, A.; Almomani, S.; et al. Innate immune checkpoint inhibitor resistance is associated with melanoma sub-types exhibiting invasive and de-differentiated gene expression signatures. Front. Immunol. 2022, 13, 955063. [Google Scholar] [CrossRef]
- Pieniazek, M.; Matkowski, R.; Donizy, P. Macrophages in skin melanoma-the key element in melanomagenesis. Oncol. Lett. 2018, 15, 5399–5404. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Yang, W.; Huang, Y.; Cui, R.; Li, X.; Li, B. Evolving Roles for Targeting CTLA-4 in Cancer Immunotherapy. Cell. Physiol. Biochem. Int. J. Exp. Cell. Physiol. Biochem. Pharmacol. 2018, 47, 721–734. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.T.; Lee, S.H.; Heo, Y.S. Molecular Interactions of Antibody Drugs Targeting PD-1, PD-L1, and CTLA-4 in Immuno-Oncology. Molecules 2019, 24, 1190. [Google Scholar] [CrossRef] [PubMed]
- Eddy, K.; Chen, S. Overcoming Immune Evasion in Melanoma. Int. J. Mol. Sci. 2020, 21, 8984. [Google Scholar] [CrossRef] [PubMed]
- Bowyer, S.; Lee, R.; Fusi, A.; Lorigan, P. Dabrafenib and its use in the treatment of metastatic melanoma. Melanoma Manag. 2015, 2, 199–208. [Google Scholar] [CrossRef]
- Hoffner, B.; Benchich, K. Trametinib: A Targeted Therapy in Metastatic Melanoma. J. Adv. Pract. Oncol. 2018, 9, 741–745. [Google Scholar]
- Subbiah, V.; Baik, C.; Kirkwood, J.M. Clinical Development of BRAF plus MEK Inhibitor Combinations. Trends Cancer 2020, 6, 797–810. [Google Scholar] [CrossRef]
- Larkin, J.; Ascierto, P.A.; Dréno, B.; Atkinson, V.; Liszkay, G.; Maio, M.; Mandalà, M.; Demidov, L.; Stroyakovskiy, D.; Thomas, L.; et al. Combined vemurafenib and cobimetinib in BRAF-mutated melanoma. N. Engl. J. Med. 2014, 371, 1867–1876. [Google Scholar] [CrossRef]
- Davis, J.; Wayman, M. Encorafenib and Binimetinib Combination Therapy in Metastatic Melanoma. J. Adv. Pract. Oncol. 2022, 13, 450–455. [Google Scholar] [CrossRef]
- Ascierto, P.A.; Ferrucci, P.F.; Fisher, R.; Del Vecchio, M.; Atkinson, V.; Schmidt, H.; Schachter, J.; Queirolo, P.; Long, G.V.; Di Giacomo, A.M.; et al. Dabrafenib, trametinib and pembrolizumab or placebo in BRAF-mutant melanoma. Nat. Med. 2019, 25, 941–946. [Google Scholar] [CrossRef]
- Gutzmer, R.; Stroyakovskiy, D.; Gogas, H.; Robert, C.; Lewis, K.; Protsenko, S.; Pereira, R.P.; Eigentler, T.; Rutkowski, P.; Demidov, L.; et al. Atezolizumab, vemurafenib, and cobimetinib as first-line treatment for unresectable advanced BRAF(V600) mutation-positive melanoma (IMspire150): Primary analysis of the randomised, double-blind, placebo-controlled, phase 3 trial. Lancet 2020, 395, 1835–1844. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Durand, S.; Dalle, S.; Caramel, J. EMT-Inducing Transcription Factors, Drivers of Melanoma Phenotype Switching, and Resistance to Treatment. Cancers 2020, 12, 2154. [Google Scholar] [CrossRef] [PubMed]
- Min, C.; Eddy, S.F.; Sherr, D.H.; Sonenshein, G.E. NF-kappaB and epithelial to mesenchymal transition of cancer. J. Cell Biochem. 2008, 104, 733–744. [Google Scholar] [CrossRef] [PubMed]
- Caramel, J.; Papadogeorgakis, E.; Hill, L.; Browne, G.J.; Richard, G.; Wierinckx, A.; Saldanha, G.; Osborne, J.; Hutchinson, P.; Tse, G.; et al. A switch in the expression of embryonic EMT-inducers drives the development of malignant melanoma. Cancer Cell 2013, 24, 466–480. [Google Scholar] [CrossRef]
- Denecker, G.; Vandamme, N.; Akay, O.; Koludrovic, D.; Taminau, J.; Lemeire, K.; Gheldof, A.; De Craene, B.; Van Gele, M.; Brochez, L.; et al. Identification of a ZEB2-MITF-ZEB1 transcriptional network that controls melanogenesis and melanoma progression. Cell Death Differ. 2014, 21, 1250–1261. [Google Scholar] [CrossRef]
- Alonso, S.R.; Tracey, L.; Ortiz, P.; Pérez-Gómez, B.; Palacios, J.; Pollán, M.; Linares, J.; Serrano, S.; Sáez-Castillo, A.I.; Sánchez, L.; et al. A high-throughput study in melanoma identifies epithelial-mesenchymal transition as a major determinant of metastasis. Cancer Res. 2007, 67, 3450–3460. [Google Scholar] [CrossRef]
- Eichhoff, O.M.; Weeraratna, A.; Zipser, M.C.; Denat, L.; Widmer, D.S.; Xu, M.; Kriegl, L.; Kirchner, T.; Larue, L.; Dummer, R.; et al. Differential LEF1 and TCF4 expression is involved in melanoma cell phenotype switching. Pigment. Cell Melanoma Res. 2011, 24, 631–642. [Google Scholar] [CrossRef]
- Larue, L.; Delmas, V. The WNT/Beta-catenin pathway in melanoma. Front. Biosci. 2006, 11, 733–742. [Google Scholar] [CrossRef]
- Mehta, A.; Kim, Y.J.; Robert, L.; Tsoi, J.; Comin-Anduix, B.; Berent-Maoz, B.; Cochran, A.J.; Economou, J.S.; Tumeh, P.C.; Puig-Saus, C.; et al. Immunotherapy Resistance by Inflammation-Induced Dedifferentiation. Cancer Discov. 2018, 8, 935–943. [Google Scholar] [CrossRef]
- Rambow, F.; Marine, J.C.; Goding, C.R. Melanoma plasticity and phenotypic diversity: Therapeutic barriers and opportunities. Genes Dev. 2019, 33, 1295–1318. [Google Scholar] [CrossRef]
- Mort, R.L.; Jackson, I.J.; Patton, E.E. The melanocyte lineage in development and disease. Development 2015, 142, 620–632. [Google Scholar] [CrossRef] [PubMed]
- Delmas, V.; Beermann, F.; Martinozzi, S.; Carreira, S.; Ackermann, J.; Kumasaka, M.; Denat, L.; Goodall, J.; Luciani, F.; Viros, A.; et al. Beta-catenin induces immortalization of melanocytes by suppressing p16INK4a expression and cooperates with N-Ras in melanoma development. Genes Dev. 2007, 21, 2923–2935. [Google Scholar] [CrossRef] [PubMed]
- Webster, M.R.; Kugel, C.H., 3rd; Weeraratna, A.T. The Wnts of change: How Wnts regulate phenotype switching in melanoma. Biochim. Et Biophys. Acta 2015, 1856, 244–251. [Google Scholar] [CrossRef] [PubMed]
- O’Connell, M.P.; Weeraratna, A.T. Hear the Wnt Ror: How melanoma cells adjust to changes in Wnt. Pigment. Cell Melanoma Res. 2009, 22, 724–739. [Google Scholar] [CrossRef]
- Dissanayake, S.K.; Olkhanud, P.B.; O’Connell, M.P.; Carter, A.; French, A.D.; Camilli, T.C.; Emeche, C.D.; Hewitt, K.J.; Rosenthal, D.T.; Leotlela, P.D.; et al. Wnt5A regulates expression of tumor-associated antigens in melanoma via changes in signal transducers and activators of transcription 3 phosphorylation. Cancer Res. 2008, 68, 10205–10214. [Google Scholar] [CrossRef]
- Orgaz, J.L.; Sanz-Moreno, V. Emerging molecular targets in melanoma invasion and metastasis. Pigment. Cell Melanoma Res. 2013, 26, 39–57. [Google Scholar] [CrossRef]
- Xu, X.Z.; Garcia, M.V.; Li, T.Y.; Khor, L.Y.; Gajapathy, R.S.; Spittle, C.; Weed, S.; Lessin, S.R.; Wu, H. Cytoskeleton alterations in melanoma: Aberrant expression of cortactin, an actin-binding adapter protein, correlates with melanocytic tumor progression. Mod. Pathol. Off. J. U. S. Can. Acad. Pathol. Inc 2010, 23, 187–196. [Google Scholar] [CrossRef] [PubMed]
- Wong, S.Y.; Hynes, R.O. Lymphatic or hematogenous dissemination: How does a metastatic tumor cell decide? Cell Cycle 2006, 5, 812–817. [Google Scholar] [CrossRef]
- Arozarena, I.; Bischof, H.; Gilby, D.; Belloni, B.; Dummer, R.; Wellbrock, C. In melanoma, beta-catenin is a suppressor of invasion. Oncogene 2011, 30, 4531–4543. [Google Scholar] [CrossRef]
- Gallagher, S.J.; Rambow, F.; Kumasaka, M.; Champeval, D.; Bellacosa, A.; Delmas, V.; Larue, L. Beta-catenin inhibits melanocyte migration but induces melanoma metastasis. Oncogene 2013, 32, 2230–2238. [Google Scholar] [CrossRef]
- Kim, I.S.; Heilmann, S.; Kansler, E.R.; Zhang, Y.; Zimmer, M.; Ratnakumar, K.; Bowman, R.L.; Simon-Vermot, T.; Fennell, M.; Garippa, R.; et al. Microenvironment-derived factors driving metastatic plasticity in melanoma. Nat. Commun. 2017, 8, 14343. [Google Scholar] [CrossRef] [PubMed]
- Papaccio, F.; Kovacs, D.; Bellei, B.; Caputo, S.; Migliano, E.; Cota, C.; Picardo, M. Profiling Cancer-Associated Fibroblasts in Melanoma. Int. J. Mol. Sci. 2021, 22, 7255. [Google Scholar] [CrossRef] [PubMed]
- Altorki, N.K.; Markowitz, G.J.; Gao, D.; Port, J.L.; Saxena, A.; Stiles, B.; McGraw, T.; Mittal, V. The lung microenvironment: An important regulator of tumour growth and metastasis. Nat. Rev. Cancer 2019, 19, 9–31. [Google Scholar] [CrossRef] [PubMed]
- Spranger, S.; Gajewski, T.F. Tumor-intrinsic oncogene pathways mediating immune avoidance. Oncoimmunology 2016, 5, e1086862. [Google Scholar] [CrossRef] [PubMed]
- Kovacs, D.; Migliano, E.; Muscardin, L.; Silipo, V.; Catricalà, C.; Picardo, M.; Bellei, B. The role of Wnt/β-catenin signaling pathway in melanoma epithelial-to-mesenchymal-like switching: Evidences from patients-derived cell lines. Oncotarget 2016, 7, 43295–43314. [Google Scholar] [CrossRef]
- Yang, L.; Li, A.; Lei, Q.; Zhang, Y. Tumor-intrinsic signaling pathways: Key roles in the regulation of the immunosuppressive tumor microenvironment. J. Hematol. Oncol. 2019, 12, 125. [Google Scholar] [CrossRef]
- Lee, H.; Jeong, A.J.; Ye, S.K. Highlighted STAT3 as a potential drug target for cancer therapy. BMB Rep. 2019, 52, 415–423. [Google Scholar] [CrossRef]
- Jung, J.E.; Kim, H.S.; Lee, C.S.; Shin, Y.J.; Kim, Y.N.; Kang, G.H.; Kim, T.Y.; Juhnn, Y.S.; Kim, S.J.; Park, J.W.; et al. STAT3 inhibits the degradation of HIF-1alpha by pVHL-mediated ubiquitination. Exp. Mol. Med. 2008, 40, 479–485. [Google Scholar] [CrossRef]
- Wang, T.; Niu, G.; Kortylewski, M.; Burdelya, L.; Shain, K.; Zhang, S.; Bhattacharya, R.; Gabrilovich, D.; Heller, R.; Coppola, D.; et al. Regulation of the innate and adaptive immune responses by Stat-3 signaling in tumor cells. Nat. Med. 2004, 10, 48–54. [Google Scholar] [CrossRef]
- Fujiwara, Y.; Takeya, M.; Komohara, Y. A novel strategy for inducing the antitumor effects of triterpenoid compounds: Blocking the protumoral functions of tumor-associated macrophages via STAT3 inhibition. BioMed. Res. Int. 2014, 2014, 348539. [Google Scholar] [CrossRef]
- Milella, M.; Falcone, I.; Conciatori, F.; Matteoni, S.; Sacconi, A.; De Luca, T.; Bazzichetto, C.; Corbo, V.; Simbolo, M.; Sperduti, I.; et al. PTEN status is a crucial determinant of the functional outcome of combined MEK and mTOR inhibition in cancer. Sci. Rep. 2017, 7, 43013. [Google Scholar] [CrossRef] [PubMed]
- Crane, C.A.; Panner, A.; Murray, J.C.; Wilson, S.P.; Xu, H.; Chen, L.; Simko, J.P.; Waldman, F.M.; Pieper, R.O.; Parsa, A.T. PI(3) kinase is associated with a mechanism of immunoresistance in breast and prostate cancer. Oncogene 2009, 28, 306–312. [Google Scholar] [CrossRef] [PubMed]
- Mittendorf, E.A.; Philips, A.V.; Meric-Bernstam, F.; Qiao, N.; Wu, Y.; Harrington, S.; Su, X.; Wang, Y.; Gonzalez-Angulo, A.M.; Akcakanat, A.; et al. PD-L1 expression in triple-negative breast cancer. Cancer Immunol. Res. 2014, 2, 361–370. [Google Scholar] [CrossRef] [PubMed]
- Lastwika, K.J.; Wilson, W., 3rd; Li, Q.K.; Norris, J.; Xu, H.; Ghazarian, S.R.; Kitagawa, H.; Kawabata, S.; Taube, J.M.; Yao, S.; et al. Control of PD-L1 Expression by Oncogenic Activation of the AKT-mTOR Pathway in Non-Small Cell Lung Cancer. Cancer Res. 2016, 76, 227–238. [Google Scholar] [CrossRef]
- Guo, W.; Wang, H.; Li, C. Signal pathways of melanoma and targeted therapy. Signal Transduct. Target. Ther. 2021, 6, 424. [Google Scholar] [CrossRef] [PubMed]
- Carr, S.; Smith, C.; Wernberg, J. Epidemiology and Risk Factors of Melanoma. Surg. Clin. N. Am. 2020, 100, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Lambert, A.W.; Pattabiraman, D.R.; Weinberg, R.A. Emerging Biological Principles of Metastasis. Cell 2017, 168, 670–691. [Google Scholar] [CrossRef] [PubMed]
- Fares, J.; Fares, M.Y.; Khachfe, H.H.; Salhab, H.A.; Fares, Y. Molecular principles of metastasis: A hallmark of cancer revisited. Signal Transduct. Target. Ther. 2020, 5, 28. [Google Scholar] [CrossRef] [PubMed]
- Ruocco, M.R.; Avagliano, A.; Granato, G.; Vigliar, E.; Masone, S.; Montagnani, S.; Arcucci, A. Metabolic flexibility in melanoma: A potential therapeutic target. Semin. Cancer Biol. 2019, 59, 187–207. [Google Scholar] [CrossRef] [PubMed]
- Wessely, A.; Steeb, T.; Berking, C.; Heppt, M.V. How Neural Crest Transcription Factors Contribute to Melanoma Heterogeneity, Cellular Plasticity, and Treatment Resistance. Int. J. Mol. Sci. 2021, 22, 5761. [Google Scholar] [CrossRef]
- Takebe, N.; Harris, P.J.; Warren, R.Q.; Ivy, S.P. Targeting cancer stem cells by inhibiting Wnt, Notch, and Hedgehog pathways. Nat. Rev. Clin. Oncol. 2011, 8, 97–106. [Google Scholar] [CrossRef] [PubMed]
- Cornell, R.A.; Eisen, J.S. Notch in the pathway: The roles of Notch signaling in neural crest development. Semin. Cell Dev. Biol. 2005, 16, 663–672. [Google Scholar] [CrossRef] [PubMed]
- McCubrey, J.A.; Steelman, L.S.; Chappell, W.H.; Abrams, S.L.; Wong, E.W.; Chang, F.; Lehmann, B.; Terrian, D.M.; Milella, M.; Tafuri, A.; et al. Roles of the Raf/MEK/ERK pathway in cell growth, malignant transformation and drug resistance. Biochim. Et Biophys. Acta 2007, 1773, 1263–1284. [Google Scholar] [CrossRef]
- Kolch, W. Meaningful relationships: The regulation of the Ras/Raf/MEK/ERK pathway by protein interactions. Biochem. J. 2000, 351, 289–305. [Google Scholar] [CrossRef] [PubMed]
- Long, G.V.; Menzies, A.M.; Nagrial, A.M.; Haydu, L.E.; Hamilton, A.L.; Mann, G.J.; Hughes, T.M.; Thompson, J.F.; Scolyer, R.A.; Kefford, R.F. Prognostic and clinicopathologic associations of oncogenic BRAF in metastatic melanoma. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2011, 29, 1239–1246. [Google Scholar] [CrossRef]
- Balan, V.; Leicht, D.T.; Zhu, J.; Balan, K.; Kaplun, A.; Singh-Gupta, V.; Qin, J.; Ruan, H.; Comb, M.J.; Tzivion, G. Identification of novel in vivo Raf-1 phosphorylation sites mediating positive feedback Raf-1 regulation by extracellular signal-regulated kinase. Mol. Biol. Cell 2006, 17, 1141–1153. [Google Scholar] [CrossRef]
- Brummer, T.; Naegele, H.; Reth, M.; Misawa, Y. Identification of novel ERK-mediated feedback phosphorylation sites at the C-terminus of B-Raf. Oncogene 2003, 22, 8823–8834. [Google Scholar] [CrossRef]
- Catalanotti, F.; Reyes, G.; Jesenberger, V.; Galabova-Kovacs, G.; de Matos Simoes, R.; Carugo, O.; Baccarini, M. A Mek1-Mek2 heterodimer determines the strength and duration of the Erk signal. Nat. Struct. Mol. Biol. 2009, 16, 294–303. [Google Scholar] [CrossRef]
- Dougherty, M.K.; Muller, J.; Ritt, D.A.; Zhou, M.; Zhou, X.Z.; Copeland, T.D.; Conrads, T.P.; Veenstra, T.D.; Lu, K.P.; Morrison, D.K. Regulation of Raf-1 by direct feedback phosphorylation. Mol. Cell 2005, 17, 215–224. [Google Scholar] [CrossRef]
- Roskoski, R., Jr. ERK1/2 MAP kinases: Structure, function, and regulation. Pharmacol. Res. 2012, 66, 105–143. [Google Scholar] [CrossRef]
- Dumaz, N. Mechanism of RAF isoform switching induced by oncogenic RAS in melanoma. Small GTPases 2011, 2, 289–292. [Google Scholar] [CrossRef] [PubMed]
- Ji, Z.; Flaherty, K.T.; Tsao, H. Molecular therapeutic approaches to melanoma. Mol. Asp. Med. 2010, 31, 194–204. [Google Scholar] [CrossRef] [PubMed]
- Balmanno, K.; Cook, S.J. Tumour cell survival signalling by the ERK1/2 pathway. Cell Death Differ. 2009, 16, 368–377. [Google Scholar] [CrossRef] [PubMed]
- Lin, K.; Baritaki, S.; Militello, L.; Malaponte, G.; Bevelacqua, Y.; Bonavida, B. The Role of B-RAF Mutations in Melanoma and the Induction of EMT via Dysregulation of the NF-κB/Snail/RKIP/PTEN Circuit. Genes Cancer 2010, 1, 409–420. [Google Scholar] [CrossRef] [PubMed]
- Kudo-Saito, C.; Shirako, H.; Takeuchi, T.; Kawakami, Y. Cancer metastasis is accelerated through immunosuppression during Snail-induced EMT of cancer cells. Cancer Cell 2009, 15, 195–206. [Google Scholar] [CrossRef]
- Rawlings, J.S.; Rosler, K.M.; Harrison, D.A. The JAK/STAT signaling pathway. J. Cell Sci. 2004, 117, 1281–1283. [Google Scholar] [CrossRef]
- Derynck, R.; Zhang, Y.E. Smad-dependent and Smad-independent pathways in TGF-beta family signalling. Nature 2003, 425, 577–584. [Google Scholar] [CrossRef]
- Zavadil, J.; Böttinger, E.P. TGF-beta and epithelial-to-mesenchymal transitions. Oncogene 2005, 24, 5764–5774. [Google Scholar] [CrossRef]
- Morita, T.; Mayanagi, T.; Sobue, K. Dual roles of myocardin-related transcription factors in epithelial mesenchymal transition via slug induction and actin remodeling. J. Cell Biol. 2007, 179, 1027–1042. [Google Scholar] [CrossRef]
- Huang, S.; Hölzel, M.; Knijnenburg, T.; Schlicker, A.; Roepman, P.; McDermott, U.; Garnett, M.; Grernrum, W.; Sun, C.; Prahallad, A.; et al. MED12 controls the response to multiple cancer drugs through regulation of TGF-β receptor signaling. Cell 2012, 151, 937–950. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Bergami, P.; Fitchman, B.; Ronai, Z. Understanding signaling cascades in melanoma. Photochem. Photobiol. 2008, 84, 289–306. [Google Scholar] [CrossRef]
- Kahn, M. Can we safely target the WNT pathway? Nat. Rev. Drug Discov. 2014, 13, 513–532. [Google Scholar] [CrossRef]
- Dissanayake, S.K.; Wade, M.; Johnson, C.E.; O’Connell, M.P.; Leotlela, P.D.; French, A.D.; Shah, K.V.; Hewitt, K.J.; Rosenthal, D.T.; Indig, F.E.; et al. The Wnt5A/protein kinase C pathway mediates motility in melanoma cells via the inhibition of metastasis suppressors and initiation of an epithelial to mesenchymal transition. J. Biol. Chem. 2007, 282, 17259–17271. [Google Scholar] [CrossRef] [PubMed]
- Platanias, L.C. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat. Rev. Immunol. 2005, 5, 375–386. [Google Scholar] [CrossRef] [PubMed]
- Carbone, D.P.; Reck, M.; Paz-Ares, L.; Creelan, B.; Horn, L.; Steins, M.; Felip, E.; van den Heuvel, M.M.; Ciuleanu, T.E.; Badin, F.; et al. First-Line Nivolumab in Stage IV or Recurrent Non-Small-Cell Lung Cancer. N. Engl. J. Med. 2017, 376, 2415–2426. [Google Scholar] [CrossRef]
- DeNardo, D.G.; Andreu, P.; Coussens, L.M. Interactions between lymphocytes and myeloid cells regulate pro- versus anti-tumor immunity. Cancer Metastasis Rev. 2010, 29, 309–316. [Google Scholar] [CrossRef]
- Galluzzi, L.; Yamazaki, T.; Kroemer, G. Linking cellular stress responses to systemic homeostasis. Nat. Rev. Mol. Cell Biol. 2018, 19, 731–745. [Google Scholar] [CrossRef]
- Yu, J.L.; Rak, J.W.; Carmeliet, P.; Nagy, A.; Kerbel, R.S.; Coomber, B.L. Heterogeneous vascular dependence of tumor cell populations. Am. J. Pathol. 2001, 158, 1325–1334. [Google Scholar] [CrossRef] [PubMed]
- Giancotti, F.G. Mechanisms governing metastatic dormancy and reactivation. Cell 2013, 155, 750–764. [Google Scholar] [CrossRef] [PubMed]
- Sosa, M.S.; Bragado, P.; Aguirre-Ghiso, J.A. Mechanisms of disseminated cancer cell dormancy: An awakening field. Nat. Rev. Cancer 2014, 14, 611–622. [Google Scholar] [CrossRef]
- Milotti, E.; Fredrich, T.; Chignola, R.; Rieger, H. Oxygen in the Tumor Microenvironment: Mathematical and Numerical Modeling. Adv. Exp. Med. Biol. 2020, 1259, 53–76. [Google Scholar] [CrossRef] [PubMed]
- Reinfeld, B.I.; Madden, M.Z.; Wolf, M.M.; Chytil, A.; Bader, J.E.; Patterson, A.R.; Sugiura, A.; Cohen, A.S.; Ali, A.; Do, B.T.; et al. Cell-programmed nutrient partitioning in the tumour microenvironment. Nature 2021, 593, 282–288. [Google Scholar] [CrossRef] [PubMed]
- Wouters, J.; Kalender-Atak, Z.; Minnoye, L.; Spanier, K.I.; De Waegeneer, M.; Bravo González-Blas, C.; Mauduit, D.; Davie, K.; Hulselmans, G.; Najem, A.; et al. Robust gene expression programs underlie recurrent cell states and phenotype switching in melanoma. Nat. Cell Biol. 2020, 22, 986–998. [Google Scholar] [CrossRef]
- Khoja, L.; Shenjere, P.; Hodgson, C.; Hodgetts, J.; Clack, G.; Hughes, A.; Lorigan, P.; Dive, C. Prevalence and heterogeneity of circulating tumour cells in metastatic cutaneous melanoma. Melanoma Res. 2014, 24, 40–46. [Google Scholar] [CrossRef] [PubMed]
- Maurus, K.; Hufnagel, A.; Geiger, F.; Graf, S.; Berking, C.; Heinemann, A.; Paschen, A.; Kneitz, S.; Stigloher, C.; Geissinger, E.; et al. The AP-1 transcription factor FOSL1 causes melanocyte reprogramming and transformation. Oncogene 2017, 36, 5110–5121. [Google Scholar] [CrossRef]
- McGranahan, N.; Swanton, C. Biological and therapeutic impact of intratumor heterogeneity in cancer evolution. Cancer Cell 2015, 27, 15–26. [Google Scholar] [CrossRef]
- Quail, D.F.; Joyce, J.A. Microenvironmental regulation of tumor progression and metastasis. Nat. Med. 2013, 19, 1423–1437. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hossain, S.M.; Eccles, M.R. Phenotype Switching and the Melanoma Microenvironment; Impact on Immunotherapy and Drug Resistance. Int. J. Mol. Sci. 2023, 24, 1601. https://doi.org/10.3390/ijms24021601
Hossain SM, Eccles MR. Phenotype Switching and the Melanoma Microenvironment; Impact on Immunotherapy and Drug Resistance. International Journal of Molecular Sciences. 2023; 24(2):1601. https://doi.org/10.3390/ijms24021601
Chicago/Turabian StyleHossain, Sultana Mehbuba, and Michael R. Eccles. 2023. "Phenotype Switching and the Melanoma Microenvironment; Impact on Immunotherapy and Drug Resistance" International Journal of Molecular Sciences 24, no. 2: 1601. https://doi.org/10.3390/ijms24021601
APA StyleHossain, S. M., & Eccles, M. R. (2023). Phenotype Switching and the Melanoma Microenvironment; Impact on Immunotherapy and Drug Resistance. International Journal of Molecular Sciences, 24(2), 1601. https://doi.org/10.3390/ijms24021601







