A Novel Mutation Associated with Neonatal Lethal Cardiomyopathy Leads to an Alternative Transcript Expression in the X-Linked Complex I NDUFB11 Gene
Abstract
1. Introduction
2. Results
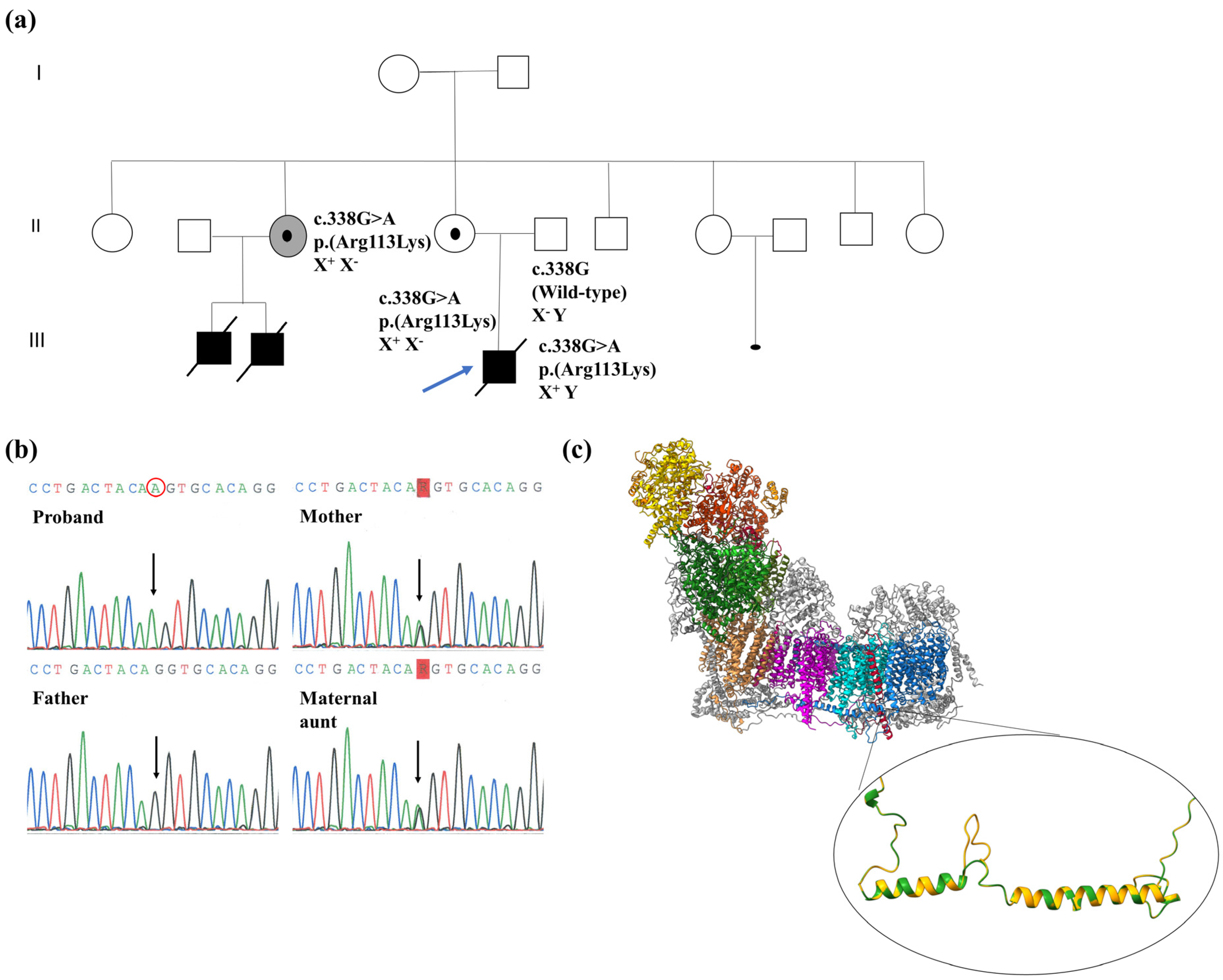
2.1. Biochemical, Genetic and Molecular Characterization of the NDUFB11:c.338G>A Variant
2.2. cDNA Analysis of NDUFB11
2.3. X Chromosome Inactivation Pattern by HUMARA Assay
2.4. Western Blot of NDUFB11 and Subunits of MRC Complexes in Proband’s Tissues
2.5. Steady-State Levels of Mitochondrial Supercomplexes and CI In-Gel Activity (CI-IGA)
3. Discussion
4. Materials and Methods
4.1. Patient and Family
4.2. Mitochondrial Respiratory Chain Analysis
4.3. Molecular Genetic Studies
4.3.1. Whole Mitochondrial Genome Next-Generation Sequencing
4.3.2. Next-Generation Sequencing (NGS)—OXPHOS Panel
4.3.3. Sanger Sequencing of the NDUFB11 Gene
4.3.4. mRNA Analysis of NDUFB11
4.3.5. Analysis of X Chromosome Inactivation (XCI)
4.4. Western Blot Analysis of NDUFB11 and Other MRC Complexes Subunits
4.5. Blue-Native PAGE for MRC and Supercomplexes and CI In-Gel Activity (CI-IGA) Assay
4.6. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fiedorczuk, K.; Sazanov, L.A. Mammalian Mitochondrial Complex I Structure and Disease-Causing Mutations. Trends Cell Biol. 2018, 28, 835–867. [Google Scholar] [CrossRef] [PubMed]
- Formosa, L.E.; Dibley, M.G.; Stroud, D.A.; Ryan, M.T. Building a Complex Complex: Assembly of Mitochondrial Respiratory Chain Complex I. Semin. Cell Dev. Biol. 2018, 76, 154–162. [Google Scholar] [CrossRef] [PubMed]
- González-Quintana, A.; García-Consuegra, I.; Belanger-Quintana, A.; Serrano-Lorenzo, P.; Lucia, A.; Blázquez, A.; Docampo, J.; Ugalde, C.; Morán, M.; Arenas, J.; et al. Novel NDUFA13 Mutations Associated with OXPHOS Deficiency and Leigh Syndrome: A Second Family Report. Genes 2020, 11, 855. [Google Scholar] [CrossRef] [PubMed]
- Fiedorczuk, K.; Letts, J.A.; Degliesposti, G.; Kaszuba, K.; Skehel, M.; Sazanov, L.A. Atomic Structure of the Entire Mammalian Mitochondrial Complex i. Nature 2016, 538, 406–410. [Google Scholar] [CrossRef]
- Guerrero-Castillo, S.; Baertling, F.; Kownatzki, D.; Wessels, H.J.; Arnold, S.; Brandt, U.; Nijtmans, L. The Assembly Pathway of Mitochondrial Respiratory Chain Complex I. Cell Metab. 2017, 25, 128–139. [Google Scholar] [CrossRef]
- Sánchez-Caballero, L.; Guerrero-Castillo, S.; Nijtmans, L. Unraveling the Complexity of Mitochondrial Complex i Assembly: A Dynamic Process. Biochim. Biophys. Acta Bioenerg. 2016, 1857, 980–990. [Google Scholar] [CrossRef]
- Fernandez-Vizarra, E.; Zeviani, M. Mitochondrial Disorders of the OXPHOS System. FEBS Lett. 2021, 595, 1062–1106. [Google Scholar] [CrossRef]
- Mimaki, M.; Wang, X.; McKenzie, M.; Thorburn, D.R.; Ryan, M.T. Understanding Mitochondrial Complex I Assembly in Health and Disease. Biochim. Biophys. Acta Bioenerg. 2012, 1817, 851–862. [Google Scholar] [CrossRef]
- Morán, M.; Rivera, H.; Sánchez-Aragó, M.; Blázquez, A.; Merinero, B.; Ugalde, C.; Arenas, J.; Cuezva, J.M.; Martín, M.A. Mitochondrial Bioenergetics and Dynamics Interplay in Complex I-Deficient Fibroblasts. Biochim. Biophys. Acta Mol. Basis Dis. 2010, 1802, 443–453. [Google Scholar] [CrossRef]
- Koene, S.; Rodenburg, R.J.; van der Knaap, M.S.; Willemsen, M.A.A.P.; Sperl, W.; Laugel, V.; Ostergaard, E.; Tarnopolsky, M.; Martin, M.A.; Nesbitt, V.; et al. Natural Disease Course and Genotype-Phenotype Correlations in Complex I Deficiency Caused by Nuclear Gene Defects: What We Learned from 130 Cases. J. Inherit. Metab. Dis. 2012, 35, 737–747. [Google Scholar] [CrossRef]
- Bugiani, M.; Invernizzi, F.; Alberio, S.; Briem, E.; Lamantea, E.; Carrara, F.; Moroni, I.; Farina, L.; Spada, M.; Donati, M.A.; et al. Clinical and Molecular Findings in Children with Complex I Deficiency. Biochim. Biophys. Acta Bioenerg. 2004, 1659, 136–147. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Moreira, D.; Ugalde, C.; Smeets, R.; Rodenburg, R.J.T.; Lopez-Laso, E.; Ruiz-Falco, M.L.; Briones, P.; Martin, M.A.; Smeitink, J.A.M.; Arenas, J. X-Linked NDUFA1 Gene Mutations Associated with Mitochondrial Encephalomyopathy. Ann. Neurol. 2007, 61, 73–83. [Google Scholar] [CrossRef] [PubMed]
- Miyauchi, A.; Osaka, H.; Nagashima, M.; Kuwajima, M.; Monden, Y.; Kohda, M.; Kishita, Y.; Okazaki, Y.; Murayama, K.; Ohtake, A.; et al. Leigh Syndrome with Spinal Cord Involvement Due to a Hemizygous NDUFA1 Mutation. Brain Dev. 2018, 40, 498–502. [Google Scholar] [CrossRef] [PubMed]
- Stenton, S.L.; Prokisch, H. Genetics of Mitochondrial Diseases: Identifying Mutations to Help Diagnosis. EBioMedicine 2020, 56, 102784. [Google Scholar] [CrossRef] [PubMed]
- Shehata, B.M.; Cundiff, C.A.; Lee, K.; Sabharwal, A.; Lalwani, M.K.; Davis, A.K.; Agrawal, V.; Sivasubbu, S.; Iannucci, G.J.; Gibson, G. Exome Sequencing of Patients with Histiocytoid Cardiomyopathy Reveals a de Novo NDUFB11 Mutation That Plays a Role in the Pathogenesis of Histiocytoid Cardiomyopathy. Am. J. Med. Genet. A 2015, 167, 2114–2121. [Google Scholar] [CrossRef]
- Rea, G.; Homfray, T.; Till, J.; Roses-Noguer, F.; Buchan, R.J.; Wilkinson, S.; Wilk, A.; Walsh, R.; John, S.; McKee, S.; et al. Histiocytoid Cardiomyopathy and Microphthalmia with Linear Skin Defects Syndrome: Phenotypes Linked by Truncating Variants in NDUFB11. Mol. Case Stud. 2017, 3, a001271. [Google Scholar] [CrossRef]
- Lichtenstein, D.A.; Crispin, A.W.; Sendamarai, A.K.; Campagna, D.R.; Schmitz-Abe, K.; Sousa, C.M.; Kafina, M.D.; Schmidt, P.J.; Niemeyer, C.M.; Porter, J.; et al. A Recurring Mutation in the Respiratory Complex 1 Protein NDUFB11 Is Responsible for a Novel Form of X-Linked Sideroblastic Anemia. Blood 2016, 128, 913–917. [Google Scholar] [CrossRef]
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hegde, M.; Lyon, E.; Spector, E.; et al. Standards and Guidelines for the Interpretation of Sequence Variants: A Joint Consensus Recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015, 17, 405–424. [Google Scholar] [CrossRef]
- Guo, R.; Zong, S.; Wu, M.; Gu, J.; Yang, M. Architecture of Human Mitochondrial Respiratory Megacomplex I2III2IV2. Cell 2017, 170, 1247–1257.e12. [Google Scholar] [CrossRef]
- Migeon, B.R. X-Linked Diseases: Susceptible Females. Genet. Med. 2020, 22, 1156–1174. [Google Scholar] [CrossRef]
- Fassone, E.; Rahman, S. Complex I Deficiency: Clinical Features, Biochemistry and Molecular Genetics. J. Med. Genet. 2012, 49, 578–590. [Google Scholar] [CrossRef] [PubMed]
- Valente, L.; Piga, D.; Lamantea, E.; Carrara, F.; Uziel, G.; Cudia, P.; Zani, A.; Farina, L.; Morandi, L.; Mora, M.; et al. Identification of Novel Mutations in Five Patients with Mitochondrial Encephalomyopathy. Biochim. Biophys. Acta Bioenerg. 2009, 1787, 491–501. [Google Scholar] [CrossRef] [PubMed]
- Panelli, D.; Lorusso, F.P.; Papa, F.; Panelli, P.; Stella, A.; Caputi, M.; Sardanelli, A.M.; Papa, S. The Mechanism of Alternative Splicing of the X-Linked NDUFB11 Gene of the Respiratory Chain Complex I, Impact of Rotenone Treatment in Neuroblastoma Cells. Biochim. Biophys. Acta Gene Regul. Mech. 2013, 1829, 211–218. [Google Scholar] [CrossRef] [PubMed]
- van Rahden, V.A.; Fernandez-Vizarra, E.; Alawi, M.; Brand, K.; Fellmann, F.; Horn, D.; Zeviani, M.; Kutsche, K. Mutations in NDUFB11, Encoding a Complex i Component of the Mitochondrial Respiratory Chain, Cause Microphthalmia with Linear Skin Defects Syndrome. Am. J. Hum. Genet. 2015, 96, 640–650. [Google Scholar] [CrossRef] [PubMed]
- Petruzzella, V.; Tessa, A.; Torraco, A.; Fattori, F.; Dotti, M.T.; Bruno, C.; Cardaioli, E.; Papa, S.; Federico, A.; Santorelli, F.M. The NDUFB11 Gene Is Not a Modifier in Leber Hereditary Optic Neuropathy. Biochem. Biophys. Res. Commun. 2007, 355, 181–187. [Google Scholar] [CrossRef]
- Barros, M.H.; McStay, G.P. Modular Biogenesis of Mitochondrial Respiratory Complexes. Mitochondrion 2020, 50, 94–114. [Google Scholar] [CrossRef]
- Anna, A.; Monika, G. Splicing Mutations in Human Genetic Disorders: Examples, Detection, and Confirmation. J. Appl. Genet. 2018, 59, 253–268. [Google Scholar] [CrossRef]
- Reinson, K.; Kovacs-Nagy, R.; Õiglane-Shlik, E.; Pajusalu, S.; Nõukas, M.; Wintjes, L.T.; van den Brandt, F.C.A.; Brink, M.; Acker, T.; Ahting, U.; et al. Diverse Phenotype in Patients with Complex I Deficiency Due to Mutations in NDUFB11. Eur. J. Med. Genet. 2019, 62, 103572. [Google Scholar] [CrossRef]
- Kohda, M.; Tokuzawa, Y.; Kishita, Y.; Nyuzuki, H.; Moriyama, Y.; Mizuno, Y.; Hirata, T.; Yatsuka, Y.; Yamashita-Sugahara, Y.; Nakachi, Y.; et al. A Comprehensive Genomic Analysis Reveals the Genetic Landscape of Mitochondrial Respiratory Chain Complex Deficiencies. PLoS Genet. 2016, 12, e1005679. [Google Scholar] [CrossRef]
- Stroud, D.A.; Surgenor, E.E.; Formosa, L.E.; Reljic, B.; Frazier, A.E.; Dibley, M.G.; Osellame, L.D.; Stait, T.; Beilharz, T.H.; Thorburn, D.R.; et al. Accessory Subunits Are Integral for Assembly and Function of Human Mitochondrial Complex i. Nature 2016, 538, 123–126. [Google Scholar] [CrossRef]
- Dang, Q.C.L.; Phan, D.H.; Johnson, A.N.; Pasapuleti, M.; Alkhaldi, H.A.; Zhang, F.; Vik, S.B. Analysis of Human Mutations in the Supernumerary Subunits of Complex i. Life 2020, 10, 296. [Google Scholar] [CrossRef] [PubMed]
- Patrat, C.; Ouimette, J.F.; Rougeulle, C. X Chromosome Inactivation in Human Development. Development 2020, 147, dev183095. [Google Scholar] [CrossRef] [PubMed]
- Bujan, N.; Morén, C.; García-García, F.J.; Blázquez, A.; Carnicer, C.; Cortés, A.B.; González, C.; López-Gallardo, E.; Lozano, E.; Moliner, S.; et al. Multicentric Standardization of Protocols for the Diagnosis of Human Mitochondrial Respiratory Chain Defects. Antioxidants 2022, 11, 741. [Google Scholar] [CrossRef] [PubMed]
- González-Quintana, A.; Trujillo-Tiebas, M.J.; Fernández-Perrone, A.L.; Blázquez, A.; Lucia, A.; Morán, M.; Ugalde, C.; Arenas, J.; Ayuso, C.; Martín, M.A. Uniparental Isodisomy as a Cause of Mitochondrial Complex I Respiratory Chain Disorder Due to a Novel Splicing NDUFS4 Mutation. Mol. Genet. Metab. 2020, 131, 341–348. [Google Scholar] [CrossRef] [PubMed]
- Weissensteiner, H.; Pacher, D.; Kloss-Brandstätter, A.; Forer, L.; Specht, G.; Bandelt, H.J.; Kronenberg, F.; Salas, A.; Schönherr, S. HaploGrep 2: Mitochondrial Haplogroup Classification in the Era of High-Throughput Sequencing. Nucleic Acids Res. 2016, 44, W58–W63. [Google Scholar] [CrossRef]
- Wang, K.; Li, M.; Hakonarson, H. ANNOVAR: Functional Annotation of Genetic Variants from High-Throughput Sequencing Data. Nucleic Acids Res. 2010, 38, e164. [Google Scholar] [CrossRef]
- Desai, R.; Frazier, A.E.; Durigon, R.; Patel, H.; Jones, A.W.; Rosa, I.D.; Lake, N.J.; Compton, A.G.; Mountford, H.S.; Tucker, E.J.; et al. ATAD3 Gene Cluster Deletions Cause Cerebellar Dysfunction Associated with Altered Mitochondrial DNA and Cholesterol Metabolism. Brain 2017, 140, 1595–1610. [Google Scholar] [CrossRef]
- Comertpay, S.; Pastorino, S.; Tanji, M.; Mezzapelle, R.; Strianese, O.; Napolitano, A.; Baumann, F.; Weigel, T.; Friedberg, J.; Sugarbaker, P.; et al. Evaluation of Clonal Origin of Malignant Mesothelioma. J. Transl. Med. 2014, 12, 301. [Google Scholar] [CrossRef]
- Kubota, T.; Nonoyama, S.; Tonoki, H.; Masuno, M.; Imaizumi, K.; Kojima, M.; Wakui, K.; Shimadzu, M.; Fukushima, Y. A New Assay for the Analysis of X-Chromosome Inactivation Based on Methylation-Specific PCR. Hum. Genet. 1999, 104, 49–55. [Google Scholar] [CrossRef]
- Torres, R.J.; Puig, J.G. Skewed X Inactivation in Lesch-Nyhan Disease Carrier Females. J. Hum. Genet. 2017, 62, 1079–1083. [Google Scholar] [CrossRef]
- Nijtmans, L.G.J.; Henderson, N.S.; Holt, I.J. Blue Native Electrophoresis to Study Mitochondrial and Other Protein Complexes. Methods 2002, 26, 327–334. [Google Scholar] [CrossRef] [PubMed]


| MRC Complex | Patient 1 | Controls 2 |
|---|---|---|
| CI (NADH-Decylubiquinone oxidoreductase) | 3.4 | 15–64 |
| CII (Succinate deshydrogenase) | 29 | 26–65 |
| CIII (Decylubiquinol-cytochrome c oxidoreductase) | 126 | 40–89 |
| CIV (Cytochrome c oxidase) | 90 | 70–228 |
| I + III (NADH cytochrome c reductase) | 3.2 | 8–72 |
| II + III (Succinate cytochrome c reductase) | 14 | 11–25 |
| CS (Citrate synthase) | 268 | 105–350 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Amate-García, G.; Ballesta-Martínez, M.J.; Serrano-Lorenzo, P.; Garrido-Moraga, R.; González-Quintana, A.; Blázquez, A.; Rubio, J.C.; García-Consuegra, I.; Arenas, J.; Ugalde, C.; et al. A Novel Mutation Associated with Neonatal Lethal Cardiomyopathy Leads to an Alternative Transcript Expression in the X-Linked Complex I NDUFB11 Gene. Int. J. Mol. Sci. 2023, 24, 1743. https://doi.org/10.3390/ijms24021743
Amate-García G, Ballesta-Martínez MJ, Serrano-Lorenzo P, Garrido-Moraga R, González-Quintana A, Blázquez A, Rubio JC, García-Consuegra I, Arenas J, Ugalde C, et al. A Novel Mutation Associated with Neonatal Lethal Cardiomyopathy Leads to an Alternative Transcript Expression in the X-Linked Complex I NDUFB11 Gene. International Journal of Molecular Sciences. 2023; 24(2):1743. https://doi.org/10.3390/ijms24021743
Chicago/Turabian StyleAmate-García, Guillermo, María Juliana Ballesta-Martínez, Pablo Serrano-Lorenzo, Rocío Garrido-Moraga, Adrián González-Quintana, Alberto Blázquez, Juan C. Rubio, Inés García-Consuegra, Joaquín Arenas, Cristina Ugalde, and et al. 2023. "A Novel Mutation Associated with Neonatal Lethal Cardiomyopathy Leads to an Alternative Transcript Expression in the X-Linked Complex I NDUFB11 Gene" International Journal of Molecular Sciences 24, no. 2: 1743. https://doi.org/10.3390/ijms24021743
APA StyleAmate-García, G., Ballesta-Martínez, M. J., Serrano-Lorenzo, P., Garrido-Moraga, R., González-Quintana, A., Blázquez, A., Rubio, J. C., García-Consuegra, I., Arenas, J., Ugalde, C., Morán, M., Guillén-Navarro, E., & Martín, M. A. (2023). A Novel Mutation Associated with Neonatal Lethal Cardiomyopathy Leads to an Alternative Transcript Expression in the X-Linked Complex I NDUFB11 Gene. International Journal of Molecular Sciences, 24(2), 1743. https://doi.org/10.3390/ijms24021743









