Two-Step Identification of N-, S-, R- and T-Cytoplasm Types in Onion Breeding Lines Using High-Resolution Melting (HRM)-Based Markers
Abstract
:1. Introduction
2. Results
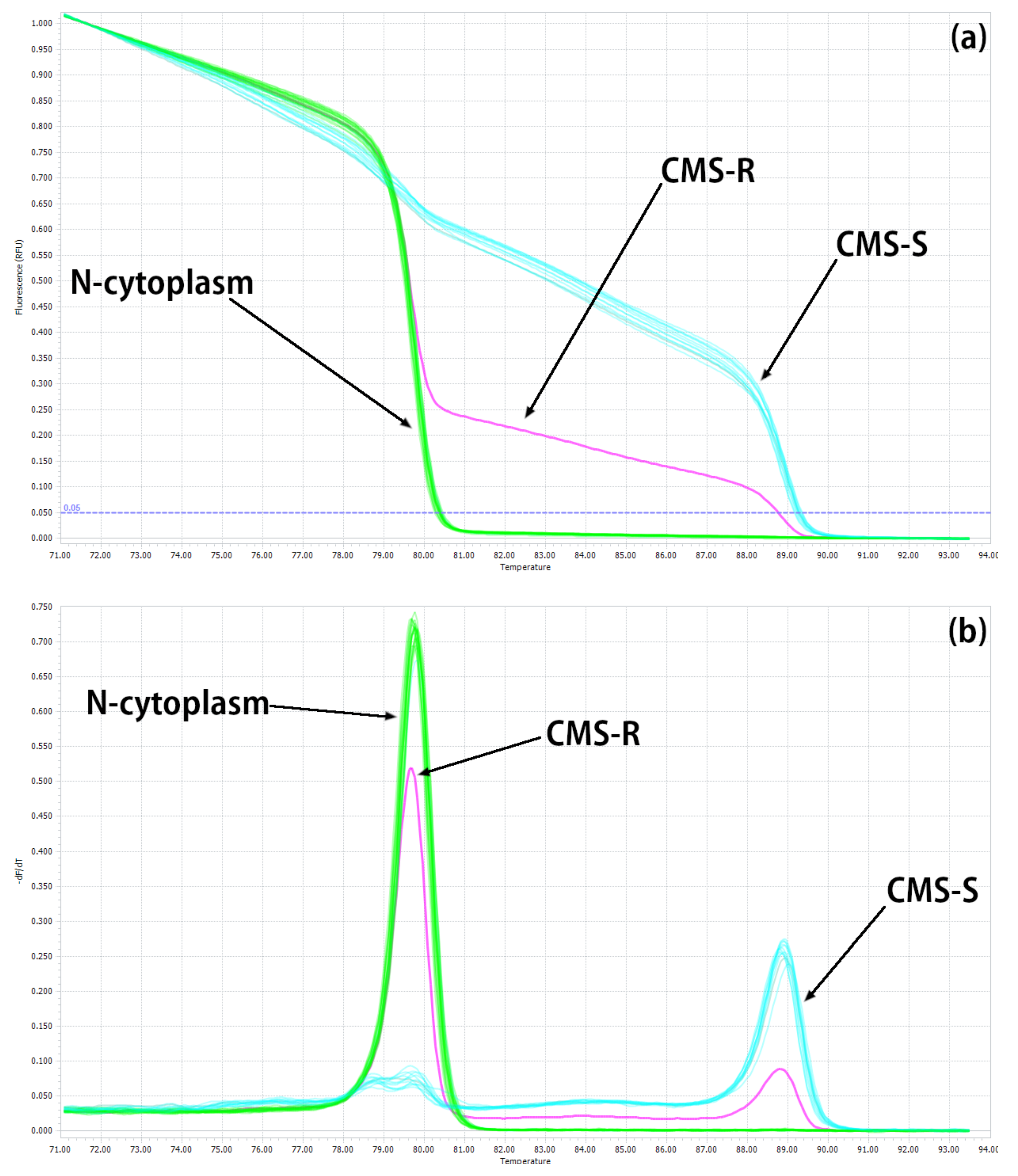
2.1. Development of HRM Markers for the Identification of N-, S- and R-Cytoplasms (First Step)
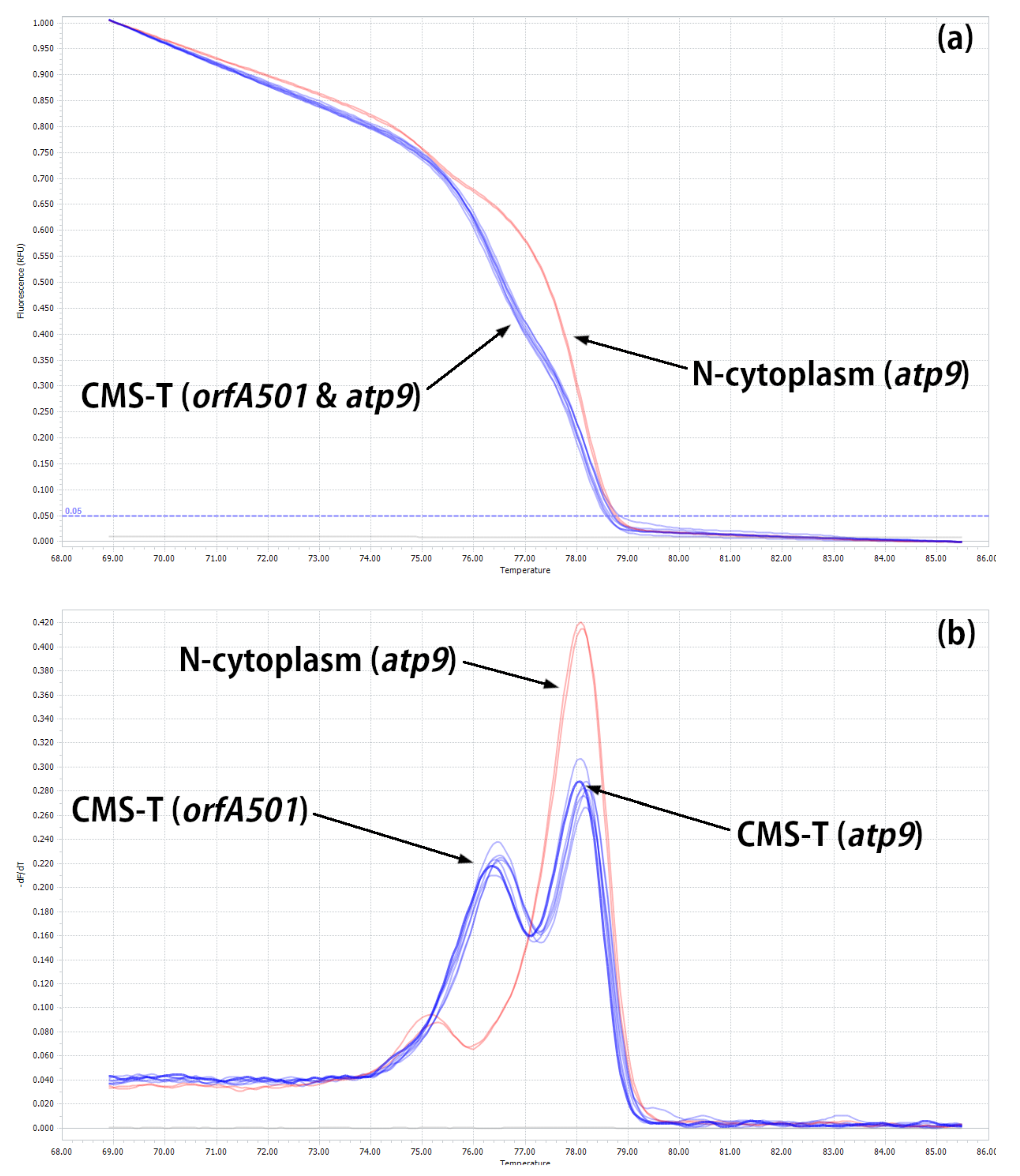
2.2. Developing HRM Markers for Distinguishing between N- and T-Cytoplasm Types (Second Steps)
2.3. Validation of the Reliability of the Two-Step HRM Marker System by Comparison with Existing PCR Marker Systems
2.4. Analysis of the Cytoplasm Types and Ms Locus in Breeding Lines Used by Russian Breeders
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Total DNA Extraction
4.3. Partial Sequencing of cox1 and orf725
4.4. AcM-HRM Analysis of Cytoplasm Type
4.5. T-HRM and atp9-HRM
4.6. Conventional PCR
4.7. HRM Analysis of the Ms Locus
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| HRM | High-resolution melting |
| CMS | Cytoplasmic Male Sterility |
References
- Jones, H. A male sterile onion. Proc. Am. Soc. Hort. Sci. 1936, 34, 582–585. [Google Scholar]
- Horn, R.; Gupta, K.J.; Colombo, N. Mitochondrion role in molecular basis of cytoplasmic male sterility. Mitochondrion 2014, 19, 198–205. [Google Scholar] [CrossRef] [PubMed]
- Luo, D.; Xu, H.; Liu, Z.; Guo, J.; Li, H.; Chen, L.; Fang, C.; Zhang, Q.; Bai, M.; Yao, N.; et al. A detrimental mitochondrial-nuclear interaction causes cytoplasmic male sterility in rice. Nat. Genet. 2013, 45, 573–577. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Liu, Y.G. Male sterility and fertility restoration in crops. Annu. Rev. Plant Biol. 2014, 65, 579–606. [Google Scholar] [CrossRef]
- Peng, X.; Li, F.; Li, S.; Zhu, Y. Expression of a mitochondrial gene orfH79 from the CMS-HongLian rice inhibits Saccharomyces cerevisiae growth and causes excessive ROS accumulation and decrease in ATP. Biotechnol. Lett. 2009, 31, 409–414. [Google Scholar] [CrossRef]
- Wang, K.; Gao, F.; Ji, Y.; Liu, Y.; Dan, Z.; Yang, P.; Zhu, Y.; Li, S. ORFH 79 impairs mitochondrial function via interaction with a subunit of electron transport chain complex III in H onglian cytoplasmic male sterile rice. New Phytol. 2013, 198, 408–418. [Google Scholar] [CrossRef]
- Balk, J.; Leaver, C.J. The PET1-CMS mitochondrial mutation in sunflower is associated with premature programmed cell death and cytochrome c release. Plant Cell 2001, 13, 1803–1818. [Google Scholar] [CrossRef] [Green Version]
- Ji, J.; Huang, W.; Yin, C.; Gong, Z. Mitochondrial cytochrome c oxidase and F1Fo-ATPase dysfunction in peppers (Capsicum annuum L.) with cytoplasmic male sterility and its association with orf507 and Ψatp6-2 genes. Int. J. Mol. Sci. 2013, 14, 1050–1068. [Google Scholar] [CrossRef] [Green Version]
- Wan, C.; Li, S.; Wen, L.; Kong, J.; Wang, K.; Zhu, Y. Damage of oxidative stress on mitochondria during microspores development in Honglian CMS line of rice. Plant Cell Rep. 2007, 26, 373–382. [Google Scholar] [CrossRef]
- Hu, J.; Huang, W.; Huang, Q.; Qin, X.; Yu, C.; Wang, L.; Li, S.; Zhu, R.; Zhu, Y. Mitochondria and cytoplasmic male sterility in plants. Mitochondrion 2014, 19, 282–288. [Google Scholar] [CrossRef]
- Gualberto, J.M.; Newton, K.J. Plant mitochondrial genomes: Dynamics and mechanisms of mutation. Annu. Rev. Plant Biol. 2017, 68, 225–252. [Google Scholar] [CrossRef] [PubMed]
- Chevigny, N.; Schatz-Daas, D.; Lotfi, F.; Gualberto, J.M. DNA repair and the stability of the plant mitochondrial genome. Int. J. Mol. Sci. 2020, 21, 328. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kozik, A.; Rowan, B.A.; Lavelle, D.; Berke, L.; Schranz, M.E.; Michelmore, R.W.; Christensen, A.C. The alternative reality of plant mitochondrial DNA: One ring does not rule them all. PLoS Genet. 2019, 15, e1008373. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hanson, M.R.; Bentolila, S. Interactions of mitochondrial and nuclear genes that affect male gametophyte development. Plant Cell 2004, 16, S154–S169. [Google Scholar] [CrossRef] [Green Version]
- Chen, Z.; Zhao, N.; Li, S.; Grover, C.E.; Nie, H.; Wendel, J.F.; Hua, J. Plant mitochondrial genome evolution and cytoplasmic male sterility. Crit. Rev. Plant Sci. 2017, 36, 55–69. [Google Scholar] [CrossRef]
- Tuteja, R.; Saxena, R.K.; Davila, J.; Shah, T.; Chen, W.; Xiao, Y.L.; Fan, G.; Saxena, K.; Alverson, A.J.; Spillane, C.; et al. Cytoplasmic male sterility-associated chimeric open reading frames identified by mitochondrial genome sequencing of four Cajanus genotypes. DNA Res. 2013, 20, 485–495. [Google Scholar] [CrossRef] [Green Version]
- Berninger, B. Contribution to the study of male sterility in the onion (Allium cepa L.). Ann. Amelior. Plant. 1965, 15, 183–199. [Google Scholar]
- Havey, M. A putative donor of S-cytoplasm and its distribution among open-pollinated populations of onion. Theor. Appl. Genet. 1993, 86, 128–134. [Google Scholar] [CrossRef]
- Havey, M.J.; Kim, S. Molecular marker characterization of commercially used cytoplasmic male sterilities in onion. J. Am. Soc. Hortic. Sci. 2021, 146, 351–355. [Google Scholar] [CrossRef]
- Jones, H.A. Inheritance of male sterility in the onion and the production of hybrid seed. Proc. Am. Soc. Hort. Sci. 1943, 43, 189–194. [Google Scholar]
- Havey, M. The cytoplasms of sterile lines used to produce commercial hybrid-onion seed. Allium Improv. Nwsl 1994, 4, 25–27. [Google Scholar]
- De Courcel, A.; Vedel, F.; Boussac, J. DNA polymorphism in Allium cepa cytoplasms and its implications concerning the origin of onions. Theor. Appl. Genet. 1989, 77, 793–798. [Google Scholar] [CrossRef] [PubMed]
- Havey, M. Identification of cytoplasms using the polymerase chain reaction to aid in the extraction of maintainer lines from open-pollinated populations of onion. Theor. Appl. Genet. 1995, 90, 263–268. [Google Scholar] [CrossRef] [PubMed]
- Sato, Y. PCR amplification of CMS-specific mitochondrial nucleotide sequences to identify cytoplasmic genotypes of onion (Allium cepa L.). Theor. Appl. Genet. 1998, 96, 367–370. [Google Scholar] [CrossRef] [PubMed]
- Engelke, T.; Terefe, D.; Tatlioglu, T. A PCR-based marker system monitoring CMS-(S), CMS-(T) and (N)-cytoplasm in the onion (Allium cepa L.). Theor. Appl. Genet. 2003, 107, 162–167. [Google Scholar] [CrossRef]
- Kim, S.; Lee, E.T.; Cho, D.Y.; Han, T.; Bang, H.; Patil, B.S.; Ahn, Y.K.; Yoon, M.K. Identification of a novel chimeric gene, orf725, and its use in development of a molecular marker for distinguishing among three cytoplasm types in onion (Allium cepa L.). Theor. Appl. Genet. 2009, 118, 433–441. [Google Scholar] [CrossRef]
- Kim, S. A codominant molecular marker in linkage disequilibrium with a restorer-of-fertility gene (Ms) and its application in reevaluation of inheritance of fertility restoration in onions. Mol. Breed. 2014, 34, 769–778. [Google Scholar] [CrossRef]
- Engelke, T.; Tatlioglu, T. A PCR-marker for the CMS1 inducing cytoplasm in chives derived from recombination events affecting the mitochondrial gene atp9. Theor. Appl. Genet. 2002, 104, 698–702. [Google Scholar] [CrossRef]
- Logan, D.C. Plant mitochondrial dynamics. Biochim. Biophys. Acta BBA Mol. Cell Res. 2006, 1763, 430–441. [Google Scholar] [CrossRef] [Green Version]
- Kim, B.; Kim, S. Identification of a variant of CMS-T cytoplasm and development of high resolution melting markers for distinguishing cytoplasm types and genotyping a restorer-of-fertility locus in onion (Allium cepa L.). Euphytica 2019, 215, 1–11. [Google Scholar] [CrossRef]
- Mackay, J.F.; Wright, C.D.; Bonfiglioli, R.G. A new approach to varietal identification in plants by microsatellite high resolution melting analysis: Application to the verification of grapevine and olive cultivars. Plant Methods 2008, 4, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Vossen, R.H.; Aten, E.; Roos, A.; den Dunnen, J.T. High-Resolution Melting Analysis (HRMA)—More than just sequence variant screening. Hum. Mutat. 2009, 30, 860–866. [Google Scholar] [CrossRef]
- Kim, S.; Kim, C.W.; Park, M.; Choi, D. Identification of candidate genes associated with fertility restoration of cytoplasmic male-sterility in onion (Allium cepa L.) using a combination of bulked segregant analysis and RNA-seq. Theor. Appl. Genet. 2015, 128, 2289–2299. [Google Scholar] [CrossRef] [PubMed]
- Melgar, S.; Havey, M.J. The dominant Ms allele in onion shows reduced penetrance. J. Am. Soc. Hortic. Sci. 2010, 135, 49–52. [Google Scholar] [CrossRef]
- Timin, N.; Pishnaya, O.; Agafonov, A.; Mamedov, M.; Titova, I.; Kan, L.; Logunova, V.; Romanov, V.; Shmykova, N.; Timina, L.; et al. Interspecific Hybridization of Vegetable Crops (Onion—Allium L., Carrot—Daucus L., Pepper—Capsicum L.); All-Russian Research Institute of Vegetable Breeding and Seed Production: Moscow, Russia, 2013; ISBN 978-5-901695-59-3. [Google Scholar]
- Schweisguth, B. Étude d’un nouveau type de stérilité male chez l’oignon, Allium cepa L. Ann. Amélior Plant 1973, 23, 221–233. [Google Scholar]
- Havey, M. Diversity among male-sterility-inducing and male-fertile cytoplasms of onion. Theor. Appl. Genet. 2000, 101, 778–782. [Google Scholar] [CrossRef]
- De Vries, J.; Wietsma, W. Allium roylei Stearn restores cytoplasmic male sterility of Rijnsburger onion (A. cepa L.). J. Genet. Breed. 1992, 46, 379–382. [Google Scholar]
- Rogers, S.O.; Bendich, A.J. Extraction of DNA from milligram amounts of fresh, herbarium and mummified plant tissues. Plant Mol. Biol. 1985, 5, 69–76. [Google Scholar] [CrossRef]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.; Clamp, M.; Barton, G.J. Jalview Version 2-a multiple sequence alignment editor and analysis workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [CrossRef]


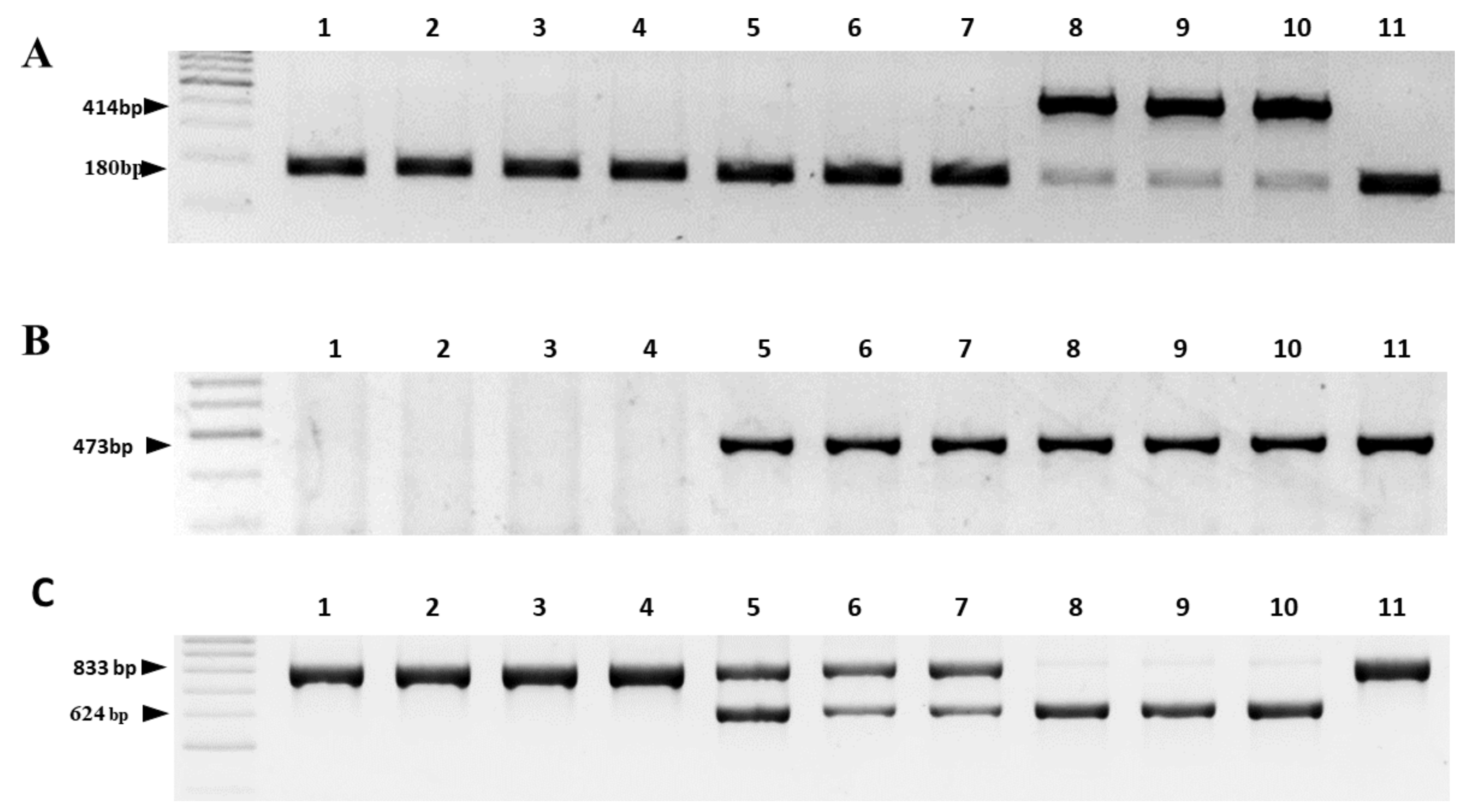
| Reference | Marker on the Gene | Cytoplasm 1/Amplicon Size, bp | |||
|---|---|---|---|---|---|
| N | S | R | T | ||
| [24] | cob | 180 | 414 | 180 | 180 |
| [25] | orfA501 | absent | 473 | 473 | 473 |
| [26] | cox1 | 833 | absent | 833 | 833 |
| orf725 | absent | 628 | 628 | absent | |
| Breeding Line | Markers | Cytoplasm Type | ||||
|---|---|---|---|---|---|---|
| cob | orfA501 | cox1 | orf725 | PCR | AcM-HRM | |
| Banko | 180 | absent | 833 | absent | N | N |
| Sibirskiy | 180 | absent | 833 | absent | N | N |
| Derek-8 | 180 | absent | 833 | absent | N | N |
| Odintsovets-28 | 180 | absent | 833 | absent | N | N |
| LFK | 180 | 473 | 833 | 628 | R | R |
| CM Banko | 180 | 473 | 833 | 628 | R | R |
| Ivashka | 180 | 473 | 833 | 628 | R | R |
| Rawhide-17 | 414 | 473 | absent | 628 | S | S |
| Sandra-276 | 414 | 473 | absent | 628 | S | S |
| Derek-3 | 414 | 473 | absent | 628 | S | S |
| Odintsovets-37 | 180 | 473 | 833 | absent | T | T |
| Cytoplasm Type | Genotype of Ms Locus | |||
|---|---|---|---|---|
| MsMs | Msms | msms | Total | |
| N | 12 | 15 | 73 | 100 |
| S | 5 | 32 | 72 | 109 |
| R | 2 | 2 | 3 | 7 |
| T | 0 | 1 | 29 | 30 |
| Total | 19 | 50 | 177 | 246 |
| Percentage | 7.7% | 20.3% | 72.0% | |
| Breeding Line | Fertility/Sterility 1 | Cytoplasm Type 2 | Ms Locus AcPMS1 |
|---|---|---|---|
| Banko | fertile | N | msms |
| Sibirskiy | fertile | N | Msms |
| Derek-8 | fertile | N | msms |
| Odintsovets-28 | fertile | N | msms |
| LFK | sterile | R | msms |
| CM Banko | sterile | R | msms |
| Ivashka | fertile | R | MsMs |
| Rawhide-17 | sterile | S | msms |
| Sandra-276 | sterile | S | msms |
| Derek-3 | sterile | S | msms |
| Odintsovets-37 | fertile | T | msms |
| Primer Names | Primer Sequence (5’-3’) | The Marker Based on Genes | Reference |
|---|---|---|---|
| ACM-HRM-F | GCTATCAAAGAGACGAAAAGCT | cox1 & orf725 | This study |
| ACM-HRM-R1 | GCTGCTTATTTATGCATTCTCCGT | ||
| ACM-HRM-R2 | AATCCTAGTGTCCGGGGTTT | ||
| MK-F | CATAGGCGGGCTCACAGGAATA | cox1 & orf725 | [26] |
| MK-R1 | AATCCTAGTGTCCGGGGTTTCT | ||
| MK-R2 | CAGCGAACTTTCATTCTTTCGC | ||
| ATGGCTCGCCTTGAAAGAGAGC | orfA501 | [25] | |
| CCAAGCATTTGGCGCTGAC | |||
| Forward primer | CTTTTCTATGGTGACAACTCCTCTT | cob | [24] |
| (S)-specific | GTCCAGTTCCTATAGAACCTATCACT | ||
| (N)-specific | TCTAGATGTCGCATCAGTGGAATCC | ||
| EX-cox1-F | TATCCAGATGCTTACGCCGG | cox1 & orf725 | This study |
| EX-cox1-R | ACTCGAACCTGCACTTCTGG | ||
| EX-orf725-F | TTACGCCGGATGGAATGCTC | ||
| EX-orf725-R | ACTGGGCGAATCACCACTTT | ||
| T-HRM-F | TTTCCGTTGCCATGGATTTT | orfA501 | This study |
| T-HRM-R | CCGAACTCCAACTCAAAAGG | ||
| atp9-F | TGGAAACGTGTTCAGTTCTTTGA | atp9 | |
| atp9-R | GCATAACCAAATGATTGTTTTGCCA | ||
| Rf-HRM7-F | CCTATTCAATCCCTGGACATTT | AcPMS1 | [30] |
| Rf-HRM7-R | GAGTTTGAAGGGCTATCTTTACTTG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khrustaleva, L.; Nzeha, M.; Ermolaev, A.; Nikitina, E.; Romanov, V. Two-Step Identification of N-, S-, R- and T-Cytoplasm Types in Onion Breeding Lines Using High-Resolution Melting (HRM)-Based Markers. Int. J. Mol. Sci. 2023, 24, 1605. https://doi.org/10.3390/ijms24021605
Khrustaleva L, Nzeha M, Ermolaev A, Nikitina E, Romanov V. Two-Step Identification of N-, S-, R- and T-Cytoplasm Types in Onion Breeding Lines Using High-Resolution Melting (HRM)-Based Markers. International Journal of Molecular Sciences. 2023; 24(2):1605. https://doi.org/10.3390/ijms24021605
Chicago/Turabian StyleKhrustaleva, Ludmila, Mais Nzeha, Aleksey Ermolaev, Ekaterina Nikitina, and Valery Romanov. 2023. "Two-Step Identification of N-, S-, R- and T-Cytoplasm Types in Onion Breeding Lines Using High-Resolution Melting (HRM)-Based Markers" International Journal of Molecular Sciences 24, no. 2: 1605. https://doi.org/10.3390/ijms24021605
APA StyleKhrustaleva, L., Nzeha, M., Ermolaev, A., Nikitina, E., & Romanov, V. (2023). Two-Step Identification of N-, S-, R- and T-Cytoplasm Types in Onion Breeding Lines Using High-Resolution Melting (HRM)-Based Markers. International Journal of Molecular Sciences, 24(2), 1605. https://doi.org/10.3390/ijms24021605






