1. Introduction
Macroautophagy (hereafter autophagy) is an evolutionary highly conserved cellular process among eukaryotes. Autophagy is involved in the maintenance of intracellular homeostasis through the degradation of most long-lived proteins and entire organelles. When canonical autophagy is activated in yeast and mammalian cells, a membrane cisterna called phagophore expands and encloses a portion of cytoplasm, resulting in the formation of the autophagosome with a double bilayer membrane surrounding the cargo. In the following step, the outer membrane of the autophagosome fuses with the lysosome, resulting in a degradative structure, termed as autolysosome or autophagolysosome, where hydrolytic enzymes, supplied by the lysosome, degrade the cytoplasm-derived materials together with the inner membrane of the autophagosome [
1,
2]. Depending on the cellular component degraded, it results in the removal of damaged organelles, the generation of free amino acids and energy sources, the removal of intracellular pathogens, the capture and processing of self-antigens for endogenous antigen presentation specific context, and others [
2,
3,
4].
Phagophore, autophagosome, and autophagolysosome formation is finely regulated by at least 30 autophagy-related proteins (Atg). Atg genes were originally described in yeast, and their orthologs have been isolated and functionally characterized in mammals [
1,
2,
5]. Atg1 and Beclin-1 (mammalian ortholog of Atg 6) participate in the early stages of this process. Further, protein associations among Atg5, Atg12, Atg16, and the lipidation of Atg8 induce the autophagosome formation [
2].
Microtubule-associated protein light chain 3 (LC3) is the mammalian equivalent of yeast Atg8 and exists in two forms, LC3I and LC3II, being the second processed form of the first. LC3I is a 16–18 kDa polypeptide normally found in cytosol, whereas LC3II (12–14 kDa), which is conjugated with phosphatidylethanolamine (PE) by the action of ATG3/4/7 proteins, is present on membranes and autophagosomes and much less in autolysosomes [
6]. LC3II plays crucial roles in autophagy, such as the elongation and completion of the autophagosome and the transfer of cargo to these structures by interacting with other signaling proteins, including the complex p62/SQSTM1 (sequestosome 1) [
1,
2]. Although the molecular weight of LC3II is higher than that of LC3I due to the addition of PE, LC3II migrates faster than LC3I in SDS-PAGE, probably because of its extreme hydrophobicity [
7]. LC3II has been widely used to study canonical autophagy in somatic cells and gametes, and it has been considered as an autophagosomal marker in mammals and is used to monitor autophagy progression [
6,
7].
LC3 can also experiment with alternative processing forms and different regulatory mechanisms during the non-canonical forms of apoptosis [
8]. For example, in addition to the canonical C-terminal lipidation of LC3 with PE, in some situations, LC3 could be conjugated with phosphatidylserine (PS). This is typically associated with LC3-associated phagocytosis (LAP), where LC3 conjugation to phagosomes involves PS conjugation [
9].
In animal and human reproduction, autophagy-related processes seem to exert important roles [
10]. One of those is played during gametogenesis, in which canonical autophagy is involved in the regulation of cell maturation and cell renewal during spermatogenesis, acting as a partner of apoptosis and phagocytosis [
11,
12]. Autophagy markers, such as LC3II, Atg7, and autophagosomes, have been described in rat and mice spermatogenic cells [
10,
11,
13,
14] and increased considerably during critical steps of gamete maturation [
15]. In some of these studies, autophagy, measured as the ratio of LC3II/LC3I, increased significantly after 12 h of culture under metabolic stress [
13], after testes heat treatment [
11], or after electromagnetic radiation [
14], showing that autophagy is not only activated by physiological conditions in the spermatogenesis, but it is also playing a supporting role in gamete production under stressful situations.
Interestingly, a number of reports have shown a critical function for Atg7 and other related components in the process of acrosome biogenesis in different species during spermiogenesis [
16,
17,
18], supporting the autolysosome origination hypothesis for acrosome formation. Autophagy is also required for spermatozoa flagella biogenesis and cytoplasm removal during spermiogenesis [
10].
In post-fertilization events, autophagy seems to also have an important role in the zygote and embryo and is likely involved in the elimination of paternal mitochondrial and other sperm derived organelles [
19,
20,
21], although contradictory reports exist [
22].
We also dedicated some studies to investigating autophagy in ejaculated mammalian spermatozoa, showing that autophagy, mitophagy, and autophagic-like molecular mechanisms are involved in regulating critical processes of sperm function, such as motility and viability [
23], and they display a protective role in sperm during sperm cryopreservation and other biotechnological procedures [
24,
25].
In summary, autophagy components and processes have been described in spermatocytes, ejaculated spermatozoa, and the embryo after fertilization, but it has not been investigated so far whether autophagy-related processes are involved in the regulation of capacitation and the acrosome reaction in ejaculated mammalian spermatozoa. Therefore, in the present research work, we aimed: (1) to identify autophagy-related proteins in the acrosome of the stallion spermatozoa; (2) to study whether incubation in a capacitating media or the acrosome reaction can induce in these cells autophagy-like mechanisms, such as LC3 processing; and (3) to investigate the effect of autophagy inducers or inhibitors on sperm capacitation and acrosome reaction.
3. Discussion
In previous reports, we demonstrated that human and stallion ejaculated spermatozoa express a battery of proteins related to autophagy initiation and progression, such as Atg5, Atg16, Beclin-1, and LC3 [
23,
24,
25,
30]. In these reports, we showed that autophagy, mitophagy, and autophagic-like molecular mechanisms are activated ex vivo and are involved in the regulation of critical processes of sperm function, such as motility and viability [
23], and they exert a protective role in sperm during cryopreservation and other biotechnological procedures [
24,
25,
30]. These seminal reports were confirmed and extended by posterior studies in other laboratories [
31,
32,
33,
34]. Therefore, today, it seems clear that autophagy is not only essential for sperm development during spermatogenesis, but it also might play critical roles ex vivo in adult, ejaculated spermatozoa once the spermatogenesis and the epidydimal maturation has been completed (reviewed in [
10]).
In this report, we investigated additional functions and cellular processes that could be regulated by the molecular machinery associated with autophagy. Specifically, we investigated the putative participation of the autophagy component LC3 in the acrosome reaction in stallion spermatozoa. LC3 is the autophagosomal ortholog of yeast Atg8 and is present in somatic cells in two forms, LC3I and LC3II. The unprocessed form LC3I is localized in cytosol and, after the canonical autophagy activation, is conjugated to phosphatidylethanolamine to form LC3-phosphatidylethanolamine conjugate (Atg-8-PE, LC3-PE or LC3II), which is recruited to autophagosomal membranes [
35]. LC3II is considered an autophagosomal marker in mammals, and it is widely used for monitoring canonical autophagy flux in mammalian somatic cells [
1,
2,
6,
7], as well as in mammalian spermatozoa [
23,
25,
32]. LC3 is also crucial in regulating pathways leading to non-canonical functions of autophagy machinery, including LC3-associated phagocytosis and LC3-associated endocytosis [
8].
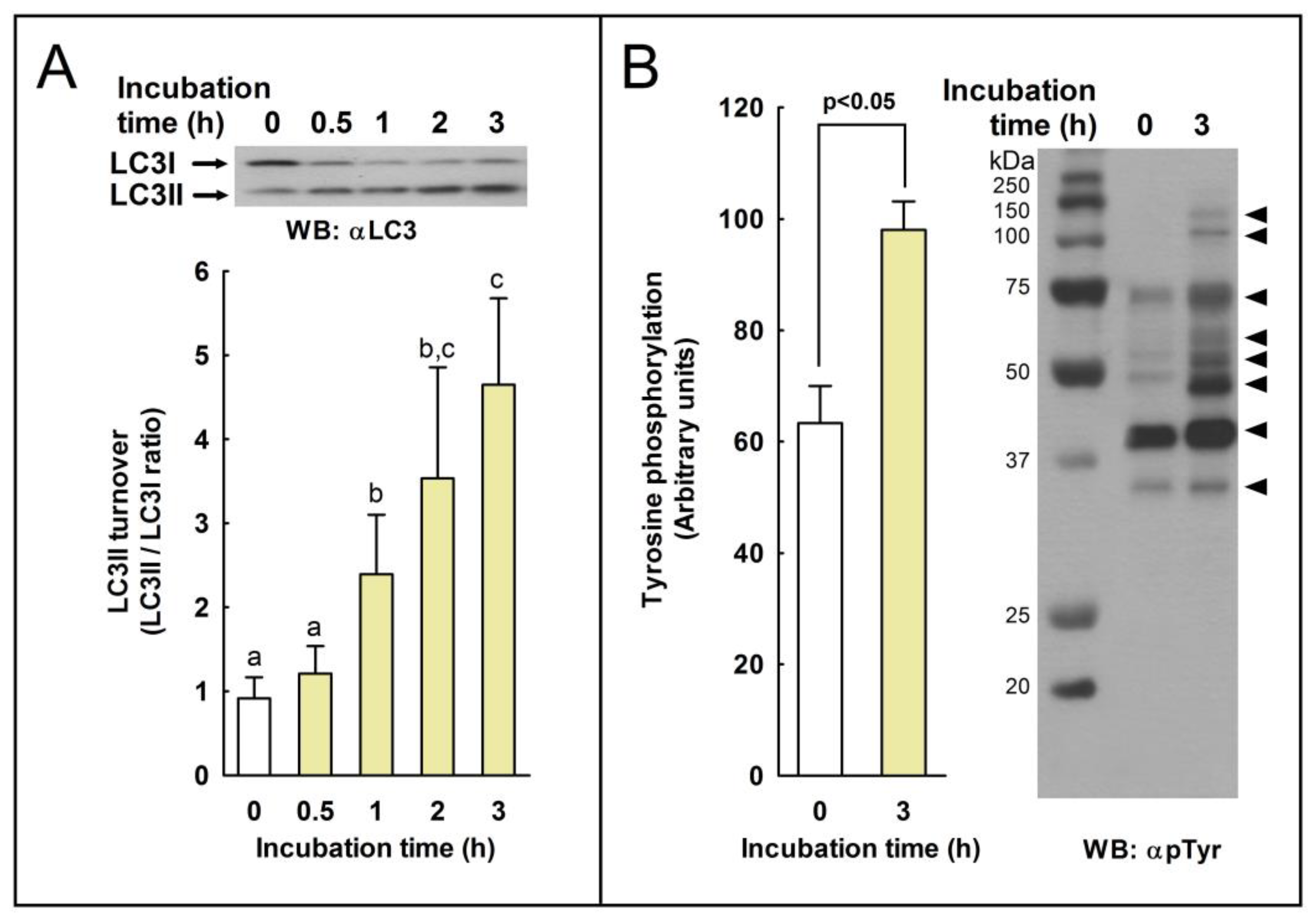
We studied whether autophagy-related processes are active or not in these cells during sperm capacitation by studying by western blotting the conversion of LC3I to LC3II (LC3II/LC3I ratio). In somatic cells, autophagy is induced mainly by the deprivation of nutrients over time by oxidative stress, by virus infection and replication, or by many other stressful situations in the cell as a survival mechanism and in response to a number of cellular stresses [
2,
11,
36]. In stallion spermatozoa, we observed a significant increase in LC3 processing only after 1 h of incubation in capacitating conditions (
Figure 1), indicating that LC3 processing might be achieved in the sperm cell ex vivo upon normal physiological situations. Capacitation is a prerequisite of the acrosome reaction [
26,
37]. Our next objective was to investigate whether the activation of the pathways leading to LC3 processing during capacitation could also be activated during the acrosome reaction or, in opposition, were independent mechanisms. To address this question, we challenged capacitated stallion spermatozoa with the calcium ionophore A23187, a well-known inductor of the acrosome reaction in these cells [
26]. We found that this treatment increased both the percentage of acrosome-reacted cells, as expected, but also caused a striking increment in the LC3II/LCI ratio compared to the untreated cells (
Figure 2).
Next, we aimed to investigate whether the pharmacological activation or inhibition of the autophagy pathways could also stimulate or avoid the acrosome reaction in the spermatozoa. To address these questions, we employed compounds widely used in somatic cells as canonical autophagy activators (STF 62247) [
29,
38] or inhibitors (E-64d, chloroquine, and 3-MA) [
29,
39]. In stallion spermatozoa, STF-62247 induced a significant increment in LC3I processing to LC3II and in the precent of acrosome-reacted cells. On the other hand, the inhibition of autophagy with E 64d also significantly increased the processing of LC3I to LC3II, whereas chloroquine increased LC3 turnover as well and also the percentage of acrosome-reacted cells. In somatic cells, chloroquine inhibited autophagy as it raised the lysosomal pH, whereas STF-62247 stimulated autophagy because it acidified the pH in the same organelle [
29]. These results, with STF-62247 and chloroquine increasing the ratio of LC3II/LC3I and the acrosome reaction, are not easily interpretable since both inhibitors display opposing mechanisms of action. However, the increment in the LC3II/LC3I ratio is possible because it accelerated the transition from LC3I to LC3II, or because LC3II is not degraded by proteolysis and accumulates [
7]. One way or another, it seems that variations in the pH of the acrosomal lumen are not enough to block or slow down LC3 processing induced by calcium ionophore or to block or reduce the percentage of cells experiencing the acrosome reaction. It seems also clear that, since both processes take place simultaneously, the processing of LC3II might be an important step in the development of the acrosome reaction. This was further clarified by the results with 3-MA, a compound that was able to inhibit the LC3 turnover concomitantly with the almost complete inhibition of the acrosome exocytosis induced by calcium ionophore. All these results likely indicate that a relationship between LC3 and the acrosome reaction exists and that this turnover (LC3 lipidation) is probably essential to trigger the acrosome exocytosis in mammalian spermatozoa.
As has been described in somatic cells, the inhibition of the proteolytic degradation of LC3II seems to be responsible for the increased LC3 ratio in the samples treated with E-64d, a membrane-permeable inhibitor of acid proteases [
29], very abundant in lysosomes in somatic cells and in acrosome in sperm cells [
40]. The accumulation of LC3II upon the inhibition of acrosomal proteases could indicate that LC3II is proteolytically degraded in the acrosome during the acrosome reaction, similarly to the mechanism described in the autophagosome in somatic cells, where LC3 is cleaved by lysosomal proteases during canonical autophagy [
1,
2].
After we defined the ability of capacitation and calcium ionophore to accelerate the processing of LC3, we investigated the subcellular distribution of LC3 and its transitions during such processes in the spermatozoa. We found that this protein has at least three different patterns of distribution along the sperm head, which change during sperm activation and might suggest that LC3 translocates to be recruited to target membranes [
35]. In mammalian spermatozoa, the translocation of proteins seems completely necessary for its interaction with other proteins and, therefore, for triggering a proper signaling cascade [
41,
42]. In fresh uncapacitated spermatozoa, where the LC3II/LC3I ratio is low, most of the sperm cells were stained in the postacrosomic region. After the incubation of spermatozoa in capacitating conditions, we detected a redistribution of LC3 proteins from the postacrosomic to the acrosomic region, indicating that LC3 is actively processed in normal ejaculated spermatozoa after a physiological challenge and that LC3 processing is not a mere reminiscence from spermatogenesis. In the next step, we saw that LC3 colocalized with PNA-FITC (
Figure 4 and
Figure 5). PNA-FITC and PSA-FITC are the most routinely used lectins to label the acrosome. PNA lectin was shown to bind Galß(1–3)Gal NAC residues located on the outer acrosomal membrane, whilst PSA was reported to recognize α-methyl mannoside residues from complex oligosaccharide structures, localized within the acrosome contents [
43]. At working concentrations, they do not bind to intact acrosomes unless cells have been previously permeabilized, giving a specific comparison between unreacted and reacted sperm [
44]. Results in our experiments, where PNA-FITC and LC3 signals clearly colocalized, indicate that, when LC3 is associated with PE, it can be recruited to the acrosomal membrane or to the apical plasmalemma membrane that is surrounding the acrosome. In somatic cells, LC3 lipidation (PE conjugation, or the transition of LC3I to LC3II) is a critical and necessary step to associate to the autophagosome membranes, and only LC3II is associated with the membranes during canonical [
1,
2,
35] as well as non-canonical autophagy [
8].
The immunoreactivity of additional components related to autophagy that were investigated in our study was mainly detected in the tail and in the head, associated or not to the acrosome (
Figure 3). In somatic cells, it was described that LC3 processing (i.e., the conversion from LC3I to LC3II) requires several Atg and related proteins, which form multimolecular complexes involved in the processing of LC3I to LC3II, which ultimately associates with biological membranes (typically with autophagosomes) [
35,
45]. In our study, Beclin-1 colocalized with LC3 in the acrosome, whereas Atg16 was mainly located in the tail and postacrosome in the head. In mature cells, LC3 was located in the middle piece but primarily can be found in the head. In most of the cells in the untreated population, LC3 was located in the postacrosomal region, where it colocalized with Atg16. Upon capacitation, LC3 was mainly detected in the acrosome, where it colocalized with Beclin-1. It is worth mentioning that, upon its association with PI3K, Beclin-1 is also associated with the membrane [
45,
46], and the final step in LC3 processing, namely PA conjugation and membrane association, is mediated, among other proteins, by Atg7, which is also related to acrosome membranes and LC3 processing in murine precursor sperm cells [
16]. Therefore, when considering that Beclin-1/PI3K complexes perform their functions in somatic cells after the Atg16 nucleated complexes, it can be easily speculated that LC3 translocation from the postacrosome region to the acrosome upon sperm activation, where more than likely LC3 is processed to LC3II and is associated with the acrosome or sperm apical membranes, is mediated by the sequential activation of Atg16- and Beclin-1-bearing complexes. Such a hypothesis involves an earlier step requiring Atg16 (postacrosome) and a subsequent step involving Beclin-1/PI3K (acrosome) and, based on the literature [
2,
16], possibly also Atg7. To completely elucidate whether those mechanisms effectively take place during LC3 processing in stallion spermatozoa obviously requires additional studies, but so far, the localization of proteins, both in basal as well in activated sperm cells, is fully compatible with the described mechanisms in somatic cells.
In conclusion, we described additional processes that could be regulated by the autophagy components in sperm cells ex vivo. The LC3 ratio rapidly increases during sperm capacitation and, more intensely, during the acrosome reaction. During capacitation, LC3 translocate in the sperm head from the proacrosomal region to the acrosome, and later, during the acrosome reaction, LC3 seems to colocalize with the apical plasmalemma or with the acrosome membrane. Given these results, it can be speculated that, in stallion sperm, a form of noncanonical autophagy utilizes some components of autophagy machinery to facilitate the acrosome reaction. In support of this hypothesis, there is the fact that the acrosome is an organelle that shows a high resemblance to lysosomes [
40], and accumulating evidence implicates LC3 and other autophagy-related proteins in cellular secretion [
47,
48]. Therefore, it can be speculated that autophagy proteins and related pathways could play a role in the fusion of the acrosome, a lysosome-derived vacuole, to the plasmalemma during the acrosome reaction, participating in a process that has been described as a secretory lysosome [
40,
49]. Considering that, in the spermatozoa, the process of canonical autophagy (the formation of an autophagolysosome) is only possible in a sizeable environment of the cytoplasmic droplet and given the origin and functional organization of the acrosome [
50], the implication of LC3 in acrosome exocytosis is perhaps one of the functions more easily attributable to these autophagy components in mammalian sperm cells. However, the participation or not of a non-canonical form of autophagy in the regulation of these processes deserves further studies.
4. Materials and Methods
4.1. Reagents
Chemical salts (NaCl, KCl, MgSO4, KPO4, sodium pyruvate, sodium lactate, CaCl2, NaHCO3, Na3VO4), HEPES, EGTA, EDTA, deoxycholate, Triton X-100, glucose, bovine serum albumin (BSA), formaldehyde, phosphate buffered saline (PBS), chloroquine diphosphate salt (Ref# C6628), sterile-filtered dimethyl sulfoxide (DMSO; Ref# D2650), E-64d (Aloxistatin) (Ref# E8640), and anti-LC3B polyclonal antibody (Ref# L7543) were purchased from Sigma-Aldrich (Madrid, Spain). 3-Methyladenine (Ref# S2767) was purchased from Selleck Chemicals (Planegg, Germany). Anti-Beclin-1 pAb (Ref# PD017Y), Anti-Atg16 pAb (Ref# PM040Y), and a positive control for anti-LC3 (Ref# PM036-PNY) were purchased from MBL International (Woburn, MA, USA). EthD-1 and the anti-rabbit IgG antibody labeled with Alexa 546 were obtained from Molecular Probes (Thermo Fisher Scientific, Madrid, Spain). Anti-rabbit IgG horseradish peroxidase (HRP)-conjugated secondary antibody and enhanced chemiluminescence detection reagents were from Pierce Protein Biology (Thermo Fisher Scientific, Madrid, Spain). A complete, EDTA-free, protease inhibitor cocktail was from Roche Diagnostics (Barcelona, Spain). Bradford reagent, Tris/Glycine/SDS buffer (10×), and Tris/Glycine buffer (10×) were from Bio-Rad (Madrid, Spain). STF-62247 (Ref# 189497) was from Merck Millipore (Darmstadt, Germany).
4.2. Collection of Equine Semen
Semen was obtained from a total of 14 Andalusian horses aged between 4 and 12 years that were housed in individual stables at the Veterinary Teaching Hospital of the University of Extremadura, Caceres, Spain. Stallions were maintained according to institutional and European regulations. All animals were healthy sperm donors of proven fertility and were collected on a regular basis (three collections per week) during the breeding season for AI. Ejaculates were collected using a Missouri model artificial vagina with an inline filter to eliminate the gel fraction, and it was lubricated and warmed to 45–50 °C. Semen was immediately evaluated and processed.
4.3. Incubation Media and Treatments
To perform the incubation of semen samples, for each stallion’s ejaculate, aliquots were centrifuged at 800× g for 10 min at RT. Seminal plasma was removed and the sperm pellet resuspended to 100 × 106 spermatozoa mL-1 in modified Biggers, Whitten, and Whittingham (BWW) media consisting of: 91.6 mM NaCl, 4.6 mM KCl, 2.44 mM MgSO4, 1.2 mM KPO4, 20 mM HEPES, 5.6 mM glucose (anhydrous), 0.27 mM sodium pyruvate, 44 mM sodium lactate, 1.7 mM CaCl2, 25 mM, NaHCO3, and 7 mg/mL BSA. The pH of the solution was adjusted to 7.4, and osmotic concentration was kept at 320 mOsm/kg.
4.4. Autophagy Regulation
Samples were incubated 3 h at 37 °C in the presence of autophagy modulators, namely STF-62247 (50 µM), chloroquine (50 µM), and E-64d (10 µg/mL; 29 µM) or for 4 h at 37 °C with 3-MA (5 mM). STF-62247 was described as a selective autophagy activator, whereas chloroquine, 3-MA, and E-64d inhibit autophagy [
29]. Chloroquine raises the lysosomal pH, which leads to the inhibition of both fusion of autophagosome with lysosomes and lysosomal protein degradation; 3-MA inhibits class III Pi3K, impairing the signaling leading to LC3 processing; and E-64d is a membrane-permeable and irreversible inhibitor of cathepsins [
29]. The vehicle for STF and E-64d was DMSO, which was systematically added to the controlled sperm population at the maximal concentration used (0.004%;
v/
v). Chloroquine is water-soluble; therefore, the control population was incubated in the absence of vehicle; 3-MA is also water soluble at the working concentration and was dissolved directly in the incubating medium (BWW), prewarmed at 45 °C to facilitate its solubilization. The 3-MA-containing medium was cooled down to 37 °C before use.
4.5. Western Blotting
To separate the proteins according to their apparent molecular masses, SDS-PAGE was performed as previously described [
51] with minor modifications. Samples (1 mL) were washed with Phosphate-buffered saline (PBS), resuspended in lysis buffer (50 mM Trizma base, 150 mM NaCl, 1% Triton X-100, 1% deoxycholate, 1 mM EGTA, 0.4 mM EDTA), supplemented with a protease inhibitor cocktail and phosphatase inhibitor (0.2 mM Na3VO4), and sonicated for 5 s at 4 °C. The homogenate was centrifuged for 15 min at 10,000×
g at 4 °C, and the protein concentration was estimated in the supernatant, containing soluble proteins in non-ionic and ionic detergents. Proteins were mixed with loading buffer supplemented with 5% mercaptoethanol and further denatured by heating for 10 min at 70 °C. Ten to fifteen micrograms of protein extract from mammalian spermatozoa or positive controls were loaded and resolved by SDS PAGE on a 12% polyacrylamide gel. The proteins were then transferred to a nitrocellulose membrane, which was subsequently blocked with blocking buffer (5% non-fat dry milk) in a Tris buffered saline with Tween-20 (TBST) (10 mM Trizma base, 100 mM NaCl, and 0.05% Tween 20) for 1 h at RT. Immunoblotting was performed by incubating the membranes in blocking buffer overnight at 4 °C with the primary antibodies against LC3 and tyrosine-phosphorylated proteins. Membranes were then washed in TBST and incubated with goat anti-rabbit horseradish peroxidase-conjugated secondary antibody for 45 min at RT. Following 3 washes with TBST for 10 min each, the signal was visualized using a SuperSignal West Pico Chemiluminescent Substrate Kit according to the manufacturer’s instructions. Band intensity was quantified using the software Scion Image for Windows, version 4.02 (Scion Corp., Frederick, MD, USA).
4.6. Assessment of Acrosome-Reacted Spermatozoa and Cell Viability
One mL of sperm suspension (5 × 10
6 spermatozoa mL
−1) was loaded with Hoechst 33342 (0.5 μM). After thorough mixing, the sperm suspension was incubated at RT in the dark for 25 min. The suspension was then washed and resuspended with PBS and stained with PNA-FITC (25 µM). After thorough mixing, the sperm suspension (1 mL) was incubated at 37 °C in the dark for 15 min. Finally, ethidium homodimer-1 (EthD-1) (1.167 mM) was added to the sperm suspension during the last 5 min of incubation. During the analysis, this staining allowed us to exclude debris and non-sperm events (Hoechst, see below) and distinguished four sperm subpopulations [
27]. The subpopulation of unstained spermatozoa was considered alive with intact acrosomes (unreacted cells). Biological membranes in these cells are impermeable to PNA and EthD-1; therefore, these cells remained unlabeled. The acrosome membrane was only available in acrosome-reacted cells, whose plasmalemma fused with the external acrosome membrane, allowing the PNA to enter to the acrosome. Consequently, the PNA-FITC positive subpopulation, emitting green fluorescence, corresponded to the acrosome-reacted spermatozoa. Finally, two additional subpopulations were easily detected, corresponding to spermatozoa stained with both EthD-1 and PNA-FITC (emitting green and red fluorescence) and spermatozoa stained with EthD-1 (emitting only red fluorescence). Both subpopulations were dead, unviable spermatozoa. Flow cytometric analyses were performed using an MACSQuant Analyser 10 (Miltenyi Biotech) flow cytometer, equipped with 3 lasers emitting at 405, 488, and 635 nm. The system was controlled using MACSQuantify Software. Sperm subpopulations were divided by quadrants to quantify the frequency of each subpopulation. Forward and sideways light scatter were recorded for a total of 30,000 events per sample. Non-sperm events were eliminated by gating the sperm population after Hoechst 33342 staining. All positive events for Hoechst 33342 within a certain range of the side scatter (SCC) distribution were considered spermatozoa. The compensation of spectral overlap was performed before each experiment using negative (unstained) and positive (single stained) controls for each single-stained compensation control sample. Additionally, thresholds for each quadrant were determined using unstained, isotype controls and single-stained control samples for each of the probes used in each individual experiment.
4.7. Indirect Immunofluorescence with Classical Microscopy and with Confocal Microscopy
After the incubation under the required conditions, spermatozoa were washed twice with PBS and then processed for immunofluorescence. Fifteen µL of pretreated semen samples were spread on poly-L-lysine-coated glass-bottom chambers (Nunc Ref#155411) and allowed to attach for 10 min. Spermatozoa were fixed with 4% formaldehyde in PBS for 15 min at RT and permeabilized with 0.2% Triton X-100 (v/v) in PBS for 5 min. Cells were washed three times for 10 min with PBS and incubated in PBS supplemented with 5% BSA (w/v) for 90 min to block nonspecific sites. After blocking, slides were incubated with primary antibodies (LC3) overnight at 4 °C, diluted in PBS containing 5% BSA (w/v). The following day, samples were extensively washed with PBS and further incubated for 45 min at RT with the secondary antibody diluted to 1/500 in PBS containing 5% BSA (w/v), consisting of an anti-rabbit antibody conjugated with the Alexa 546 probe. In the colocalization studies, the Alexa 546-labeled anti-rabbit secondary antibody was used simultaneously with PNA-FITC, for which the incubation conditions for the latter were identical to those used in flow cytometry (25 µM; 15 min at RT). Finally, the slides were thoroughly washed with PBS and examined. The images were obtained with a Nikon Eclipse TE200 inverted fluorescent microscope equipped with a 100× objective in oil immersion, and the appropriate filter was set to acquire green and red images. Bright-field images (BFI) were also acquired. In the colocalization experiments, the slides were also examined in a Bio-Rad MRC1024 confocal microscope with 60× and 100× objectives, both in oil immersion. The absence of nonspecific immunolabeling was assessed by processing the samples without primary antibodies.
4.8. Colocalization Study
The images were acquired with a Bio-Rad MRC1024 confocal microscope, as described above, with the appropriate filter set to obtain green and red channels for each acquisition, corresponding, respectively, with PNA-FITC and LC3-Alexa 546. Differential interference contrast images (DCI) were also obtained. A superposition of the red and green images was initially constructed with the image processing software ImageJ [
52], by using the
merge channels function. The images were further analyzed with the ImageJ software with the addition of two plugins, namely “Colocalization” and “JACoP”, which were obtained in the plugin repository page (
https://imagej.nih.gov/ij/plugins) (last access on 20 December 2022). The first one was used to generate a qualitative visualization analysis, which highlighted the colocalized points of red and green 8-bit images, appearing as white areas in the constructed image. The second plugin is a compilation of co-localization tools, which calculated a set of commonly used co-localization indicators [
28]. In this study, we used Pearson’s correlation coefficient (PCC), Mander’s overlap coefficient (MOC), and Mander’s colocalization coefficients for channel 1 (M1) and channel 2 (M2) for each obtained green/red image pair, without modifying the automatic threshold in any channel or applying ROIs. The values for PCC ranged from +1 to −1. A value of +1 represents perfect correlation, whereas −1 theoretically represents perfect exclusion, and 0 (zero) represents random localization in a situation where the labeling of both fluorochromes was proportional to the other and the detection of both was assessed in a linear range [
28]. Low (close to zero) and especially negative PCC values for fluorescent images can be difficult to interpret in terms of colocalization. However, a value close to 1 does indicate reliable colocalization and constitutes the first good quantitative estimation of this process [
28]. MOC is based on PCC, with average intensity values being taken out of the mathematical expression [
28]. This coefficient varies from 0 to 1, the first corresponding to non-overlapping images and the latter reflecting 100% colocalization between both images. Finally, M1 and M2 are the split coefficients of the Mander’s colocalization coefficients for channel 1 and 2, respectively, indicating the fraction of green channel overlapping red (M1) and the fraction of red channel overlapping green (M2). These split coefficients avoid issues relating to absolute intensities of the signal, since they are normalized against total pixel intensity, and also provide information about how well each channel overlaps the other [
28]. The obtained data for each coefficient, namely PCC, MOC, M1, and M2, were tabled accordingly with their acquisition conditions and treatments, and the mean ± SEM was calculated and used as a quantitative estimation of the LC3/PNA colocalization map after the induction of the acrosome reaction.
4.9. Statistical Analysis
Data were first examined using the Kolmogorov–Smirnov test to determine their distribution. Multivariate analysis of variance was performed (ANOVA) followed by post hoc Tukey’s test. If data did not adjust to a normal distribution, the nonparametric Mann–Whitney U-test was used to compare pairs of values directly. All analyses were performed using SPSS version 17.0 for Windows (SPSS Inc., Chicago, IL, USA). Statistical significance was set at p < 0.05.